Abstract
Purpose
Corticosteroids (CS) induce apoptosis in the malignant lymphoid cells and are critical component of combination therapy for acute lymphoblastic leukemia (ALL). Several genome-wide microarray studies demonstrated major implication of proapoptotic Bim in mediating CS-related resistance in leukemia cells.
Experimental design
We investigated Bim gene polymorphisms and their association with childhood ALL outcome, and the mechanism underlying the observed finding.
Results
Lower overall survival (OS) was associated with Bim C29201T located in BH3 domain (p=0.01). An association remained significant in multivariate model (p=0.007), was more apparent in high risk (HR) patients (p=0.004) and patients treated with dexamethasone (p=0.009), and was subsequently confirmed in the replication patient cohort (p=0.03). RNA analysis revealed that C29201T affects generation of gamma isoforms (gamma1) that lack pro-apoptotic BH3 domain. The phenotypic effect was minor suggesting the influence of additional factors that may act in conjunction with Bim genotype. Combined analysis with Mcl gene polymorphism (G -486T) revealed profound reduction in OS in individuals with both risk genotypes (p<0.0005 in discovery and p=0.002 in replication cohort) and particularly in HR patients (p≤0.008).
Conclusions
Increased expression of pro-survival Mcl1 and presence of Bim isoforms lacking pro-apoptotic function might explain marked reduction of OS in a disease and dose dependent manner in ALL patients carrying Bim and Mcl1 risk genotypes.
Keywords: corticosteroids, polymorphisms, Bim, Mcl1, apoptosis, pharmacogenetics, childhood leukemia, treatment, outcome
Introduction
Corticosteroids (CS) induce apoptosis and cell cycle arrest in the majority of malignant lymphoid cells(1). Consequently they are critical component of combination chemotherapy regimens used in the treatment of lymphoid malignancies, including childhood acute lymphoblastic leukemia (ALL)(2);(3). Despite their clinical importance, the mechanism underlying molecular basis of CS-induced apoptosis and CS-related resistance are not fully understood. CSs mediate their effect via glucocorticoid receptor (GR) (4);(5); the resulting complex recruits either co-activator or co-repressor proteins thereby inducing or repressing the expression of a large number of target genes(4, 6–8). A subpopulation of childhood ALL cases fail to respond to CS treatment and one of the underlying mechanisms is a change in GR expression (5);(9);(10). Other mechanisms are also involved in the resistance to cell death induced by these drugs. A number of studies including genome-wide expression profiling have attempted to identify critical glucocorticoid-regulated genes, which may undergo altered expression before the onset of apoptosis(11–14). The proapoptotic protein that was up-regulated by glucocorticoids in several models of CS-induced apoptosis is Bim, the Bcl-2 homology 3 (BH3)–only molecule (11);(15);(16). Bim is expressed in hematopoietic, epithelial, neuronal, and germ cells and was found frequently up-regulated in childhood leukemia samples following CS exposure. (11);(13) It is a member of Bcl-2 family that induces the mitochondrial apoptosis pathway by either opposing the pro-survival proteins of this family or by binding to the pro-apoptotic Bcl2 members directly activating their pro-apoptotic functions (17);(18). Change in Bim expression seems to influence sensitivity to CS in ALL and, as shown in primary ALL lymphoblasts, cell lines and ALL xenografts, resistance to CS is associated with attenuated induction of Bim (15, 19–21).
Bim is encoded by gene BCL2L11 and three major mRNA isoforms and Bim protein products (BimEL, BimS, and BimL) exist(15). Moreover, it is extensively regulated at transcriptional and post-transcriptional level. The changes in expression may be also affected by genetic polymorphisms.
Here we report the analysis of Bim polymorphisms and their association with ALL disease outcomes in two patient populations. Further functional analysis based on mRNA and cellular viability assays was also performed to give an insight into the underlying mechanism of significant association.
Patients and methods
Study population and endpoints in the analysis
The study population consisted of 348 Caucasian children (97.5% of patients are of French-Canadian origin from the similar geographical region) diagnosed with ALL at the Hospital Sainte-Justine (QcALL group or test group) between January 1989 and July 2005. The consecutively accrued patients underwent treatment with the Dana-Farber Cancer Institute ALL Consortium protocols DFCI 87–01 (n=34), 91–01 (n=65), 95–01 (n=125), or 2000–01 (n=124, Table 1)(2);(22);(23) Considering corticosteroid treatment, all patients received prednisone during the induction phase (40 mg/m2/day); CS were administered during the intensification and continuation phases as 5-day pulses every 3 weeks, until the completion of therapy. On Protocols 87–01 and 95–01, prednisone was used during these treatment phases, on Protocol 91–01 dexamethasone was used instead of prednisone, and on Protocol 2000–01, patients were randomized to receive either prednisone or dexamethasone. Standard-risk (SR) patients received dexamethasone at a dose of 6 mg/m2/day or prednisone at a dose of 40 mg/m2/day and high-risk (HR) patients received doses 3 times higher than those received by SR patients during both the intensification and continuation phases, except on protocol 2000–01 when HR patients received the same dose as SR patients during the continuation phase.
Table 1.
Characteristic of ALL patients in the test (QcALL) and validation (DFCI) cohorts
| No of subjects and frequency (%) | ||||
|---|---|---|---|---|
| Characteristic | QcALL | DFCI | ||
| Sex | ||||
| Female | 155 | (44.5) | 128 | (45.9) |
| Male | 193 | ( 55.5) | 151 | (54.1) |
| Age, y | ||||
| <10 | 276 | (79.3) | 228 | (81.7) |
| ≥10 | 72 | (20.7) | 51 | (18.3) |
| WBC, ×109/L | ||||
| <50 | 286 | (82.2) | 226 | (81.0) |
| >50 | 62 | (17.8) | 53 | (19.0) |
| Cell type | ||||
| B | 319 | (91.7) | 256 | (91.8) |
| T | 29 | (8.3) | 23 | (8.2) |
| Risk groups | ||||
| Standard | 164 | (47.1) | 171 | (61.3) |
| High | 184 | (52.9) | 108 | (38.7) |
| Treatment protocol | ||||
| 87–01 | 34 | (9.8) | ||
| 91–01 | 65 | (18.7) | ||
| 95–01 | 125 | (35.9) | 95 | (34.1) |
| 2000–01 | 124 | (35.6) | 184 | (65.9) |
| Event | ||||
| yes | 68 | (19.5) | 54 | (19.4) |
| no | 280 | (80.5) | 216 | (80.6) |
| Death | ||||
| yes | 31 | (8.9) | 22 | (7.9) |
| no | 317 | (91.1) | 257 | (92.1) |
| total | 348 | (100) | 279 | (100) |
An association of genotypes/haplotypes with ALL outcome was assessed by EFS, and OS analysis(24). Children who had an induction failure, relapsed after achieving complete remission, or died, were defined to have had an event.
A replication set of Caucasian patients called the Dana-Farber Cancer Institute (DFCI) group is composed of a subset of patients who underwent treatment on DFCI 95–01 and 2000–01 protocol in nine remaining consortium institutions(2);(23);(25). This group was composed of 306 cases (not consecutively accrued) whose samples provided sufficient DNA to allow genotyping. To minimize confounding due to the population stratification, similar to discovery cohort, only Caucasians (self-reported, n=279, DFCI 95–01, n=95 and 2000–01, n=184) were included in the analysis.
