Abstract
X-ray crystallography has evolved into a very powerful tool to determine the three-dimensional structure of macromolecules and macromolecular complexes. The major bottleneck in structure determination by X-ray crystallography is the preparation of suitable crystalline samples. This unit outlines steps for the crystallization of a macromolecule, starting with a purified, homogeneous sample. The first protocols describe preparation of the macromolecular sample (i.e., proteins, nucleic acids, and macromolecular complexes). The preparation and assessment of crystallization trials is then described, along with a protocol for confirming whether the crystals obtained are composed of macromolecule as opposed to a crystallization reagent . Next, the optimization of crystallization conditions is presented. Finally, protocols that facilitate the growth of larger crystals through seeding are described.
INTRODUCTION
The establishment of The Structural Genomics Consortium (SGC) as well as other centers whose aim is high-throughput structural determination of proteins and protein complexes has drastically changed the landscape for macromolecular crystallography . Many of the techniques and machinery originally developed for use in these centers have become commonplace in almost all crystallographic labs and have proven extremely useful in expediting the crystallization process. Furthermore, continued developments in recombinant DNA technology, computational methodologies for structure prediction, crystal data collection strategies, and the ever expanding market of commercially available crystallization screens has made crystallization and structural determination of macromolecules achievable in almost any laboratory setting.
The following unit outlines the steps required for the crystallization of a macromolecule starting with a purified sample. The starting macromolecule must be purified and homogeneous (≥95% pure by SDS-PAGE; UNIT 10.1) but can be at a fairly dilute concentration (≤1 µg/ml). If possible, the macromolecule should be in a standard nonphosphate buffer between pH 5.5 and 8.0, with a fairly low salt concentration (preferably <100 mM NaCl, and concentrations higher than 250 mM should be avoided). A reducing agent, either 2-mercaptoethanol (2-ME) or dithiothreitol (DTT), should also be present at a concentration between 1 and 10 mM. The total amount of macromolecule is critical, and should be at least 1 mg, preferably >5 mg.
The first protocols describe the preparation of the macromolecular sample—i.e., proteins (see Basic Protocol 1), nucleic acids (see Alternate Protocol 1 and 1.2), and macromolecular complexes (see Alternate Protocol 2). The preparation and assessment of crystallization trials is then described (see Basic Protocol 2 and Alternate Protocol 3), along with a protocol for determining whether the crystals obtained are composed of the macromolecule of interest or a crystallization reagent ( Support Protocol 1). Next, techniques for the optimization of crystallization conditions are presented (see Basic Protocol 3 and Alternate Protocol 4 and 5). Finally, protocols that facilitate the growth of larger crystals will be presented (see Alternate Protocols 6 and 7).
BASIC PROTOCOL 1
PREPARATION OF PROTEIN FOR CRYSTALLIZATION
Before a protein can be crystallized, it needs to be concentrated to at least 5 mg/ml. An ideal concentration is 20 mg/ml; however, somewhere between 5 and 20 mg/ml is usually suitable. Large macromolecules or macromolecular complexes (> ∼ 100 KDa) come sometimes be crystallized at lower concentrations.
Materials
Soluble protein sample
15 ml to 500 µl and 2 ml to ∼50 µl Centriprep and Centricon microconcentrators (Amicon)
1.5-ml screw-cap microcentrifuge tubes
Additional reagents and equipment for measurement of dynamic light scattering (UNIT 17.10; optional)
Place 15 ml soluble protein sample in a Centriprep microconcentrator that concentrates 15 ml to ∼500 µl. Centrifuge according to manufacturer’s instructions.
Transfer the sample to a Centricon microconcentrator that concentrates 2 ml to ∼50 µl, and continue centrifugation according to manufacturer’s instructions.
-
Monitor concentration C (in M) of an appropriately diluted sample (i.e., that produces an absorbance between 0.1 and 1.0) by determining A280 with a UV spectrophotometer and applying the Beer-Lambert law:
C = A280/ εl
where l is the path length (cm) and ε, the molar absorption coefficient (M–1cm–1), is calculated as follows:
ε = (5600 × no. Trp) + (1420 × no. Tyr) + (197 × no. Phe).
For proteins that do not contain Trp or Tyr residues, a colorimetric assay such as the Bradford (UNIT 3.4) can be used.
-
Continue to concentrate the protein until it is 5 mg/ml. If this is not attainable, concentrate the protein until visible precipitate is observed.
If concentration of the protein sample leads to formation of precipitate, the remaining soluble portion should be isolated by centrifugation. Reconcentration of the soluble portion should then be attempted under different salt concentrations, pHs, or concentrations of other appropriate additives.
-
If a dynamic light scattering instrument is available (e.g., Protein Solutions model DynaProMS/X, DynaPro-801), check the protein solution for monodispersity (UNIT 17.10).
Generally, proteins that are monodisperse at high concentrations (typically a polydispersity value ≤15% of the Stokes radius) have a strong tendency to crystallize, and those that are polydisperse have an equally strong tendency not to crystallize (Ferre-D’Amare and Burley, 1994). However, there are exceptions to this rule, and crystallization attempts should not be abandoned solely because of poor dynamic light scattering measurements.
-
Divide into 20- to 50-µl aliquots in 1.5-ml screw-cap microcentrifuge tubes, and flash freeze in liquid nitrogen. Store frozen (preferably at –70°C) until use for crystallization (up to 1 month in most cases).
If possible, subject part of the concentrated protein sample to crystallization trials (see Basic Protocol 2) prior to freezing, as some proteins do not respond well to freeze-thawing. If there is an activity assay available for the protein, it is recommended that the effect of freeze-thawing on this activity be established before freezing the protein sample. If the protein responds adversely, try to freeze the protein sample in the presence of a small amount of glycerol (5% to 15%) or another cryoprotectant. In such cases, it is usually necessary to remove the cryoprotectant (preferably with a microconcentrator) prior to crystallization.
ALTERNATE PROTOCOL 1
PREPARATION OF NUCLEIC ACID FRAGMENTS FOR CRYSTALLIZATION
The increasing availability of low cost, high quality DNA oligonucleotides has eliminated much of the need for in lab synthesis of DNA oligonucleotides. Manufacturers such as IDT and Operon can synthesize and purify oligos suitable for crystallization studies. However, if it is cost-prohibative to order oligos of this quality, the oligonucleotide(s) of interest (i.e., DNA or RNA) should be purified (preferably by reversed-phase HPLC; UNIT 11.6).
Materials
DNA or RNA oligonucleotides from manufacturer or HPLC purified (e.g., UNIT 11.6)
1.5 ml screw-cap microcentrifuge tube
Beaker
Stirring hot-plate
Dissolve the dried oligonucleotide in the appropriate volume of desired buffer (typically buffer that matches protein buffer) to yield approximately 1 mM of the individual oligonucleotides.
-
Determine the nucleic acid concentration C (M) by measuring A260 and converting using the Beer-Lambert equation:
C = A260/ εl
where l is the path length (cm), and ε, the molar absorption coefficient (M–1cm–1), is calculated as follows:
ε = (15,200 × no. dATPs) + (9300 × no. dCTPs) + (13,700 × no. dGTPs) + (9600 × no. dTTP) + (9600 × no. dUTPs).
Form duplexes (optional)
Mix complementary strands in stoichiometric molar amounts in a 1.5-ml screw-cap microcentrifuge tube. Tightly seal the microcentrifuge tube.
Place tube in float and float in beaker filled with water. Bring water to a boil (∼95°C) for 5 min.
Remove the beaker from the stirring hot plate and allow beaker to come to room temperature gradually.
-
Store the renatured duplex at 4°C until use for crystallization (up to 2 weeks).
For prolonged storage (>2 weeks), store the sample frozen (preferable at –70°C) and renature again prior to use. The sample may be stored for up to 1 yr.
If both the purchasing of purified oligos and in lab sythesis and purification by HPLC are not possible, an alternate protocol for the synthesis and preparation of nucleic acid substrates using basic molecular biology techniques and resources is available (Muecke, 2008). In brief, this technique utilizes self primed amplification of a DNA fragment of interest that is flanked by cohesive end fragments of bacteriophage lambda and restriction sites. The DNA that is amplified through the detailed protocol can be further purified to homogeneity by ion exchange chromatography, and has been shown to be amenable to crystallization experiments.
ALTERNATE PROTOCOL 2
PREPARATION OF MACROMOLECULAR COMPLEXES FOR CRYSTALLIZATION
There are two types of macromolecular complexes that are generally prepared for crystallization: protein–nucleic acid and protein-protein complexes. For protein–nucleic acid complexes, the concentrated macromolecules (see Basic Protocol 1 and Alternate Protocol 1) are mixed in near stoichiometric amounts. It has been empirically determined that a slight excess of nucleic acid over protein (∼20%; Aggarwal, 1990) is best. In general, one should aim for 0.8 mM complex.
Some protein-nucleic acid complexes form precipitate when mixed at the high concentrations required for crystallization. There are two common solutions to this problem: (1) lower the concentration of complex 2- to 3-fold, or (2) add monovalent or divalent metal ions at increasing concentrations. Generally, one should first try adding monovalent ions at a concentration of 25 mM, and increase slowly to a maximum of 200 mM if the precipitate persists. For divalent ions, one should start at 10 mM increasing to a maximum of 75 mM. NaCl is the most popular monovalent ion, while MgCl2 and CaCl2 are the most commonly used divalent ions. Although divalent ions are often effective in solubilizing some protein-nucleic acid complexes, they should only be used as a last resort, because they may give rise to salt crystals (this typically occurs under crystallization conditions that contain sulfate or phosphate counterions), which may be mistaken for macromolecular crystals (to determine if this is the case, see Support Protocol 1). Additionally, DNA binding proteins may prove to be unstable in the absence of cofactors or DNA substrates, and thus purification and concentration of these proteins alone may prove problematic. The addition of the DNA substrate prior to concentration may help to enhance the stability of the protein and allow the protein to be concentrated to levels suitable for crystallization trials.
Homogeneous protein-protein complexes are generally prepared by isolating the desired complex by gel filtration prior to concentration for crystallization. Multiprotein-nucleic acid complexes can be prepared either by mixing the preformed concentrated protein complex with the concentrated nucleic acid sample in appropriate stoichiometric amounts, or by isolating the multiprotein-nucleic acid complex by gel filtration (e.g., UNIT 8.3) prior to concentration for crystallization.
Many protein complexes fail to express as individual components for assembly and purification by gel filtration. However, recent developments in bacterial and eukaryotic expression techniques have provided new platforms that allow the expression of multiple proteins from a single expression vector (e.g., Unit 5.20 and Unit 5.21). This strategy allows for assembly of the protein complex in vivo, which often times proves essential for complex formation. The complexes can be purified using the same methodologies that are employed for individual recombinant proteins (employing a cleavable tag on one of the protein subunits of the complex) and subjected to the same crystallization trials.
The field of membrane protein crystallography has grown considerably in the past decade, and the techniques for membrane protein expression, purification, and crystallization have developed greatly as well. While many of the techniques used to set up initial crystallization trials of membrane proteins are identical for to the techniques described below, certain considerations must be made during the purification process and sample preparation. It is highly recommended that specific attention be given to these methods prior to setting up initial crystallization trials of membrane proteins (e.g Unit 17.9).
BASIC PROTOCOL 2
PREPARATION AND ANALYSIS OF CRYSTALLIZATION TRIALS
UNIT 17.3 discusses parameters that effect the crystallization of soluble macromolecules. Overall, the most important parameter in obtaining well-ordered macromolecular crystals is the actual macromolecular sample (see Critical Parameters and Troubleshooting). Purity, protein monodispersity, and stability (i.e., remains intact) are the most critical factors in the ability of a protein sample to crystallize. An overloaded PAGE gel (UNIT 10.1) should be run to check for contaminants. Ideally, the protein sample should be >99% pure. Gel filtration (UNIT 8.3) or dynamic light scattering (DLS; UNIT 17.10) are suitable methods for assessing protein monodispersity. In the case of protein expression in eukaryotic expression systems, mass spectrometry (Unit 16) of the sample can be used to check for post-translational modification of the protein, which may adversely effect crystallization properties.
Primarily three components make up any crystallization solution: salt, buffer, and precipitating agent. Because there is an infinite number of combinations of these parameters (McPherson, 1982), a complete, methodical search for the crystallization conditions that give rise to macromolecular crystals would take an enormous amount of time and material. There are basically two approaches for pursuing crystallization conditions of a macromolecule which has never previously been crystallized: screening matrix and a systematic approach.
The most popular approach is the screening matrix, which is employed to quickly determine which variables influence the behavior of the protein solution. This so-called shotgun approach utilizes a rather large number of wide-ranging conditions unrelated to one another. The goal of this procedure is to narrow the conditions that yield macromolecular crystals. One would typically refine the components of the successful crystallization condition(s) by separately varying the concentration of the components. (see Basic Protocol 3.) The first widely popular screen, described by Jancarik and Kim in 1991, was heavily biased towards previously published crystallization conditions (Table 17.4.1). In 1992, the Jancarik and Kim screen became commercially available as “Crystal Screen” from Hampton Research. Since then, there has been a proliferation in the number of companies that provide sparse matrix solutions for performing initial screenings (Table 17.4.2). There has also been an expansion in the number of different crystallization screens, many dedicated to certain types of macromolecules. For instance, Natrix (Hampton) is designed for nucleic acids or nucleic acid–protein complexes, the MemStart (Molecular Dimensions) screens are suitable for membrane proteins, and the Protein Complex Suite (Qiagen) is recommended for protein-protein complexes. It should be remembered that a sparse matrix screen which does not yield promising crystals does not mean that a particular macromolecule will not crystallize. For example, lysozyme, canavalin, and many immunoglobulins will not crystallize using the commercially available kits.
Table 17.4.1.
Crystallization Matrix Components Used in Crystal Screen 1 (Hampton Research) to Crystallize Proteins
| Precipitant | Salt | Buffer |
|---|---|---|
| Ammonium phosphate | Ammonium acetate | Imidazole, pH 6.5 |
| Ammonium sulfate | Ammonium sulfate | Sodium acetate, pH 4.6 |
| Lithium sulfate | Calcium acetate | Sodium citrate, pH 5.6 |
| Magnesium formate | Calcium chloride | Sodium cacodylate, pH 6.5 |
| MPDa | Lithium sulfate | Sodium HEPES, pH 7.5 |
| PEGb (400, 1500, 4000, 8000) | Magnesium acetate | Tris·Cl, pH 8.5 |
| Potassium phosphate | Magnesium chloride | |
| Potassium tartrate | Sodium acetate | |
| 2-Propanol | Sodium citrate | |
| Sodium acetate | Zinc acetate | |
| Sodium citrate | ||
| Sodium formate | ||
| Sodium phosphate | ||
| Sodium tartrate |
MPD, 2-methyl-2,4-pentanediol.
PEG, polyethylene glycol.
Table 17.4.2.
Commercially Available Crystallization Screens
| Screens | Comment |
|---|---|
| Hampton Researcha | |
| Crystal Screen | Original Jancarik & Kim Screen |
| Crystal Screen 2 | Extension of original Jancarik & Kim screen |
| Natrix | Screen for nucleic acids or protein–nucleic acid complexes |
| Nucleic Acid Mini Screen | Screen for nucleic acids, Various salts/pH screen |
| Index | Combines classic, contemporary, new conditions |
| PEG/Ion | Sparse matrix of anions and cations mixed with various PEGs |
| Crystal Screen Cryo | Crystal Screen with glycerol added |
| PEGRx | Screen of various polymers |
| Salt Rx | Various salts/pH screen |
| MembFac | Screen for membrane proteins |
| Grid Screen ammonium sulfate | Ammonium sulfate/pH screen |
| Grid Screen PEG 6000 | PEG 6000/pH screen |
| Grid Screen MPD | MPD/pH screen |
| Grid Screen PEG/LiCl | PEG 6000/pH screen |
| Grid Screen sodium chloride | Sodium chloride/pH screen |
| Quick Screen | Sodium potassium phosphate/pH screen |
| Low Ionic Strength Screen | PEG 3350/pH screen, special protocol is required |
| Silver Bullets | Portfolio of small molecules |
| Detergent Screen | Screen of popular detergents for membrane proteins |
| Emerald Biosciencesb | |
| Wizard I | Sparse matrix |
| Wizard II | Random sparse matrix |
| Cryo I | Random sparse matrix with cryo added |
| Cryo II | Random sparse matrix with cryo added |
| JCSG+ Screen | Screen of conditions proven to yield crystals |
| Precipitant Synergy 64 | Screen of mechanistically distinct precipitating agents |
| Beta-mem screen | Sparse matrix for crystallization of beta-barrel proteins |
| Molecular Dimensionsc | |
| Crsystal Strategy Screen I | Screens salts, PEGs, and cryoprotectants |
| Crystal Strategy II | pH’s, PEGs, and cryoprotectants |
| Heavy+Lite Screen | Screens in the metastable zone |
| PACT screen | PEG, ion, and pH |
| Lipidic-Sponge Screen | Targets bacterial and eukaryotic membrane proteins |
| MemGold | Based on proven membrane conditions from PDB |
| MemStart | Screen and optimize alpha helical membrane proteins |
| ProPlex | Sparse matrix aimed at crystallizing protein complexes |
| NR-LBD | Rational approach for nuclear receptor ligand binding domain crystallization |
| Morpheus Screen | Sparse matrix that includes low MW compounds that promote crystal formation |
| PGA Screen | Systematic screen based on the poly-γ-glutamic acid polymer |
| Qiagend | |
| The Classics Suite | Sparse matrix similar to Hampton Crystal Screen |
| JCSG Core Suite | Optimized Structural Genomics Screen |
| MbClass I and II | Screens optimized for membrane protein crystallization |
| Nucleix Suite | Screen for nucleic acids |
| Cubic Phase Suite | Screen for membrane protein crystallization |
| ComPAS Suite | Screen of alcohols, salts, and polymers |
| MPD Suite | Range of conditions with MPD added. |
Refer to http://www.hamptonresearch.com for more details.