The characteristics of patients for both test and validations set are provided in Table 1.
Genotyping
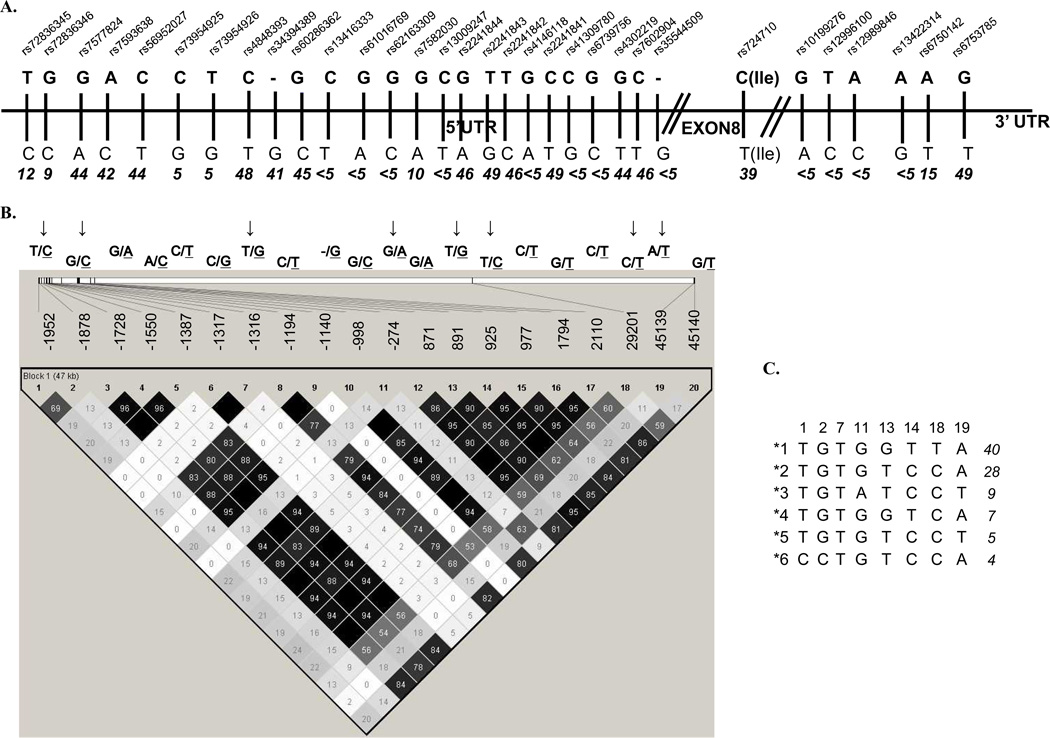
Thirty two polymorphisms located in regulatory and coding gene regions were selected from NCBI (National Center for Biotechnology Information) SNP (single nucleotide polymorphism) databases (http://www.ncbi.nlmnih.gov/SNP). Selected polymorphisms were analyzed in 60 healthy unrelated adults (of the same ethnic background as patients) to estimate allele frequency, linkage disequilibrium (LD) and haplotype phase (Figure 1). Eight tagSNPs (sufficient to define common haplotypes) with minor allele frequency (MAF) ≥ 5% were retained for analysis in patients. Primers and probes used for amplification and genotyping of these polymorphisms are shown in supplemental Table 1. dbSNP numbers along with SNP positions for the polymorphisms genotyped only in controls are given in supplemental Table 2. The subset of samples was genotyped in duplicate to ensure genotype reproducibility. Genotyping was performed by allele specific oligonucleotide hybridization as previously described(26).
Figure 1. Linear representation of Bim polymorphisms (A), Haploview LD display (B) and derived haplotypes (C).
A: Linear display of all initially selected SNPs; rs dbSNP numbers and MAF in controls is indicated for each SNP; B: LD and pair-wise r2 for SNPs with MAF higher than 5%; tagSNPs retained for the analysis in patients are indicated by arrows; C: Haplotypes with the frequency ≥ 5% derived from tagSNPs are arbitrarily named by the numbers. The frequency in controls is given next to each haplotype.
Statistics
The estimates of linkage disequilibrium (LD), and haplotype phase in control individuals were obtained by PHASE software, version 2.0(27). The tag SNPs were selected based on LD information (r2, Figure 1) using Haploview software. Association of genotypes with ALL outcome was assessed by EFS and OS. Survival differences, estimated by Kaplan-Meier analysis for patients with different genotypes, were assessed using a log-rank test. Times to event, or a death were measured as the time between diagnosis and the event of interest. For censored cases, it represented the time from diagnosis to date last known alive without an event; for longer follow-up durations all times were truncated at 5 years post-treatment. For the polymorphism associated with a disease outcome, the analyses were also performed following stratification by risk group and CS type. The hazard ratio (HR, with a 95% confidence interval, CI) for genetic variants was estimated by Cox regression analysis. Cox regression was also used to estimate multivariable HR in the presence of common prognostic factors (age, sex, presenting white blood cell (WBC) count, immunophenotype) and treatment protocol (listed in Table 1), as described previously(25).
mRNA analysis
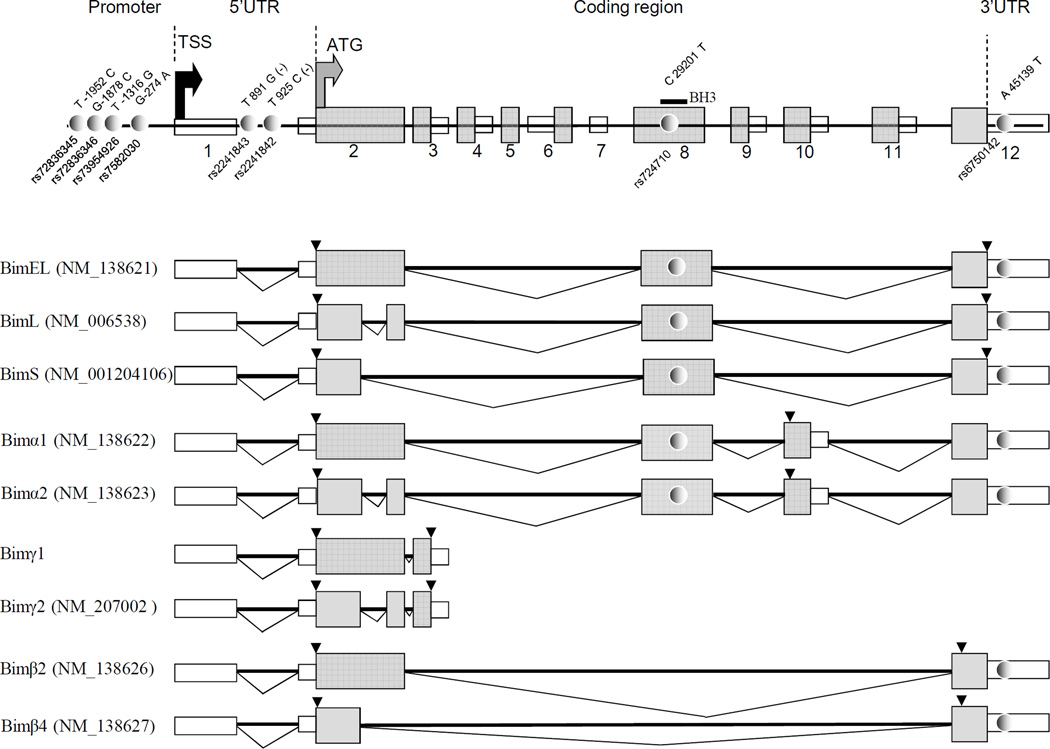
Total RNA from prednisolone (75μM Prednisolone) and dexamethasone treated (2.8X10−1µM) lymphoblastic cell lines (LCLs) of HapMap subjects of European origin (CEU) extracted with Qiagen kit was reverse transcribed using 2 µM oligo dT and random primers (mixed at 1:2 molar ratio) and M-MLV enzyme (Invitrogen) according to the protocol provided by the enzyme supplier. Quantitative PCR was carried out using the Syber Green detection system (Applied Biosystems). The expression was measured by relative quantification normalized to B2-microglobulin (primers: TACTCTCTCTTTCTGGCCTG and GGATGGATGAAACCCAGACA) and the calculation was performed using the comparative cycle threshold (CT) method. (28)Pair of primers TGACCGAGAAGGTAGACAAT and GCCATACAAATCTAAGCCAGT targeting alternatively spliced exon 3 was used to amplify non BH3-containing gamma (γ) isoforms (29, 30) (Figure 1). For remaining Bim isoforms, primers GAGATATGGATCGCCCAAGA and CAATGCATTCTCCACACCAG were used to amplify last two exons common of all isoforms except γ. When appropriate, relative mRNA levels were log transformed and the difference in relation to genotypes was analyzed by t-test or ANOVA, or expressed as quartile distribution and difference according to genotype were assessed by chi-square test.