Refer to http://www.emeraldbiosystems.com for more details.
Refer to http://www.moleculardimensions.com for more details.
Refer to http://www.qiagen.com/ProteinCrystal for more details.
The second method for macromolecular crystallization involves systematically obtaining information about the solubility of the particular macromolecule (Stura, 1994). This is done by using a matrix containing increasing concentrations of precipitating agent along one axis versus pH along the other. In this way, the solubility curve of the protein can be determined, along with the optimum pH or type or concentration of precipitating agent. The observation of dull or fibrous precipitate may permit the elimination of a particular pH or precipitating agent. On the other hand, observing a shiny precipitate or one which appears colorful (birefringence) when viewed through a polarizer may warrant further investigation (see Basic Protocol 3). Subsequent crystallization trials may then focus on the introduction of additives such as salt, alcohol (ethanol), or MPD additives to promote crystallization of the protein. This approach involves repeated and methodical probing of the solubility curve of the macromolecule until crystals are obtained.
A summary of the different screens and their applications is given in Table 17.4.2. Provided that the macromolecular sample is amenable to crystallization, these screens will generally yield directions in which to proceed in order to obtain suitable crystals for structural studies, if not diffraction-quality crystals themselves.
Although each of the commercially available premixed solutions can be prepared in the laboratory, it is convenient, at least initially, to obtain the premixed solutions. Any reagents that are not obtained from vendors should generally be of the highest-quality grade. This is especially true for PEG. This protocol focuses only on screens that are used most often for crystallizing soluble proteins, nucleic acids and macromolecular complexes.
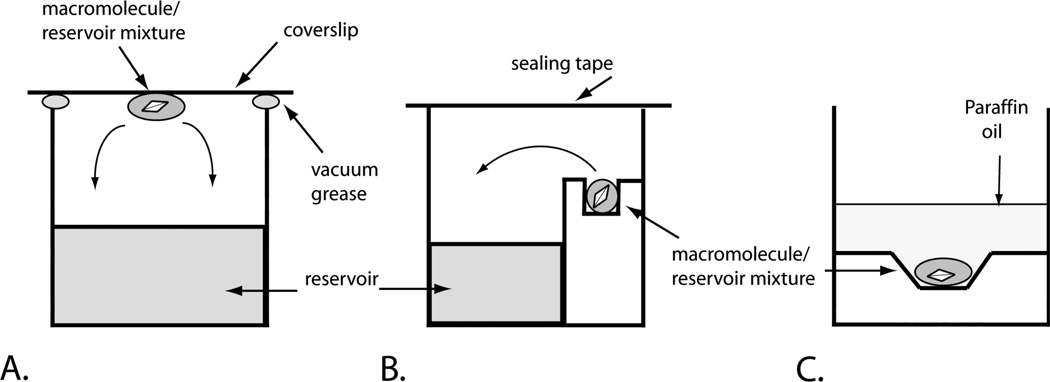
There are several different crystallization techniques that are used for macromolecules (Weber, 1997 and Rupp 2010). Our initial discussion here will describe the vapor-diffusion technique, using hanging drops (Fig. 17.4.1 A). Steps 1 to 9 should be carried out both at room temperature and 4°C. Both the Jancarik and Kim sparse crystallization screen (Crystal Screen) and the extended sparse crystallization screen (Crystal Screen 2 or its equivalent from another vendor) should be employed for optimal results with proteins, although at a minimum the sparse matrix screen can be used alone. It is recommended that macromolecular complexes (i.e. protein-nucleic acids or protein-protein complexes) also be screened using the complex specific screen (Natrix, Protein Complex, respectively), as these screens are generated from conditions shown to be useful for crystallizing the respective complexes. Crystallization droplets will typically show one of three results: clear drops, crystals, or precipitate/phase separation (Hereafter referred to as O, for clear drops; C, for crystals; and P, for precipitate or phase separation; see Anticipated Results).
Figure 17.14.1.
Typical crystallization setups. (See text for descriptions)
Materials
Crystallization screening kit(s) (Hampton Research):
Proteins and multiprotein complexes: Crystal Screens 1 (Table 17.4.3) and 2 (latter is optional)
Nucleic acids and protein–nucleic acid complexes: Natrix (Table 17.4.4)
Concentrated macromolecule sample: protein (see Basic Protocol 1), nucleic acid (see Alternate Protocol 1), or macromolecular complex (see Alternate Protocol 2)
Thomas Lubriseal Stopcock Grease
Small aluminum foil weigh dish
50 ml Erlenmeyer flask with 2.5 cm diameter opening
Hot plate
24-well crystallization trays (VDX Plate, Hampton Research)
22-mm2 plastic or siliconized coverslips (Hampton Research)
Self-closing tweezers
Microscope with ≥40× magnification, light source, and viewing platform (at least 3 × 4 in.), preferably with polarizer (e.g., Leica MZ16, Zeiss STEMI SV11, Nikon SMZ1000)
Table 17.4.3.
Components of Crystal Screen 1 (Hampton Research) for Soluble Proteinsa
| Condition | Formulationb |
|---|---|
| 1 | 30% MPD, 0.1 M sodium acetate, pH 4.6, 0.02 M calcium chloride |
| 2 | 0.4 M potassium/sodium tartrate |
| 3 | 0.4 M ammonium phosphate |
| 4 | 2.0 M ammonium sulfate, 0.1 M Tris·Cl, pH 8.5 |
| 5 | 30% MPD, 0.1 M sodium HEPES, pH 7.5, 0.2 M sodium citrate |
| 6 | 30% PEG 4000, 0.1 Tris·Cl, pH 8.5, 0.2 M magnesium chloride |
| 7 | 1.4 M sodium acetate, 0.1 M sodium cacodylate, pH 6.5 |
| 8 | 30% 2-propanol, 0.1 M sodium cacodylate, pH 6.5, 0.2 M sodium citrate |
| 9 | 30% PEG 4000, 0.1 M sodium citrate, pH 5.6, 0.2 M ammonium acetate |
| 10 | 30% PEG 4000, 0.1 M sodium acetate, pH 4.6, 0.2 M ammonium acetate |
| 11 | 1.0 M ammonium phosphate, 0.1 M sodium citrate, pH 5.6 |
| 12 | 30% 2-propanol, 0.1 M sodium HEPES, pH 7.5, 0.2 M magnesium chloride |
| 13 | 30% PEG 400, 0.1 M Tris·Cl, pH 8.5, 0.2 M sodium citrate |
| 14 | 28% PEG 400, 0.1 M sodium HEPES, pH 7.5, 0.2 M calcium chloride |
| 15 | 30% PEG 8000, 0.1 M sodium cacodylate, pH 6.5, 0.2 M ammonium sulfate |
| 16 | 1.5 M lithium sulfate, 0.1 M sodium HEPES, pH 7.5 |
| 17 | 30% PEG 4000, 0.1 M Tris·Cl pH 8.5, 0.2 M lithium sulfate |
| 18 | 20% PEG 8000, 0.1 M sodium cacodylate, pH 6.5, 0.2 M magnesium acetate |
| 19 | 30% 2-propanol, 0.1 M Tris·Cl, pH 8.5, 0.2 M ammonium acetate |
| 20 | 25% PEG 4000 |
| 21 | 30% MPD, 0.1 M sodium cacodylate, pH 6.5, 0.2 M magnesium acetate |
| 22 | 30% PEG 4000, 0.1 M Tris·Cl, pH 8.5, 0.2 M sodium acetate |
| 23 | 30% PEG 400, 0.1 M sodium HEPES, pH 7.5, 0.2 M magnesium chloride |
| 24 | 20% 2-propanol, 0.1 M sodium acetate, pH 4.6, 0.2 M calcium chloride |
| 25 | 1.0 M sodium acetate, 0.1 M imidazole, pH 6.5 |
| 26 | 30% MPD, 0.1 M sodium citrate, pH 5.6, 0.2 M ammonium acetate |
| 27 | 20% 2-propanol, 0.1 M sodium HEPES, pH 7.5, 0.2 M sodium citrate |
| 28 | 30% PEG 8000, 0.1 M sodium cacodylate, pH 6.5, 0.2 M sodium acetate |
| 29 | 0.8 M potassium/sodium tartrate, 0.1 M sodium HEPES, pH 7.5 |
| 30 | 30% PEG 8000, 0.2 M ammonium sulfate |
| 31 | 30% PEG 4000, 0.2 M ammonium sulfate |
| 32 | 2.0 M ammonium sulfate |
| 33 | 4.0 M sodium formate |
| 34 | 2.0 M sodium formate, 0.1 M sodium acetate, pH 4.6 |
| 35 | 1.6 M sodium/potassium phosphate, 0.1 M sodium HEPES, pH 7.5 |
| 36 | 8% PEG 8000, 0.1 M Tris·Cl, pH 8.5 |
| 37 | 8% PEG 4000, 0.1 M sodium acetate, pH 4.6 |
| 38 | 1.4 M sodium citrate, 0.1 M sodium HEPES, pH 7.5 |
| 39 | 2% PEG 400, 2.0 M ammonium sulfate, 0.1 M sodium HEPES, pH 7.5 |
| 40 | 20% 2-propanol, 20% PEG 4000, 0.1 M sodium citrate, pH 5.6 |
| 41 | 10% 2-propanol, 20% PEG 4000, 0.1 M sodium HEPES, pH 7.5 |
| 42 | 20% PEG 8000, 0.05 M potassium phosphate |
| 43 | 30% PEG 1500 |
| 44 | 0.2 M magnesium formate |
| 45 | 18% PEG 8000, 0.1 M sodium cacodylate, pH 6.5, 0.2 M zinc acetate |
| 46 | 18% PEG 8000, 0.1 M sodium cacodylate, pH 6.5, 0.2 M calcium acetate |
| 47 | 2.0 M ammonium sulfate, 0.1 M sodium acetate, pH 4.6 |
| 48 | 2.0 M ammonium phosphate, 0.1 M Tris·Cl, pH 8.5 |
MPD, 2-methyl-2,4-pentanediol; PEG, polyethylene glycol.
Table 17.4.4.
Components of Natrix (Hampton Research) for Nucleic Acid and Protein-Nucleic Acid Complexes Screena
| Condition | Formulationb |
|---|---|
| 1 | 0.01 M magnesium chloride, 0.05 M MES, pH 5.6, 1.8 M lithium sulfate |
| 2 | 0.01 M magnesium acetate, 0.05 M MES, pH 5.6, 2.5 M ammonium sulfate |
| 3 | 0.1 M magnesium acetate, 0.05 M MES, pH 5.6, 20% MPD |
| 4 | 0.2 M KCl, 0.01 M magnesium sulfate, 0.05 M MES, pH 5.6, 10% PEG 400 |
| 5 | 0.2 M KCl, 0.01 M magnesium chloride, 0.05 M MES, pH 5.6, 5% PEG 8000 |
| 6 | 0.1 M ammonium sulfate, 0.01 M magnesium chloride, 0.05 M MES, pH 5.6, 20% PEG 8000 |
| 7 | 0.02 M magnesium chloride, 0.05 M MES, pH 6.0, 15% isopropanol |
| 8 | 0.005 M magnesium sulfate, 0.1 M ammonium acetate, 0.05 M MES, pH 6.0, 0.6 M NaCl |
| 9 | 0.1 M KCl, 0.01 M magnesium chloride, 0.05 M MES, pH 6.0, 10% PEG 400 |
| 10 | 0.005 M magnesium sulfate, 0.05 M MES, pH 6.0, 5% PEG 4000 |
| 11 | M magnesium chloride, 0.05 M sodium cacodylate, pH 6.0, 1.0 M lithium sulfate |
| 12 | M magnesium sulfate, 0.05 M sodium cacodylate, pH 6.0, 1.8 M lithium sulfate |
| 13 | 0.015 M magnesium acetate, 0.05 M sodium cacodylate, pH 6.0, 1.7 M ammonium sulfate |
| 14 | M KCl, 0.025 M magnesium chloride, 0.05 M sodium cacodylate, pH 6.0, 15% isopropanol |
| 15 | 0.04 M magnesium chloride, 0.05 M sodium cacodylate, pH 6.0, 5% MPD |
| 16 | 0.04 M magnesium acetate, 0.05 M sodium cacodylate, pH 6.0, 30% MPD |
| 17 | M KCl, 0.01 M calcium chloride, 0.05 M sodium cacodylate, pH 6.0, 10% PEG 4000 |
| 18 | M magnesium acetate, 0.05 M sodium cacodylate, pH 6.5, 1.3 M lithium sulfate |
| 19 | M magnesium sulfate, 0.05 M sodium cacodylate, pH 6.5, 2.0 M ammonium sulfate |
| 20 | M ammonium acetate, 0.015 M magnesium acetate, 0.05 M sodium cacodylate, pH 6.5, 10% isopropanol |
| 21 | 0.2 M KCl, 0.005 M magnesium chloride, 0.05 M sodium cacodylate, pH 6.5, 10% 1,6-hexanediol |
| 22 | 0.08 M magnesium acetate, 0.05 M sodium cacodylate, pH 6.5 |
| 23 | M KCl, 0.01 M magnesium chloride, 0.05 M sodium cacodylate, pH 6.5, 10% PEG 4000 |
| 24 | 0.2 M ammonium acetate, 0.01 M calcium chloride, 0.05 M sodium cacodylate, pH 6.5, 10% PEG 4000 |
| 25 | 0.08 M magnesium acetate, 0.05 M sodium cacodylate, pH 6.5, 30% PEG 4000 |
| 26 | M KCl, 0.1 M magnesium acetate, 0.05 M sodium cacodylate, pH 6.5, 10% PEG 8000 |
| 27 | M ammonium acetate, 0.01 M magnesium acetate, 0.05 M sodium cacodylate, pH 6.5, 30% PEG 8000 |
| 28 | 0.05 M magnesium sulfate, 0.05 M sodium HEPES, pH 7.0, 1.6 M lithium sulfate |
| 29 | 0.01 M magnesium chloride, 0.05 M sodium HEPES, pH 7.0, 4.0 M lithium chloride |
| 30 | 0.01 M magnesium chloride, 0.05 M sodium HEPES, pH 7.0 |
| 31 | 0.005 M magnesium chloride, 0.05 M sodium HEPES, pH 7.0, 25% PEG monomethyl ether 550 |
| 32 | 0.2 M KCl, 0.01 M magnesium chloride, 0.05 M sodium HEPES, pH 7.0, 20% 1,6- hexanediol |
| 33 | 0.2 M ammonium chloride, 0.01 M magnesium chloride, 0.05 M sodium HEPES, pH 7.0, 30% 1,6-hexanediol |
| 34 | 0.1 M KCl, 0.005 M magnesium sulfate, 0.05 M sodium HEPES, pH 7.0, 15% MPD |
| 35 | M KCl, 0.01 M magnesium chloride, 0.05 M sodium HEPES, pH 7.0, 5% PEG 400 |
| 36 | 0.1 M KCl, 0.01 M calcium chloride, 0.05 M sodium HEPES, pH 7.0, 10% PEG 400 |
| 37 | M KCl, 0.025 M magnesium sulfate, 0.05 M sodium HEPES, pH 7.0, 20% PEG 200 |
| 38 | 0.2 M ammonium acetate, 0.15 M magnesium acetate, 0.05 M sodium HEPES, pH 7.0, 5% PEG 4000 |
| 39 | M ammonium acetate, 0.02 M magnesium chloride, 0.05 M sodium HEPES, pH 7.0, 5% PEG 8000 |
| 40 | 0.01 M magnesium chloride, 0.05 M Tris·Cl, pH 7.5, 1.6 M ammonium sulfate |
| 41 | M KCl, 0.015 M magnesium chloride, 0.05 M Tris·Cl, pH 7.5, 10% PEG monomethyl ether 550 |
| 42 | 0.01 M magnesium chloride, 0.05 M Tris·Cl, pH 7.5, 5% isopropanol |
| 43 | M magnesium chloride, 0.05 M ammonium acetate, 0.05 M Tris·Cl, pH 7.5, 10% MPD |
| 44 | 0.2 M KCl, 0.05 M magnesium chloride, 0.05 M Tris·Cl, pH 7.5, 10% PEG 4000 |
| 45 | 0.025 M magnesium sulfate, 0.05 M Tris·Cl, pH 8.5, 1.8 M ammonium sulfate |
| 46 | 0.005 M magnesium sulfate, 0.05 M Tris·Cl, pH 8.5, 35% 1,6-hexanediol |
| 47 | 0.1 M KCl, 0.01 M magnesium chloride, 0.05 M Tris·Cl, pH 8.5, 30% PEG 400 |
| 48 | M calcium chloride, 0.2 M ammonium chloride, 0.05 M Tris·Cl, pH 8.5, 30% PEG 4000 |
MES, 2-(N-morpholino)ethanesulfonic acid; MPD, 2-methyl-2,4-pentanediol; PEG, polyethylene glycol.
NOTE: Preparation and crystallization (steps 1 to 9) should be done both at 4°C and room temperature (±3°C).
Prepare crystallization trays
-
1.
Place a liberal amount of the Lubriseal grease in the aluminum dish and heat on the hot plate until liquefied. Once liquefied, turn down heat to avoid burning but retain liquid state.
-
2.