Semi-quantitative analysis of three major RNA isoform (Bim S, Bim L and Bim EL, Figure 2) was carried out by using primers described in(31), whereas γ1 and γ2 assessment was carried out by same PCR primers described above for total γ estimation. PCR condition included 0.4–1 µM for each primer, 0.2mM of each dNTP, 2,5 mM MgCl2, and 50 ng of cDNA, and amplification at 95/3min, 30–40 cycles of 94/30sec, 55–58/30sec, 72/30sec–1min. Amplified PCR products were analyzed by QIAxcel system (QIAGEN, http://www.qiagen.com); QIAxcel DNA High Resolution Kit (1200) (fragment size range of 15 bp–3 kb) was chosen to recognize size differences of amplicons; Electropherogram and gel images were generated for each lane; Biocalculator software was used for data export and analysis. Relative contribution of each isoform (expressed as ratio of normalized areas under the curve) was analysed as quartile distribution in relation to genotype using chi-square test.
Figure 2. Schematic representation of Bim gene.
The Bim isoforms and its NCBI Reference Sequence number including major and minor isoforms with and without BH3 domain are given below genomic structure. Exonic and coding sequence is represented by open and gray boxes respectively. SNPs are represented by gray dots. The black and gray arrows indicate the transcription and translation start site, respectively. The figure is not to scale.
The sequence of the two gamma isoforms (gamma 1 and 2) were confirmed by Sanger sequencing. Genotyping of LCLs was performed as described above.
Cellular proliferation assay
LCLs were cultured in RPMI medium supplemented with 15% fetal bovine serum and antibiotics (100 IU/ml penicillin; 100 μg/ml streptomycin). The experiments were carried out to determine the concentrations lethal to 50% of the cells (IC50) during 48 h incubations time by curve fitting the percentage of cell death growth versus controls. Around 5×104 cells/well were seeded and submitted to seven different concentrations of prednisolone ranging from 0.75µM to 750µM and eight different concentrations of dexamethasone ranging from 2.8X10−3µM to 560µM and tested in duplicate. Cell Proliferation Reagent WST-1 (Roche, 10µl/well) was added to the cells and the optical density was measured at 450nm with a Spectra MAX 180 (Molecular Devices). After correction for the culture media by using a background control in duplicate the relative cell survival was calculated as (OD drug-exposed well)/(mean OD control wells)×100%. Log-transformed values or quartile distribution was compared in relation to genotypes.
Results
Thirty two polymorphisms located in the region of potential functional importance (regulatory and coding regions) were genotyped in 60 controls of Caucasian origin to estimate allelic frequency (Figure 1A); Polymorphisms with MAF ≥5% were analyzed for pairwise LD (Figure 1B); eight tagSNPs were identified (sufficient to define six common haplotypes, arbitrarily named *1 to *6, Figure 1C) and retained for the analysis in patients.
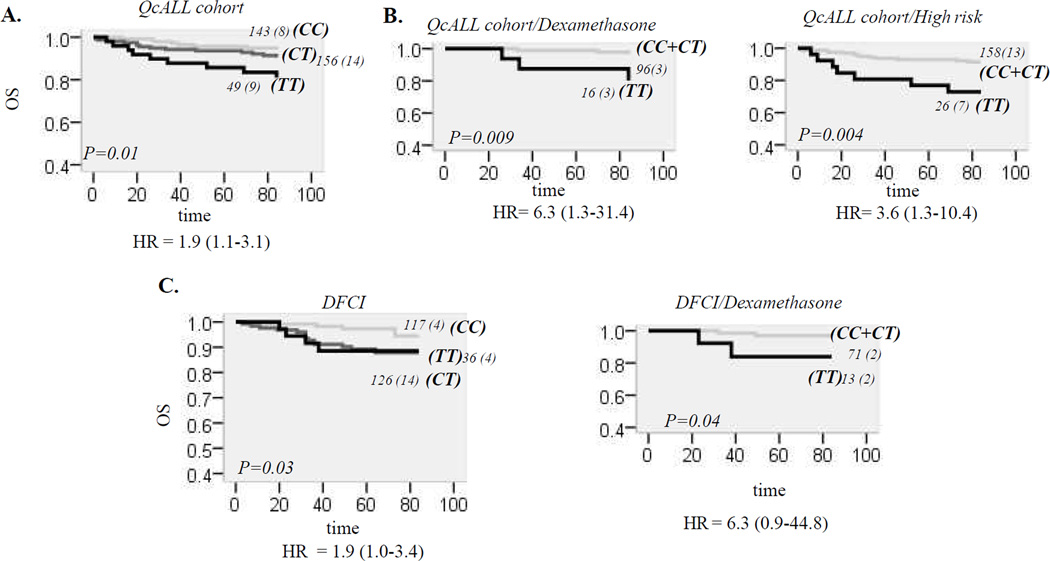
The analysis in the discovery cohort revealed no association with EFS (Table 2). In contrast, the analysis showed significant association between OS and synonymous C29201T replacement (Ile65Ile) located in BH3 domain of exon 8 (p=0.01, HR=1.9, 95% CI, 1.1–3.1, Figure 3A and Table 2) with lowest OS seen in patients with TT29201 genotype. The association remained significant (HR=2.1, 95% CI, 1.2–3.7, p=0.007) with inclusion of typical prognostic factor in Cox regression model (see methods). Given that patients were administered either prednisone or dexamethasone during the intensification and continuation treatment phases and that doses differed between standard risk (SR) and high risk (HR) patients, C29201T was subsequently analyzed following stratification by CS type and risk groups. Lower OS for TT29201 individuals was noted in patients who received dexamethasone (p=0.009, HR=6.3, 95% CI, 1.3–31.4. Figure 3B) as well in patients who were assigned to HR group irrespective of type of CS received (p=0.004, HR=3.6, 95% CI, 1.3–10.4, Figure 3B). The T29201 allele defines haplotype *1, and the same result as for the T allele, was obtained when this haplotype was analyzed for an association with OS (not shown). The analysis for C29201T variation was subsequently performed in the replication cohort (DFCI group of patients). The significant association between genotype groups and OS was seen (p=0.03, HR= 1.9, 95% CI, 1.1–3.4, Figure 3C), and remained significant in multivariable analysis (HR= 2.4, 95% CI, 1.2–4.7, p=0.01). Reduction in OS was observed for both CT and TT individuals; the TT29201 individuals had nevertheless the lowest EFS among patients who received dexamethasone (p=0.04, Figure 2C).