Dip the opening of the flask into the liquid grease, and then touch the flask to the rim of each well on the 24-well plate, leaving behind a complete ring of grease. (Dip flask back into grease on hot-plate as needed. Grease at least two 24-well plates (48 conditions) to set up the Crystal Screen 1. (see Figure 17.4.4for representation)
-
3.
Mix the solutions (reservoir mixes) in the appropriate crystallization screening kit according to manufacturer’s instructions.
-
4.
Add 500 µl of each reservoir mix to a separate well, using a new pipet tip for each transfer.
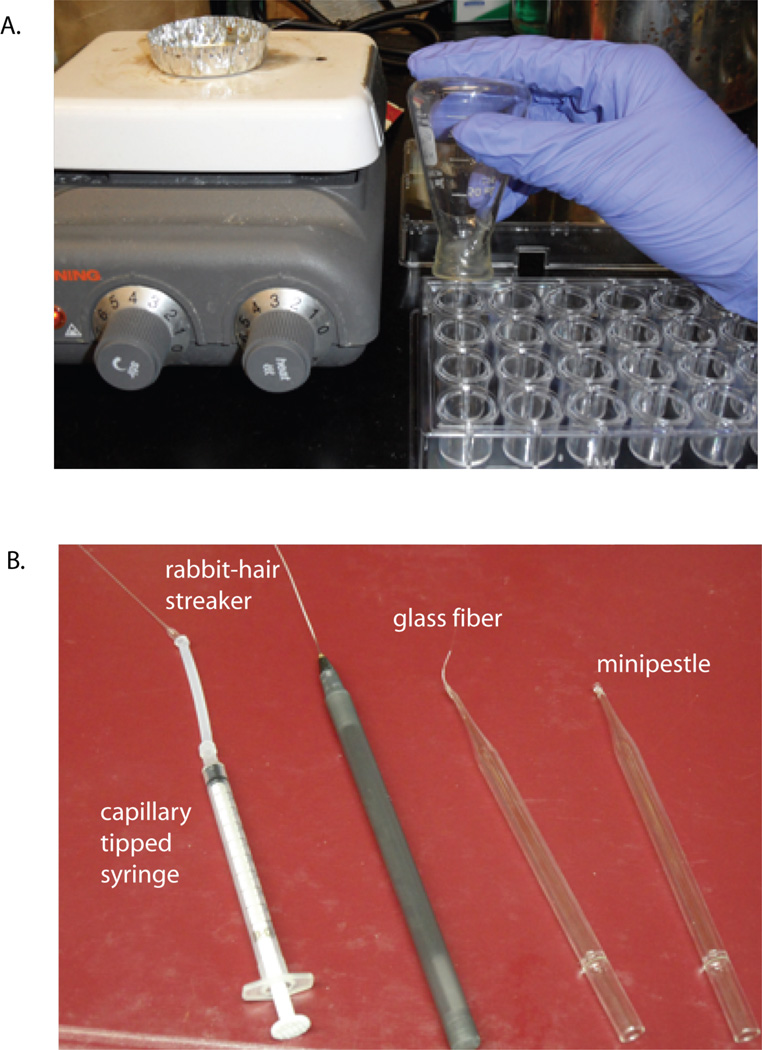
Figure 17.14.4.
Crystal tray greasing setup and typical home-made crystal manipulation tools
Set up hanging drops
-
5.
Place 1 µl concentrated macromolecule sample onto the center of a 22-mm2 plastic or siliconized coverslip. Keep stock macromolecule sample on ice.
-
6.
With a new tip, withdraw 1 µl from the reservoir mix in the first well. Add this to the 1-µl sample aliquot and pipet up and down several times to mix, taking care not to introduce air bubbles into the droplet.
-
7.
Flip the coverslip over with self-closing tweezers so that the drop is hanging down from the bottom of the coverslip. Place the coverslip over the appropriate well solution. To seal the coverslip-well interface, gently tap the edges of the coverslip into the grease using the bottom of the tweezers.
Figure 17.4.1 A illustrates the components of a completed hanging drop vapor-diffusion setup. Arrows represent path of diffusion that drives crystallization.
-
8.
Repeat steps 5 to 7 for the rest of the well solutions.
Incubate
-
9.
After all of the hanging drops have been prepared, tape together the top and bottom of each tray. Transfer the trays to a location that is free from vibrations and is maintained at a fairly constant temperature (±3°C). Leave the trays undisturbed for at least 2 days.
In general, crystallization trays should not be discarded for at least 6 months. Some proteins have been known to undergo time-dependent modifications that promote crystallization.
Analyze results
-
10.
Use a microscope with ≥40× magnification, light source, and viewing platform (preferably with polarizer) to view the crystallization drops. Note whether the drops are clear (O), show crystals (C), or show precipitate or phase separation (P).
-
11.
For those drops that produce crystals, note the crystallization conditions and see if there is a common parameter (i.e., precipitant, pH, or ionic strength). Also note the crystal size, shape, and ability to polarize light.
In general, large (≥0.4-mm3) crystals that have sharp edges and that polarize light are preferable.
-
12.
Continue to observe the crystallization trials every 2 days until no further change is noted in the drops.
Alternate Protocol 3
Crystallization screening using high-throughput robotics
The development of new crystallographic technologies have revolutionized the way in which we initiate and pursue crystal structures. The automated liquid handling devices manufactured by several different companies (i.e. Art Robbins Instruments and TTP Labtech) allow us to set up crystallization conditions using nanoliter (nl) volumes of protein samples and crystallization reagents, as well as provide an accurate and reproducible source of liquid dispensing that is far superior than could be achieved by hand. This allows us to screen a much larger set of conditions with a much smaller amount of macromolecular sample and have the confidence that all the conditions were set up in the same manner. Depending of the type of crystallization robot used, the crystallization setup can be altered as well, allowing for not only hanging drop vapor diffusion experiments but sitting drop vapor diffusion (Figure 17.4.1 B) and microbatch under oil (Figure 17.4.1 C). Our discussion here will briefly describe the set up of the sitting drop vapor diffusion method using the Art Robbins Crystal Phoenix robot highlighting the benefits of using the robotic system. However, this description provides only a very general overview and specific attention should be given to the manufacturers detailed protocols if one uses these systems.
Materials
Crystal Screen 1 and 2
Intelli-Plate 96 (Art Robbins Instruments)
MASTERBLOCK 96 Deep Well Plate (Hampton Research)
Crystal Clear Sealing Tape (Hampton Research)
Microplate adhesive film (USA Scientific or other manufacturer)
Concentrated protein or macromolecular complex (Protocol 1)
Art Robbins Crystal Phoenix robot
Preparing deep well block
Pipette 500 ul of each of the 98 conditions from Crystal Screen 1 and 2 into the Deep well plate starting at position A1 and ending at H12.
- Use Microplate adhesive seal to top of the block. Ensure that the conditions do not mix with each other, and store block in a dark temperature stable (approx. room temperature) compartment.If kept properly sealed and in a temperature stable environment, screen should be good for several months. Before use, the block should be observed to make sure conditions have not dried out or precipitated.
Setting up sitting drops
Place the deep well block, Intelli-Plate, and protein sample in the proper docks on the robot stage.
Program the 96 syringe head to withdraw 90 ul of each of the crystallization conditions from the deep well block and then dispense 80 ul into the reservoirs of the Intelli-Plate.
Program the 96 syringe head to dispense 200 nl of each other the crystallization conditions into the well of the corresponding reservoir.
Program the nano-dispenser to place 200 nl of protein into each of the 96 wells with the crystallization conditions. (This set up is outlined in Figure 17.4.1 B. Arrows represent path of diffusion that drives crystallization)
Seal the top of the plate with sealing tape and place in temperature stable environment where it will not be disturbed.
Follow the same protocol above for incubating and analyzing plates. As mentioned previously, the use of an automated liquid handler will greatly increase the number of trays you will be able to set up with a purchased screen (80 ul (robot) vs. 500 ul (by hand, Protocol 1)), as well as the number of trays you will be able to set up with your sample (200 nl (robot) vs. 1 ul (by hand)). If these resources are available it is highly recommended that they be utilized to increase your output and control experimental variables.
SUPPORT PROTOCOL 1
ESTABLISHING WHETHER CRYSTALS ARE MACROMOLECULE OR CRYSTALLIZATION REAGENT USING A CRYOLOOP
Often one obtains crystals that are salt or another crystallization reagent rather than macromolecule. This occurs notoriously when the crystallization conditions contain divalent ions such as magnesium or calcium with phosphate or sulfate counterions. If crystals are suspected to be salt, prepare a control crystallization trial in which everything but the macromolecule is included. For example, after concentrating in a microconcentrator, the flow-through can be used instead of the protein sample. In preparing this control, substitute the macromolecule sample with the storage buffer that is identical to that used for the macromolecule in the drops. If this test does not show crystals, one can analyze a washed crystal directly by SDS-PAGE (UNIT 10.1). The procedure for preparing an SDS-PAGE sample from a crystal is described here. A crystal that is at least 0.2 mm3 is usually large enough to wash with a cryoloop and dissolve for visualization by SDS-PAGE and silver staining (UNIT 10.5; both proteins and nucleic acids are stained with silver reagent). The procedure involves extensively washing the crystal surface, so that it is free of macromolecule from the hanging drop, before dissolving the crystal for gel analysis.
Materials
Crystallization tray(s) containing 0.2-mm3 (or larger) crystal(s) and reservoir solutions (see Basic Protocol 2)
Macromolecule storage solution (i.e., solution in which the macromolecule is soluble)
Purified macromolecule
2× SDS sample buffer (UNIT 10.1)
Microscope with ≥40× magnification, light source, and viewing platform (at least 3 × 4 in.), preferably with polarizer (e.g., Leica MZ16, Zeiss STEMI SV11, Nikon SMZ1000)
Cryoloops (Hampton Research)
Plastic petri dish
Additional reagents and equipment for SDS-PAGE (UNIT 10.1) and silver staining (UNIT 10.5)
Invert the coverslip harboring 0.2-mm3 crystals from the crystallization tray and place on a flat surface. Pipet 100 µl of the respective reservoir solution directly onto the drop.
-
View the drop under the microscope with ≥40× magnification, use a cryoloop to mix the solution in the drop, and gently move the crystal to make sure it is detached from the coverslip surface.
(Cracking of the crystal, while normally bad, can be a positive indicator at this point that you have obtained protein crystals. Salt or other small molecule crystals are extremely durable and are generally resistant to cracking when poked.)
Pipet three separate 100-µl drops of reservoir solution into a plastic petri dish.
-
Place the cryoloop under the crystal such that the crystal is hanging half inside and half outside the circle, then lift the loop swiftly upward. Transfer the crystal to the first 100-µl drop, shake slightly to pry the crystal off the loop, and mix the solution slightly.
When the crystal is “lassoed” in this manner, it is held in the loop by the surface tension of the liquid that is attached to the rim of the loop.
Transfer the crystal to the second drop, mix slightly by swirling the loop in the reservoir solution, and finally transfer the crystal to the third drop.
Use the cryoloop to transfer the crystal to a microcentrifuge tube containing 5 µl macromolecule storage solution, and shake the cryoloop to release the crystal. Vortex the solution to fully dissolve the crystal.
-
Set up the following controls:
For a negative control, transfer 5 µl from the third washing drop to a microcentrifuge tube containing 5 µl of the macromolecule storage solution.
For a positive control, add 1 µg purified macromolecule in a final volume of 5 µl macromolecule storage solution to another microcentrifuge tube.
The negative control ensures that all of the free macromolecule in solution has been washed from the crystal surface.
Add 5 µl of 2× SDS sample buffer to each tube and analyze the samples by SDS-PAGE (UNIT 10.1). Visualize macromolecule(s) by silver staining the gel (UNIT 10.5).
BASIC PROTOCOL 3
OPTIMIZATION OF CRYSTALLIZATION PARAMETERS
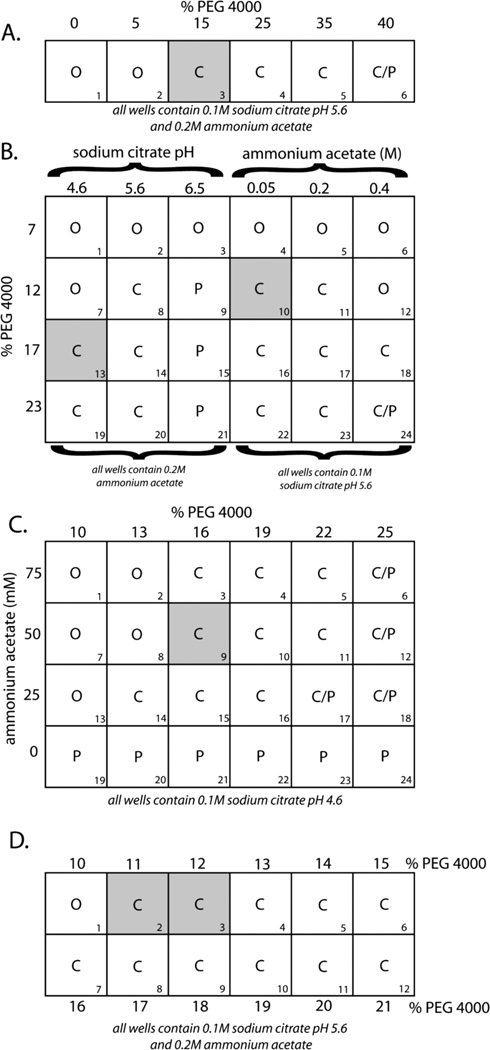
Although crystals that are obtained from sparse matrix screens are sometimes already suitable for structural studies, more often the crystals need to be optimized prior to crystallographic analysis. Common problems include crystals that are too small, too thin, clustered, or intergrown. In these cases, the optimization of precipitant concentration, pH, and ionic strength often improve crystal quality. There are different ways of optimizing crystallization parameters. The procedure generally used in the author’s laboratory is presented here. Figure 17.4.2 illustrates a typical optimization procedure, which is discussed further in the steps below.
Figure 17.14.2.
Optimization of crystallization parameters (see Basic Protocol 3 for discussion)
Materials
Thomas Lubriseal Stopcock Grease
Small aluminum foil weigh dish
50 ml Erlenmeyer flask with 2.5 cm diameter opening
Hot plate
Concentrated reagents needed to set up crystal screens (for our example described here: 1M sodium citrate pH 5.6, 4.6, and 6.5; 2M ammonium acetate; 50% PEG 4000)
Concentrated macromolecule sample: protein (see Basic Protocol 1), nucleic acid (see Alternate Protocol 1), or macromolecular complex (see Alternate Protocol 2)
24-well crystallization trays (VDX Plate, Hampton Research)
22-mm2 plastic or siliconized coverslips (Hampton Research)
Self-closing tweezers
Microscope with ≥40× magnification, light source, and viewing platform (at least 3 × 4 in.), preferably with polarizer (e.g., Leica MZ16, Zeiss STEMI SV11, Nikon SMZ1000)
Additional reagents and equipment for preparation of crystallization trials (see Basic Protocol 2)
-
For each crystallization condition that yields crystals, prepare a crystallization tray (see Basic Protocol 2, steps 1 and 2) that contains an initial one-dimensional grid to optimize the concentration of precipitant. Using the crystallization screening kit, design precipitant concentrations that range in 10% and 15% increments from zero to ∼20% higher than that used in the screen condition.
As an example (Fig. 17.4.2A), protein crystallization trials) using Crystal Screen 1 (Table 17.4.3) yielded a shower of nicely shaped but small crystals under condition 9, which contains 30% polyethylene glycol (PEG) 4000, 0.2 M ammonium acetate, 0.1 M sodium citrate, pH 5.6. This condition was used as a starting point for further optimization. The starting protein concentration in the crystallization drop was 10 mg/ml. Figure 17.4.2A shows the first grid screen that was prepared to optimize the precipitant concentration. (O, clear drops; C, crystals; P, precipitant)
Prepare reservoirs by adding the appropriate amounts of stock precipitant, buffer, and salt directly to the well and diluting with water to a final volume of 1 ml. Mix by pipetting up and down several times with a 1-ml micropipettor.
-
In each well, mix 1 µl concentrated macromolecule with 1 µl reservoir solution and equilibrate several days over the appropriate reservoir as described (see Basic Protocol 2, steps 5 to 9).
Trays should be incubated at the same temperature used to obtain the initial crystals.
Observe crystallization trays under a microscope with ≥40× magnification, light source, and viewing platform, and note the conditions that yield the best crystals (i.e., those with the sharpest edges, largest size, and most invariant form).
-
Continue to observe crystallization trays for at least 1 week.
In the example, after one week it was determined that crystals prepared from reservoir 3 (shaded gray) were bigger and larger than those prepared from reservoirs 4 and 5. The condition for this reservoir (15% PEG 4000, 0.2 M ammonium acetate, 0.1 M sodium citrate, pH 5.6) was used as a starting point to optimize pH and ionic strength.
-
Prepare a two-dimensional grid to optimize pH and ionic strength conditions as a function of precipitant concentration (e.g., Fig. 17.4.2B) in the following manner:
Vary the precipitant concentration more narrowly, based on the results of steps 1 to 5.
Vary the ionic strength in 20% to 25% increments.
Vary the pH in 1-unit increments both above and below the values used in the initial screen.
Set up and observe trials as in steps 2 to 5 above.
Note that different buffers will have to be used to establish different pH values. Generally, use the buffers that are employed in the sparse matrix screens.
In the example, observations made after 1 week determined that crystals from reservoirs 10 and 13 (shaded gray) looked the longest and most nicely shaped (Fig. 17.4.2B). Crystals in reservoir 18 looked striated with blunt edges. Therefore, the conditions from reservoirs 10 and 13 were used to further optimize the conditions at pH 4.6 and 0.05 M ammonium acetate.