Table 2.
Association of Bim polymorphisms with EFS and OS in discovery cohort
| Polymorphism | EFS | OS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pa | pa | |||||||||||
| dbSNP | Position | Genotype | Total | N event |
A | D | R | Total | N event |
A | D | R |
| rs2241842 | 925 | CC | 76 | 16 | 0.3 | 0.06 | 0.7 | 76 | 7 | 0.4 | 0.1 | 0.9 |
| CT | 166 | 25 | 166 | 11 | ||||||||
| TT | 106 | 27 | 106 | 13 | ||||||||
| rs2241843 | 891 | GG | 84 | 19 | 0.6 | 0.9 | 0.4 | 84 | 11 | 0.3 | 0.7 | 0.1 |
| GT | 164 | 30 | 164 | 12 | ||||||||
| TT | 100 | 19 | 100 | 8 | ||||||||
| rs73954926 | 1316 | GG | 2 | 1 | 0.2 | 0.3 | nc | 2 | 1 | 0.6 | 0.3 | nc |
| TG | 42 | 10 | 42 | 1 | ||||||||
| TT | 295 | 54 | 295 | 27 | ||||||||
| rs72836346 | 1878 | CC | 4 | 1 | 0.7 | 0.5 | 0.8 | 4 | 0 | 1.0 | 0.8 | 0.5 |
| GC | 38 | 5 | 38 | 4 | ||||||||
| GG | 295 | 56 | 295 | 24 | ||||||||
| rs7582030 | 274 | AA | 7 | 1 | 0.9 | 0.8 | 0.7 | 7 | 0 | 0.1 | 0.1 | 0.4 |
| GA | 77 | 14 | 77 | 4 | ||||||||
| GG | 250 | 48 | 250 | 25 | ||||||||
| rs724710 | 298 | CC | 143 | 25 | 0.5 | 0.4 | 0.3 | 143 | 8 | 0.01 | 0.08 | 0.01 |
| CT | 156 | 31 | 156 | 14 | ||||||||
| TT | 49 | 12 | 49 | 9 | ||||||||
| rs72836345 | 1952 | CC | 5 | 2 | 0.2 | 0.4 | nc | 5 | 2 | 0.2 | 0.6 | nc |
| TC | 56 | 7 | 56 | 4 | ||||||||
| TT | 276 | 55 | 276 | 21 | ||||||||
| rs6750142 | 2251 | AA | 229 | 45 | 0.9 | 0.8 | 0.7 | 229 | 23 | 0.2 | 0.2 | 0.3 |
| AT | 104 | 19 | 104 | 7 | ||||||||
| TT | 13 | 3 | 13 | 0 | ||||||||
p value estimated by log-rank test using additive (A), dominant (D) and recessive (R) model; nc, not calculated due to low genotype count
Figure 3. Relationship between Bim C29201T and OS in ALL patients.
A. OS according to C29201T genotypes in discovery group (QcALL). OS curves are given for CC, CT and TT genotype. The number of all patients in each curve (with the number of patients that died in brackets) is indicated next to the curve. OS difference between genotype groups is estimated by log-rank test and p value is given on the plot. Genotype-associated risk (univariable hazard ratio, HR, with 95% confidence interval (CI, in brackets) is indicated below plot).
B. OS curves according to Bim C29201T genotypes in high risk patients and patients treated with dexamethasone. OS for QcALL patients who received dexamethasone (left plot), or who were assigned to high risk (right plot), with (black line) and without (gray line) TT29201 genotype.
C. OS curves according to genotypes of Bim C29201T polymorphism in replication cohort. Left plot is OS for all patients of replication cohort (DFCI). Curves are represented for CC, CT and TT genotypes. Right plot is OS for patients with and without TT genotype who received dexamethasone.
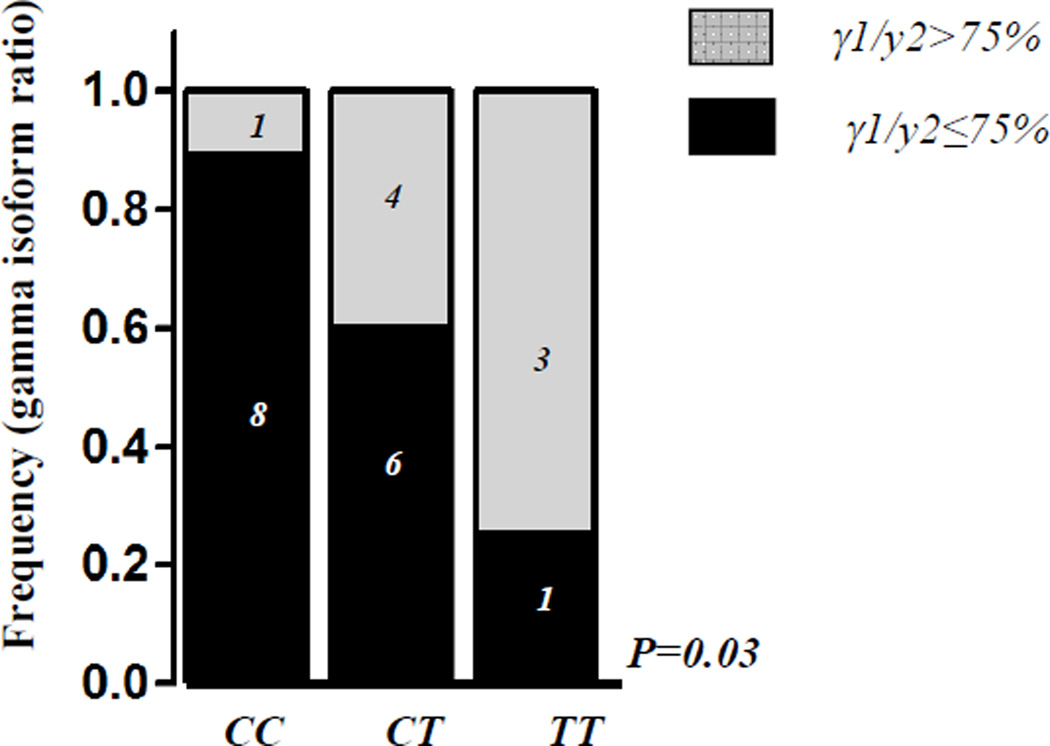
The C29201T polymorphism seems to affect exonic splicer element in BH3 domain, as based on F-SNP prediction tool(32), introducing additional binding site to serine and arginin rich (SR) protein SRp55. We therefore performed quantitative and semi-quantitative mRNA analysis in lymphoblastoid cell lines (LCL) following addition of prednisone or dexamethasone to investigate whether it may affect the total Bim mRNA levels (except gamma), levels of major (BimS, BimL and BimEL) isoforms, or levels of gamma isoforms (total and γ1/ γ2 ratio), which lack BH3 domain and correlate with reduced apoptotic function. (29, 33) Bim gene organization with respective mRNA isoforms is schematically presented in Figure 2. The influence of genotypes on sensitivity to CS was tested by in vitro cellular viability assay following addition of either prednisone or dexamethasone to LCLs. There was no relation between C29201T genotypes and mRNA levels when either total or major individual isoforms were quantified. In contrast, non-significant increase in gamma isoform formation was noted in LCLs carrying T29201 allele after addition of dexamethasone, which seems to be due to an increase in gamma1 to gamma2 ratio (Figure 4, p=0.03). There was no significant change in cellular viability in relation to C29201T genotype.