-
To more finely optimize crystal growth conditions, use smaller changes of precipitating agent, ionic strength, and pH with 2-dimensional grids. Set up and observe trials as described in steps 2 to 5 above.
In the example, observations made after 1 week determined that crystals grown from reservoirs 9 (shaded gray) (16% PEG 4000, 50 mM ammonium acetate, 0.1 M sodium acetate, pH 4.6) and 14 (13% PEG 4000, 25 mM ammonium acetate, 0.1 M sodium acetate, pH 4.6) looked best (Fig. 17.4.2C); the crystals in reservoir 9 looked marginally better.
-
Use these conditions to prepare the next experimental grid, which is a very fine optimization of the precipitant condition. Use the results of this grid as the optimal crystallization parameters.
In the example, observation of this grid shows that crystals in reservoirs 2 and 3 (shaded gray) are the best (Fig. 17.4.2D). Therefore, the best crystallization parameters were 11% PEG 4000, 50 mM ammonium acetate, and 0.1 M sodium acetate, pH 4.6.
ALTERNATE PROTOCOL 4
OPTIMIZATION OF CRYSTAL EQUILIBRATION CONDITIONS
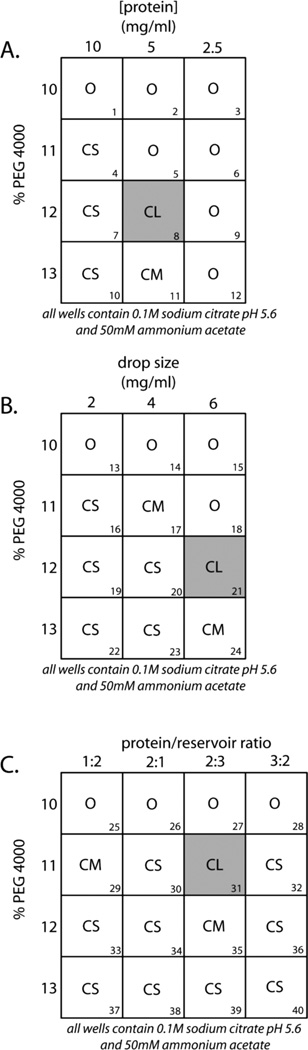
It is often the case that optimization of the chemical parameters for crystallization (see Basic Protocol 3) still yields crystals that are too small for crystallographic analysis. In such cases, it may be helpful to use a different crystallization procedure such as the sitting-drop method (Weber, 1997). Alternatively, the macromolecule concentration, the ratio of macromolecule solution to reservoir solution, and the drop size can be varied to obtain larger crystals using the vapor-diffusion technique. This latter approach is described here. Crystallization trials are performed as described previously (see Basic Protocols 2 and 3). Figure 17.4.3 illustrates a typical procedure for optimization of equilibration conditions. See Basic Protocol 3 for materials.
-
Using the optimized ionic strength, pH, and precipitant concentration (see Basic Protocol 3, step 8; 11% PEG 4000, 50mM ammonium acetate, 0.1 M sodium citrate pH 4.6), design a set of two-dimensional grids varying the parameters as follows:
Grease three 24 well crystal trays as described in Basic Protocol 2.
In each of these grids, vary the precipitant concentration (PEG 4000) finely along the 4-well axis, using four different precipitant concentrations.
Repeat this set up in each of the other trays in each of the columns in the other crystal trays.
-
Vary three different parameters in the other dimension of the grid screen (Fig. 17.4.3).
To decrease macromolecule concentration in constant drop volume (Fig 17.4.3 A): Dilute the macromolecule 2- and 4-fold relative to the concentration used in the initial screen (5 mg/ml and 2.5 mg/ml), prior to mixing with the reservoir solution (1 µl dilute macromolecule to 1 µl reservoir solution).
To increase drop volume at constant concentration (Fig 17.4.3 B): Vary the drop size by increasing the volume of both macromolecule solution and reservoir solution (i.e., 1 µl macromolecule to 1 µl reservoir, 2 to 2, and 3 to 3).
To shift the equilibration process (Fig 17.4.3 C): Vary the reservoir solution/macromolecule solution ratio (i.e., 1:2, 2:1, 2:3, 3:2) in a constant drop volume (2 µl).
-
Set up trays as described and observe them after several days.
In the example (Fig. 17.4.3A, B, and C), observations made after 1 week (O, clear drop, CS, small sized crystal; CM, medium sized crystal; CL, large sized crystal) determined that reservoirs 8, 21, and 31 (shaded gray) gave the largest crystals. Thus, the best parameters are 5 mg/ml protein mixed at a 2:3 protein/reservoir ratio in a final drop volume of 6 µl.
Figure 17.14.3.
Optimization of crystal equilibration parameters (see Alternate Protocol 4 for discussion)
ALTERNATE PROTOCOL 5
Crystal optimization using under-oil microbatch
While hanging or sitting drop vapor diffusion crystallization remains the most often used technique when setting up initial crystal trials as well as for crystal optimization, other techniques can be tried to obtain better crystals for diffraction experiments. Here we will very briefly describe one application of the under oil microbatch technique. There are several variations on this technique, which could be used to obtain more ideal crystals, however, our description will only give one example but descriptions of other protocols can be found elsewhere (Rupp, 2010). The major difference between a microbatch and a vapor-diffusion crystallization experiment is that there is little to no diffusion in a microbatch experiment. Thus, the protein to precipitant ratio does not change over time as it does in a vapor diffusion experiment.
Materials
Conentrated macromolecule or macromolecular complex
Vapor Batch 96 well plate (Hampton Research)
Eppendorf tubes
Paraffin Oil (Hampton Research or other manufacturer)
Plastic or siliconized coverslides
Pour enough Paraffin oil onto the 96 well plate to cover all of the wells. (This should mean about ¼ of an inch of oil across the entire plate, set up as outlined in Fig. 17.4.1 C)
- Using our optimized crystallization conditions from Basic Protocol 3 step 8 (0.1M sodium citrate pH 4.6, 50 mM ammonium acetate, 11% PEG 4000) as a starting point, make reservoir solutions in Eppendorf tubes with differing concentrations of PEG 4000.(With the idea that the precipitant concentration is diluted 2-fold when mixed in the vapor diffusion experiments and then allowed to increase toward equilibrium with the reservoir solution, we should prepare PEG 4000 solutions that when mixed here will yield a final concentration between 7–11% PEG 4000 (e.g 14–22% PEG 4000)
Pipette one microliter of macromolecule onto the cover slide, followed by one microliter of reservoir solution , briefly mix.
Quickly transfer protein-reservoir mix to well A1 in 96 well plate. Place tip of pipette in oil above the well. Very slowly pull pipette tip towards the surface of the oil as solution is ejected. Solution may stick to the pipette tip, but will release when tip is pulled from oil, and drop will fall into well.
Repeat with other conditions placing them in wells A2-A9 respectively.
Place tray in a temperature controlled environment where it will not be disturbed.
Monitor crystal growth using a microscope every day for a week.
Once it has been established which PEG 4000 condition is optimal, the salt and buffer condition can be optimized in a similar manner.
OPTIMIZATION BY CRYSTAL SEEDING
Many times the crystals obtained from initial crystallization trials are too small or unsuitable for X-ray diffraction studies. Along with the previously described optimization strategies, seeding techniques can yield larger, better diffracting crystals. When seeding, it is important to nucleate a crystallization drop just prior to crystal nucleation (microseeding) or crystal growth (macroseeding). More specifically, it is desirable to determine the time for introducing the seed. Since one does not typically know this ahead of time, crystal seeds are introduced into the crystallization drops at different time points (e.g., 2, 4, 8, 12, 18, and 24 hr). Ideally, one of these will result in the growth of large stable crystals.
Procedures for crystal macroseeding and microseeding are described below (see Alternate Protocols 6 and 7).
ALTERNATE PROTOCOL 6
Macroseeding
Macroseeding involves the transfer of an existing crystal through a number of stabilizing solutions into a “fresh” pre-equilibrated drop. A crystal may not grow to a larger size for a number of reasons, including macromolecule depletion and surface poisoning. During the growth phase, monomers are added to the crystal and the concentration of the macromolecule decreases. Consequently, it may be desirable to transfer the crystal to a macromolecule solution where the concentration of macromolecule is higher and the crystal can grow again. Another reason a crystal fails to continue to grow is that the surfaces have been poisoned by contaminants. As such, it is desirable to “wash” the surfaces of the contaminants by transferring the crystal through multiple stabilizing solutions.
Crystal drops can all be set up at the same time and seeded at different times after equilibration (e.g., 2, 4, 8, 12, 18 and 24 hr). The drop is created the same way as described above (see Basic Protocol 2). A typical seeding procedure is described below.