Figure 4. Bim gamma isoform in HapMap lymphoblastoid cell lines (LCLs) according to Bim C29201T genotypes.
Gamma1 to gamma2 ratio obtained by semi-quantitative estimation in individuals with CC, CT and TT genotype. Ratio is expressed as quartile distribution and the highest ratio (presence or not of the highest quartile) was compared between genotypes by chi-square test.
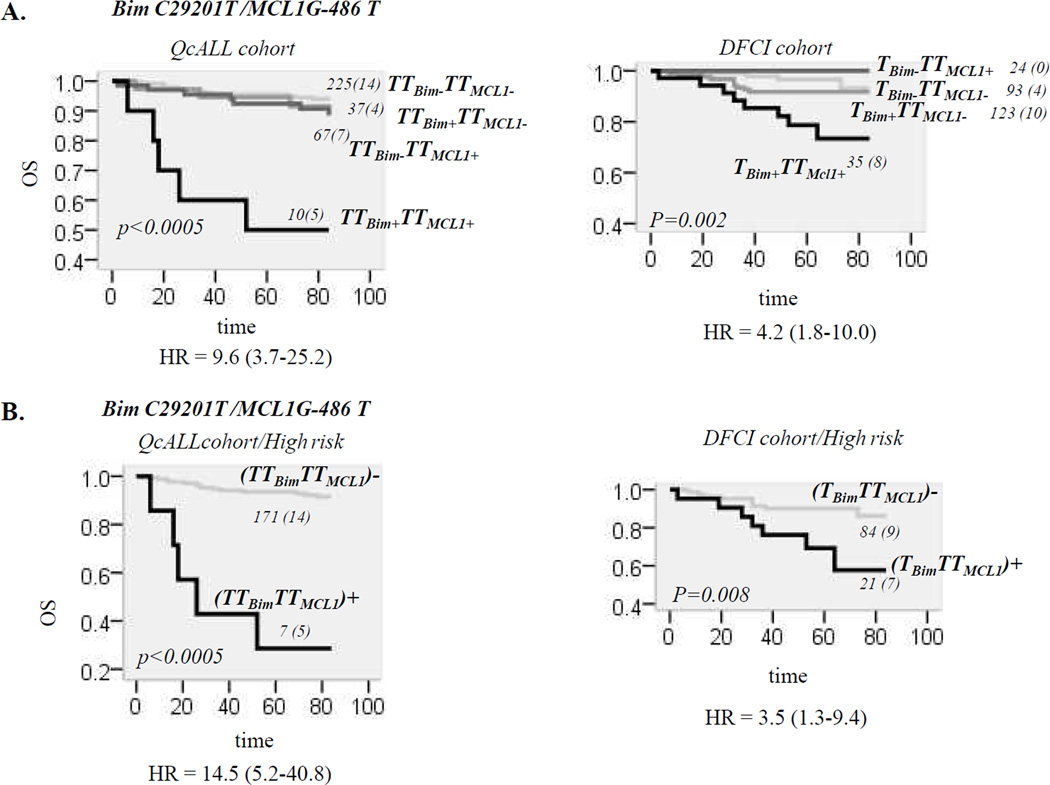
Given that the functional effect of a C29201T substitution was relatively minor, we further explored whether the effect seen in clinical setting may arise through the interaction with other genes of apoptotic pathway. Bim/Mcl1 complex plays a role of a regulatory “node,” critical for controlling cell apoptosis in response to multiple signals(34). We recently reported that three Mcl1 promoter SNPs in perfect LD were associated with higher promoter activity and lower OS in the QcALL cohort(35). The combined effect between Bim C29201T and Mcl1 G-486T variations (one of SNPs in LD) showed that the effect on ALL outcome was potentiated and present only in individuals carrying both risk genotypes. Marked reduction in OS was seen in discovery (p<0.0005, HR=9.6, 95% CI, 3.7–25.2 Figure 5A) and replication cohort (p=0.002, HR=4.2, 95% CI, 1.8–10.0, Figure 5A) in individuals with combined genotypes. The association was limited to HR patients (p<0.0005 and p=0.008, respectively, Figure 5B).
Figure 5. Association between combined Bim/Mcl1 genotypes and OS in patients with ALL.
A. Association between combined Bim/Mcl1 genotypes and OS in all patients of discovery and replication cohort. The OS curves for QcALL and DFCI patients having none genotype at risk (Bim−Mcl−), only Bim (Bim+Mcl−) or Mcl1 (Mcl1+Bim−), or both at risk genotypes (Bim+Mcl+). Risk genotype for Bim was TT29201 in discovery cohort, and presence of at least one T29101 allele, in replication cohort.
B. Association between combined Bim/Mcl1 genotypes and OS in high risk patients of discovery and replication cohort. The OS curves are given for the patents with (BimMcl1)+ and without (BimMcl1)− risk genotypes at both loci.
Discussion
We showed that C29201T substitution located within BH3 domain of Bim gene was associated with inferior ALL outcome independently from typical prognostic factors, as demonstrated by reduced overall survival in two patient cohorts. Bim plays a critical role in development and homeostasis of the lymphoid system(36). It is a critical regulator of apoptosis in normal and malignant cells, a tumor suppressor in B- cell malignancies(37) and confers resistance of normal and malignant lymphocytes to corticosteroids(38). Several line of evidence suggest that CS resistance in ALL was associated with attenuated induction of Bim: Resistance to CS was associated with a failure to induce the Bim when xenograft ALL cells were exposed to dexamethasone;(19);(20) Reduction of Bim mRNA and protein levels by RNA interference in CS-sensitive ALL cells reduced the activation of caspase-3 and increased cellular viability following CS exposure.(15) Significantly lower Bim expression correlated with poor early prednisone response and event free survival in paediatric ALL patients(39). Bim expression and function is regulated by a variety of mechanisms from transcriptional and posttranscriptional regulation to posttranslational modification and epigenetic silencing.(40–42) The Bim gene sequence alterations may as well affect mRNA splicing, the rate of transcription and protein level, affecting the cellular fate and response to CS treatment. Interestingly, C29201T v is predicted(32) to introduce splicing enhancer element for SRp55 protein (also known as SR splicing factor SRSF6). SRs have an important part in splice-site selection through association with splicing enhancers and silencers.(43, 44) It has been shown that change in BimS splicing is mediated by SRp55 in melanoma cells(45) Moreover, SRSF1 over expression promoted alternative splicing of Bim to produce gamma isoforms(29). In our study, the T29201 allele was not associated with the change of overall Bim expression or preferential transcription of any of major mRNA isoforms. However, change in gamma1 to gamma2 ratio was noted in individuals with T29201 allele after addition of dexamethasone to LCLs. Gamma isoforms include an alternatively spliced exon 3 instead of BH3 containing one, thus lacking the BH3 domain and the apoptotic function.(29) Change in Bim alternative splicing in favour of gamma isoforms generation was consistent with the delay in apoptosis during mammary epithelial cell transformation.(29); Interestingly, expression of Bim gamma1, but not gamma2, promoted a decrease in apoptosis.
The functional changes in studied LCLs were relatively minor suggesting that either subtle changes may have important phenotypic consequences or Bim may cooperate with other factors leading to phenotypic changes observed in clinical setting. Our recent study,(35) which analyzed functional SNPs in promoter region of 11 intrinsic apoptosis genes with ALL susceptibility and ALL outcome, showed an association between Mcl1 promoter polymorphisms only and reduced OS in high-risk ALL patients(35). Here we assessed combined Mcl1/Bim effect, which showed marked reduction in OS, only for individuals with both at risk genotypes. The preferential association between Bim and Mcl1 in regulating cell death signals was reported in several studies. Mutation analysis revealed that the extent to which Mcl1 mutants were able to exert their anti-apoptotic effects in hemopoietic cells correlated with their ability to associate with Bim.(46) Bim phosphorylation was associated with reduced Bim/Mcl1 binding in chronic lymphocytic leukemia (CLL)(34). Low Mcl1 expression and high Bim/Mcl1 ratio correlated with a favorable response to novel BH3 mimetic navitoclax in CLL patients.(47) Disruption of Mcl1/Bim complex is a mechanism utilized by granzyme B and TRAIL-induced caspase to induce a mitochondrial apoptotic cascade(48),(49). The Bim/Mcl1 association with ALL outcome seems to be limited to HR patients. Risk-dependent change in Bim expression was also reported by others: Significantly lower Bim expression was detected in HR childhood ALL patients who exhibited slow responses to induction regimen compared with patients who responded rapidly.(50) Similar response was noted in our cohort. Four patients of our studied cohort carrying both risk genotypes (five patients died from progressive disease among seven with such combined genotypes, see Figure 5) experienced induction failure. Higher expression of Mcl1 along with the presence of Bim isoforms lacking BH3 domain in patients with combined genotype may possibly explain association with poor outcome, particularly in HR risk patients with less favorable features of presenting disease who require higher CS doses. However, the regulation of the affinity of Bim for each prosurvival protein is complex and interaction between them in addition to expression level may play a key role in neoplastic cell survival(51).
Finally, several Bim polymorphisms were previously shown to play a role in susceptibility to non-Hodgkin lymphoma.(30) Interestingly one of these variants (intronic rs686952) was in perfect LD with C29201T variation as based on our analysis in HapMap individuals. Likewise, the deletion polymorphism resulting in BIM gamma formation, that had a profound effect on the sensitivity to tyrosine kinase inhibitors in chronic myeloid leukemia, was recently reported(33). This polymorphism is limited to East-Asian population and could not be assessed in this study, but suggests importance of Bim gene variations and alternative Bim gamma isoforms in mediating resistance to variety of drugs.
In conclusion, we reported the association between C29201T polymorphism in BH3 domain of Bim gene with reduced OS in two childhood ALL cohorts, possibly arising through the generation of Bim isoforms lacking BH3 domain. The effect seems to be dependent on interaction with the risk genotype in Mcl1 gene. It is potentiated in HR patients and thus likely influenced by more progressive disease requiring higher corticosteroid doses. The strength of the study comes from the fact that Bim induction has been shown to shape CS-related resistance in ALL. It identifies for the first time the polymorphism in the Bim gene (supported with functional assessment and analysis of two independent cohorts of reasonable sample size) and importantly, points to the role of an interaction between apoptosis gene variations in modulating therapeutic responses in ALL. There are several limitations to the study, including small number of patients with the combined gene effects, limited mechanistic insight into the Bim-related resistance in ALL and limited number of factors investigated. The other mechanism may affect the Bim function, and many other genetic(52),(53) and non-genetic factors contribute to complex pattern of responsiveness to CS. In the light of this complexity, this finding should be regarded as only one of the components of multifactorial CS resistance in paediatric ALL, which adds to our understanding how genetic variations modulate therapeutic responses in ALL.
Supplementary Material
Statement of translational relevance.
Acute lymphoblastic leukemia (ALL) is the most frequent malignancy of childhood. The treatment of pediatric ALL has greatly improved in the past four decades due to the introduction of effective combination risk-adapted therapies. However, therapy resistance in a significant number of children still is a major obstacle to successful treatment. Intensive treatment has also significant short-term side effects and long-term consequences. Identification of genetic component underlying this variability, would allow traditional treatment to be complemented by genotype-based drug dose adjustment. Genome-wide experiments pointed to Bim gene as a major pro-apoptotic gene mediating corticosteroid related effects and resistance in ALL. Here we analyzed genetic variations in the gene Bim and try to explain an observed association through functional study and gene-gene interaction with other components of apoptotic machinery. The study provides a new insight into the Bim pharmacogenetics and its role with the respect to ALL.
Acknowledgment
We thank all patients and their parents who consented to participate in genetics studies related to leukemia.
Grant Support
Canadian Institutes of Health Research, Leukemia Lymphoma Society of Canada, Charles Bruneau Foundation, Fonds de la Recherche en Santé du Québec and Centre d'excellence en Oncologie pédiatrique et en soins palliatifs supported this study. Dana-Farber Cancer Institute ALL treatment protocols are supported by the National Cancer Institute/NIH grant 5 P01CA068484.
Footnotes
Authorship
Contribution: V.G., J.R, M.L, B.S. and I.B. performed experiments; D.S., C.L., F.C., S.E.S., L.B.S., D.N. and J.L.K. contributed to sample and clinical data collection and processing; V.G., J.R, M.L. and M.K. performed the data analysis; M.K. designed the research and drafted the article; All authors contributed to the interpretation of data and revised the manuscript.
conflict of interest statement
The authors declare no competing financial interests.
References
- 1.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350(15):1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 2.Silverman LB, Stevenson KE, O'Brien JE, Asselin BL, Barr RD, Clavell L, et al. Long-term results of Dana-Farber Cancer Institute ALL Consortium protocols for children with newly diagnosed acute lymphoblastic leukemia (1985–2000) Leukemia. 2010;24(2):320–334. doi: 10.1038/leu.2009.253. Epub 2009/12/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371(9617):1030–1043. doi: 10.1016/S0140-6736(08)60457-2. Epub 2008/03/25. [DOI] [PubMed] [Google Scholar]
- 4.Geley S, Fiegl M, Hartmann BL, Kofler R. Genes mediating glucocorticoid effects and mechanisms of their regulation. Reviews of physiology, biochemistry and pharmacology. 1996;128:1–97. doi: 10.1007/3-540-61343-9_7. Epub 1996/01/01. [DOI] [PubMed] [Google Scholar]
- 5.Schlossmacher G, Stevens A, White A. Glucocorticoid receptor-mediated apoptosis: mechanisms of resistance in cancer cells. J Endocrinol. 2011;211(1):17–25. doi: 10.1530/JOE-11-0135. Epub 2011/05/24. [DOI] [PubMed] [Google Scholar]
- 6.Gaynon PS, Carrel AL. Glucocorticosteroid therapy in childhood acute lymphoblastic leukemia. Adv Exp Med Biol. 1999;457:593–605. doi: 10.1007/978-1-4615-4811-9_66. Epub 1999/09/29. [DOI] [PubMed] [Google Scholar]
- 7.Kofler R. The molecular basis of glucocorticoid-induced apoptosis of lymphoblastic leukemia cells. Histochem Cell Biol. 2000;114(1):1–7. doi: 10.1007/s004180000165. Epub 2000/08/26. [DOI] [PubMed] [Google Scholar]
- 8.Tissing WJ, Meijerink JP, den Boer ML, Pieters R. Molecular determinants of glucocorticoid sensitivity and resistance in acute lymphoblastic leukemia. Leukemia. 2003;17(1):17–25. doi: 10.1038/sj.leu.2402733. Epub 2003/01/17. [DOI] [PubMed] [Google Scholar]
- 9.Labuda M, Gahier A, Gagne V, Moghrabi A, Sinnett D, Krajinovic M. Polymorphisms in glucocorticoid receptor gene and the outcome of childhood acute lymphoblastic leukemia (ALL) Leukemia research. 2010;34(4):492–497. doi: 10.1016/j.leukres.2009.08.007. Epub 2009/09/18. [DOI] [PubMed] [Google Scholar]
- 10.Kay P, Schlossmacher G, Matthews L, Sommer P, Singh D, White A, et al. Loss of glucocorticoid receptor expression by DNA methylation prevents glucocorticoid induced apoptosis in human small cell lung cancer cells. PLoS ONE. 2011;6(10):e24839. doi: 10.1371/journal.pone.0024839. Epub 2011/10/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gautier C, Negrerie M, Wang ZQ, Lambry JC, Stuehr DJ, Collin F, et al. Dynamic regulation of the inducible nitric-oxide synthase by NO: comparison with the endothelial isoform. J Biol Chem. 2004;279(6):4358–4365. doi: 10.1074/jbc.M305048200. Epub 2003/11/05. [DOI] [PubMed] [Google Scholar]
- 12.Thompson EB, Johnson BH. Regulation of a distinctive set of genes in glucocorticoid-evoked apoptosis in CEM human lymphoid cells. Recent progress in hormone research. 2003;58:175–197. doi: 10.1210/rp.58.1.175. Epub 2003/06/11. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt S, Rainer J, Riml S, Ploner C, Jesacher S, Achmuller C, et al. Identification of glucocorticoid-response genes in children with acute lymphoblastic leukemia. Blood. 2006;107(5):2061–2069. doi: 10.1182/blood-2005-07-2853. Epub 2005/11/19. [DOI] [PubMed] [Google Scholar]
- 14.Miller AL, Komak S, Webb MS, Leiter EH, Thompson EB. Gene expression profiling of leukemic cells and primary thymocytes predicts a signature for apoptotic sensitivity to glucocorticoids. Cancer cell international. 2007;7:18. doi: 10.1186/1475-2867-7-18. Epub 2007/11/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abrams MT, Robertson NM, Yoon K, Wickstrom E. Inhibition of glucocorticoid-induced apoptosis by targeting the major splice variants of BIM mRNA with small interfering RNA and short hairpin RNA. J Biol Chem. 2004;279(53):55809–55817. doi: 10.1074/jbc.M411767200. Epub 2004/10/29. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, Malone MH, He H, McColl KS, Distelhorst CW. Microarray analysis uncovers the induction of the proapoptotic BH3-only protein Bim in multiple models of glucocorticoid-induced apoptosis. J Biol Chem. 2003;278(26):23861–23867. doi: 10.1074/jbc.M301843200. [DOI] [PubMed] [Google Scholar]
- 17.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9(1):47–59. doi: 10.1038/nrm2308. Epub 2007/12/22. [DOI] [PubMed] [Google Scholar]
- 18.Huang DC, Strasser A. BH3-Only proteins-essential initiators of apoptotic cell death. Cell. 2000;103(6):839–842. doi: 10.1016/s0092-8674(00)00187-2. Epub 2001/01/04. [DOI] [PubMed] [Google Scholar]
- 19.Bachmann PS, Gorman R, Mackenzie KL, Lutze-Mann L, Lock RB. Dexamethasone resistance in B-cell precursor childhood acute lymphoblastic leukemia occurs downstream of ligand-induced nuclear translocation of the glucocorticoid receptor. Blood. 2005;105(6):2519–2526. doi: 10.1182/blood-2004-05-2023. [DOI] [PubMed] [Google Scholar]
- 20.Bachmann PS, Gorman R, Papa RA, Bardell JE, Ford J, Kees UR, et al. Divergent mechanisms of glucocorticoid resistance in experimental models of pediatric acute lymphoblastic leukemia. Cancer Res. 2007;67(9):4482–4490. doi: 10.1158/0008-5472.CAN-06-4244. [DOI] [PubMed] [Google Scholar]
- 21.Ploner C, Rainer J, Niederegger H, Eduardoff M, Villunger A, Geley S, et al. The BCL2 rheostat in glucocorticoid-induced apoptosis of acute lymphoblastic leukemia. Leukemia. 2008;22(2):370–377. doi: 10.1038/sj.leu.2405039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moghrabi A, Levy DE, Asselin B, Barr R, Clavell L, Hurwitz C, et al. Results of the Dana-Farber Cancer Institute ALL Consortium Protocol 95–01 for children with acute lymphoblastic leukemia. Blood. 2007;109(3):896–904. doi: 10.1182/blood-2006-06-027714. Epub 2006/09/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vrooman LM, Stevenson KE, Supko JG, O'Brien J, Dahlberg SE, Asselin BL, et al. Postinduction dexamethasone and individualized dosing of Escherichia Coli L-asparaginase each improve outcome of children and adolescents with newly diagnosed acute lymphoblastic leukemia: results from a randomized study--Dana-Farber Cancer Institute ALL Consortium Protocol 00–01. J Clin Oncol. 2013;31(9):1202–1210. doi: 10.1200/JCO.2012.43.2070. Epub 2013/01/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dulucq S, St-Onge G, Gagne V, Ansari M, Sinnett D, Labuda D, et al. DNA variants in the dihydrofolate reductase gene and outcome in childhood ALL. Blood. 2008;111(7):3692–3700. doi: 10.1182/blood-2007-09-110593. [DOI] [PubMed] [Google Scholar]
- 25.Rousseau J, Gagne V, Labuda M, Beaubois C, Sinnett D, Laverdiere C, et al. ATF5 polymorphisms influence ATF function and response to treatment in children with childhood acute lymphoblastic leukemia. Blood. 2011;118(22):5883–5890. doi: 10.1182/blood-2011-05-355560. Epub 2011/10/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Labuda D, Krajinovic M, Richer C, Skoll A, Sinnett H, Yotova V, et al. Rapid detection of CYP1A1, CYP2D6, and NAT variants by multiplex polymerase chain reaction and allele-specific oligonucleotide assay. Anal Biochem. 1999;275(1):84–92. doi: 10.1006/abio.1999.4293. Epub 1999/11/05. [DOI] [PubMed] [Google Scholar]
- 27.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68(4):978–989. doi: 10.1086/319501. Epub 2001/03/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. Epub 2002/02/16. [DOI] [PubMed] [Google Scholar]
- 29.Anczukow O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, et al. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat Struct Mol Biol. 2012;19(2):220–228. doi: 10.1038/nsmb.2207. Epub 2012/01/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly JL, Novak AJ, Fredericksen ZS, Liebow M, Ansell SM, Dogan A, et al. Germline variation in apoptosis pathway genes and risk of non-Hodgkin's lymphoma. Cancer Epidemiol Biomarkers Prev. 2010;19(11):2847–2858. doi: 10.1158/1055-9965.EPI-10-0581. Epub 2010/09/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao J, Chen GG, Yun JP, Chun SY, Zheng ZZ, Ho RL, et al. Identification and characterization of BH3 domain protein Bim and its isoforms in human hepatocellular carcinomas. Apoptosis : an international journal on programmed cell death. 2007;12(9):1691–1701. doi: 10.1007/s10495-007-0093-5. Epub 2007/05/16. [DOI] [PubMed] [Google Scholar]
- 32.Lee PH, Shatkay H. F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2008;36(Database issue):D820–D824. doi: 10.1093/nar/gkm904. Epub 2007/11/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng KP, Hillmer AM, Chuah CT, Juan WC, Ko TK, Teo AS, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nature medicine. 2012;18(4):521–528. doi: 10.1038/nm.2713. Epub 2012/03/20. [DOI] [PubMed] [Google Scholar]
- 34.Paterson A, Mockridge CI, Adams JE, Krysov S, Potter KN, Duncombe AS, et al. Mechanisms and clinical significance of BIM phosphorylation in chronic lymphocytic leukemia. Blood. 2012;119(7):1726–1736. doi: 10.1182/blood-2011-07-367417. Epub 2011/12/14. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez R, St-Cyr J, Lalonde ME, Healy J, Richer C, Gagné V, et al. Impact of promoter polymorphisms in key regulators of the intrinsic apoptosis pathway in childhood acute lymphoblastic leukemia outcome. Haematologica (manuscript submitted) 2012 doi: 10.3324/haematol.2013.085340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286(5445):1735–1738. doi: 10.1126/science.286.5445.1735. Epub 1999/11/27. [DOI] [PubMed] [Google Scholar]
- 37.Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci U S A. 2004;101(16):6164–6169. doi: 10.1073/pnas.0401471101. Epub 2004/04/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L, et al. BH3-only proteins Puma and Bim are rate-limiting for gamma-radiation- and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood. 2005;106(13):4131–4138. doi: 10.1182/blood-2005-04-1595. Epub 2005/08/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang N, Koh GS, Lim JY, Kham SK, Ariffin H, Chew FT, et al. BIM is a prognostic biomarker for early prednisolone response in pediatric acute lymphoblastic leukemia. Exp Hematol. 2011;39(3):321–329. 9, e1–e3. doi: 10.1016/j.exphem.2010.11.009. Epub 2010/12/07. [DOI] [PubMed] [Google Scholar]
- 40.Hershko T, Ginsberg D. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J Biol Chem. 2004;279(10):8627–8634. doi: 10.1074/jbc.M312866200. Epub 2003/12/20. [DOI] [PubMed] [Google Scholar]
- 41.Harada M, Pokrovskaja-Tamm K, Soderhall S, Heyman M, Grander D, Corcoran M. Involvement of miR17 pathway in glucocorticoid-induced cell death in pediatric acute lymphoblastic leukemia. Leuk Lymphoma. 2012;53(10):2041–2050. doi: 10.3109/10428194.2012.678004. Epub 2012/04/06. [DOI] [PubMed] [Google Scholar]
- 42.Bachmann PS, Piazza RG, Janes ME, Wong NC, Davies C, Mogavero A, et al. Epigenetic silencing of BIM in glucocorticoid poor-responsive pediatric acute lymphoblastic leukemia, and its reversal by histone deacetylase inhibition. Blood. 2010;116(16):3013–3022. doi: 10.1182/blood-2010-05-284968. Epub 2010/07/22. [DOI] [PubMed] [Google Scholar]
- 43.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14(5):802–813. doi: 10.1261/rna.876308. Epub 2008/03/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119(6):831–845. doi: 10.1016/j.cell.2004.11.010. Epub 2004/12/21. [DOI] [PubMed] [Google Scholar]
- 45.Jiang CC, Lai F, Tay KH, Croft A, Rizos H, Becker TM, et al. Apoptosis of human melanoma cells induced by inhibition of B-RAFV600E involves preferential splicing of bimS. Cell death & disease. 2010;1:e69. doi: 10.1038/cddis.2010.48. Epub 2011/03/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jamil S, Wang SW, Bondy L, Mojtabavi S, Duronio V. Prevention of cytokine withdrawal-induced apoptosis by Mcl-1 requires interaction between Mcl-1 and Bim. Biochem Cell Biol. 2010;88(5):809–818. doi: 10.1139/o10-004. Epub 2010/10/06. [DOI] [PubMed] [Google Scholar]
- 47.Roberts AW, Seymour JF, Brown JR, Wierda WG, Kipps TJ, Khaw SL, et al. Substantial susceptibility of chronic lymphocytic leukemia to BCL2 inhibition: results of a phase I study of navitoclax in patients with relapsed or refractory disease. J Clin Oncol. 2012;30(5):488–496. doi: 10.1200/JCO.2011.34.7898. Epub 2011/12/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jie HB, Sarvetnick N. The role of NK cells and NK cell receptors in autoimmune disease. Autoimmunity. 2004;37(2):147–153. doi: 10.1080/0891693042000196174. Epub 2004/08/06. [DOI] [PubMed] [Google Scholar]
- 49.Han J, Goldstein LA, Gastman BR, Rabinowich H. Interrelated roles for Mcl-1 and BIM in regulation of TRAIL-mediated mitochondrial apoptosis. J Biol Chem. 2006;281(15):10153–10163. doi: 10.1074/jbc.M510349200. Epub 2006/02/16. [DOI] [PubMed] [Google Scholar]
- 50.Bhojwani D, Kang H, Menezes RX, Yang W, Sather H, Moskowitz NP, et al. Gene expression signatures predictive of early response and outcome in high-risk childhood acute lymphoblastic leukemia: A Children's Oncology Group Study [corrected] J Clin Oncol. 2008;26(27):4376–4384. doi: 10.1200/JCO.2007.14.4519. Epub 2008/09/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morales AA, Kurtoglu M, Matulis SM, Liu J, Siefker D, Gutman DM, et al. Distribution of Bim determines Mcl-1 dependence or codependence with Bcl-xL/Bcl-2 in Mcl-1-expressing myeloma cells. Blood. 2011;118(5):1329–1339. doi: 10.1182/blood-2011-01-327197. Epub 2011/06/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawedia JD, Kaste SC, Pei D, Panetta JC, Cai X, Cheng C, et al. Pharmacokinetic, pharmacodynamic, and pharmacogenetic determinants of osteonecrosis in children with acute lymphoblastic leukemia. Blood. 2011;117(8):2340–2347. doi: 10.1182/blood-2010-10-311969. quiz 556. Epub 2010/12/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pottier N, Yang W, Assem M, Panetta JC, Pei D, Paugh SW, et al. The SWI/SNF chromatin-remodeling complex and glucocorticoid resistance in acute lymphoblastic leukemia. J Natl Cancer Inst. 2008;100(24):1792–1803. doi: 10.1093/jnci/djn416. Epub 2008/12/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bouillet P, Zhang LC, Huang DC, Webb GC, Bottema CD, Shore P, et al. Gene structure alternative splicing, and chromosomal localization of pro-apoptotic Bcl-2 relative Bim. Mamm Genome. 2001;12(2):163–168. doi: 10.1007/s003350010242. Epub 2001/02/24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.