Additional Materials (also see Basic Protocol 2)
Microscope with ≥40× magnification, light source, and viewing platform (at least 3 × 4 in.), preferably with polarizer (e.g., Leica MZ16, Zeiss STEMI SV11, Nikon SMZ1000)
Tweezers
Glass fibers (see recipe) or crystal probe manipulators (Hampton Research)
Capillary-tipped syringe (optional; see recipe)
-
1.
Place four plastic or siliconized coverslips on top of the tray cover. Pipet 50 to 100 µl wash solutions (typically the same solution as the reservoir solution) onto the middle of each coverslip.
Placing the coverslips on the tray cover permits one to quickly perform the manipulations without having to make drastic adjustments to the focus of the microscope. The top of a 15-ml conical screw-cap tube can be placed over the wash solutions to prevent dehydration.
The wash solutions may also need to be optimized. If the crystals dissolve too quickly or visibly crack, then adjustments should be made to the precipitating agent. A typical wash solution contains the crystallization reservoir conditions with a 20% increase in the concentration of the precipitating agent. It may be necessary to omit any volatile alcohol solution (i.e., ethanol) in the wash solutions. Finally, some procedures suggest adding 0.5% to 2.0% (w/v) of a detergent, typically OG (octyl-β-D-glucopyranoside), to the wash solution.
-
2.
Using a microscope with ≥40× magnification, light source, and viewing platform, select a crystal without visible defects in the tray to macroseed.
It is important to select a crystal without visible defects. If the underlying crystal has defects, the crystal may grow to be an accumulation of multiple crystals.
-
3.
Being careful not to break the coverslip, gently break the vacuum grease seal using the tweezers. Carefully lift the coverslip, flip it over and place it on the tray cover next to the wash solutions.
-
4.
Add ∼20 to 50 µl wash solution to the drop to generate sufficient volume with which to work.
-
5.
Under the microscope, isolate a single crystal from other crystals or precipitate with a glass fiber or crystal probe manipulators.
For crystal clusters, carefully use the probe to gently touch the cluster to break it apart. For a crystal embedded in precipitate, starting at the edge of the crystal, sweep the precipitate away from all sides of the crystal.
A single crystal should be selected and swept to an edge of the drop free of other crystals or precipitate, so that the crystal can easily be picked up by a capillary-tipped syringe or cryoloop.
-
6a.
Using a capillary-tipped syringe: Under the microscope, gently use the capillary-tipped syringe to draw up the crystal with as little liquid as possible. Watching through the microscope, expel the crystal into the first wash solution.
-
6b.
Using a cryoloop: Perform crystal transfers with a cryoloop (Support Protocol 1).
It is often advantageous to wet the capillary before this step using wash solution. Also, before drawing up the crystal, it is useful to pull the plunger out half-way to be able to expel the crystal. The length of time of incubating the crystal in each wash step can be from 15 sec to 5 min. If cracks appear or the crystal starts to dissolve, shorter washes may be necessary.
-
7.
Using a new capillary, draw up the crystal and expel into the next wash solution as in step 4. Repeat for a total of 4 washes.
-
8.
Remove the equilibrated drop from above the reservoir, expel the crystal in the pre-equilibrated drop and replace this equilibrated drop (now containing a macroseeded crystal) over the reservoir. Make sure that vacuum grease forms an airtight seal with the coverslip.
ALTERNATE PROTOCOL 7
Microseeding: Streak Seeding
Microseeding involves providing a nucleation center for crystals to grow. The trick is to present only a few minute crystals per pre-equilibrated drop around which suitable crystals will form. The most popular form of microseeding, streak seeding, is described below. This and other seeding techniques are also described by Stura and Wilson (1990).
Materials (also see Basic Protocol 2)
Stabilization solution (e.g., reservoir solution with 20% higher precipitating reagent)
Glass fibers or crystal probe manipulators (Hampton Research)
Capillary-tipped syringe (see recipe)
Glass minipestle (see recipe)
Rabbit hair streaker (see recipe)
Prepare a pre-equilibrated drop the day before the experiment as described (see Basic Protocol 2). Lower the concentration of the precipitating agent so as to be conducive for the growth of crystals, but not nucleation.
-
Using a microscope with ≥40× magnification, light source, and viewing platform, select which crystal in the tray to microseed.
It is important to select a well-formed crystal without visible defects.
Being careful not to break the coverslip, gently break the vacuum grease seal using tweezers. Carefully lift the coverslip, flip it over, and place it on the tray cover.
Under the microscope, isolate the crystal from other crystals or precipitate with a glass fiber or crystal probe.
Under the microscope, use a capillary-tipped syringe gently to draw up the crystal with as little liquid as possible. Expel the crystal into a microcentrifuge tube containing the stabilization solution.
Using a minipestle, break the crystal apart in the stabilization solution.
Centrifuge the microcentrifuge tube containing the solution of crushed crystal 10 min at low speed (100 × g), at the temperature used to grow the crystals.
Transfer 10 µl crushed crystal solution into a new microcentrifuge tube containing 90 µl stabilization solution. Repeat to obtain a serial dilution of 10–4 to 10–8.
Gently remove the coverslip from the pre-equilibrated drop, submerge the rabbit hair streaker into various dilutions of the microcrystal solution and streak through the pre-equilibrated drop.
-
Replace the coverslip over the reservoir solution, making sure that the vacuum grease forms an airtight seal with the coverslip.
Typically a grid of dilution versus time of equilibration is used to optimize the choice of adding just a few nucleation sites at the appropriate time for nucleation.
REAGENTS AND SOLUTIONS
Use Milli-Q-purified water or equivalent for the preparation of all buffers. For common stock solutions, see APPENDIX 2E; for suppliers, see SUPPLIERS APPENDIX.
Capillary-tipped syringe
A capillary-tipped syringe is used to draw up the crystal in the liquid. To make this useful instrument, place silicone tubing over the tip of a syringe. Next obtain a glass capillary. Note that glass capillaries have two different ends: one side is sealed off, the other opens up to a larger diameter. Score the glass capillary with a diamond pencil near the tip of the sealed end and carefully break it off. Place the capillary with the large end snugly into the silicone tubing (Fig. 17.4.4 B). Small amounts of vacuum grease may be applied to the inside of the tubing to provide a sealed fit.
Glass fibers
Glass fibers are useful for manipulating crystals, breaking up crystal clusters and separating crystals from precipitate (Fig. 17.4.4 B). To prepare, hold both ends of a Pasteur pipet with each hand and place the middle part of the pipet in the flame of a Bunsen burner. Rotate the pipet in the flame until the glass becomes orange. After the pipet becomes orange, remove it from the flame, and with one quick motion pull the two ends apart. Wait for Pasteur pipet to cool and separate into 10- to 20-cm sizes. It is important to not allow holes in the glass fiber or it will draw the liquid up through capillary action.
Minipestle
The minipestle is made from a Pasteur pipet using a Bunsen burner. Using a diamond pen (Hampton Research), score the circumference of the pipet on the small diameter side, ∼1 in. away from the neck, and break it off. Put the smaller side gradually in the flame and rotate the pipet until a ball is formed. Then let the minipestle cool. Make sure the minipestle will fit in the size microcentrifuge tube containing the stabilization solution.
Rabbit-hair streaker
Many crystallographers avow that only rabbit hair works to seed crystals. However, human hair or mouse whiskers have also proven successful. For easier manipulation, make the streaker by gluing the hair onto the end of an unused pen.
Many companies also sell crystal manipulation kits that come with a variety of tools for measuring, dissecting, and moving crystals.
COMMENTARY
Background Information
In the past ten years the technology that supports x-ray data collection and structure determination has grown significantly, and in order to keep pace with this end of the crystallographic process, crystallographers have developed machinery that has greatly expedited the process of obtaining and optimizing crystals. These new high-throughput robotics have allowed researchers to screen very large sets of crystallization conditions while using small amounts of macromolecular sample. The introduction of these machines into almost every crystallography lab has greatly diminished the time and resources necessary to initially screen proteins for the ability to crystallize, and has afforded other scientists the opportunity to collaborate with crystallographers and pursue new and exciting crystal structures. However, regardless of these great technological advances almost all crystals need to be optimized by hand in order to be useful for diffraction experiments. The basic protocols explained above provide the essential methodologies that can be used by any researcher to obtain and optimize quality crystals to be used for the determination of macromolecular structures.
Macromolecular crystallization is the process by which macromolecules are coaxed out of solution in the form of crystals. The induction of crystal formation is facilitated by a precipitating agent that functions to drive equilibration between the small macromolecule-containing solution and large reservoir solution. A volatile precipitating agent, such as 2-methyl-2,4-pentanediol (MPD) or isopropanol, transfers by vapor-diffusion from the reservoir to the drop, thus increasing the precipitant concentration in the drop and driving the macromolecule out of solution (in the form of precipitate or crystals). Alternatively, a nonvolatile precipitating agent, such as salt or large polyethylene glycol (PEG), functions to promote the diffusion of water from the drop to the reservoir, causing the precipitant concentration to increase in the drop, thus driving the macromolecule out of solution. A more detailed discussion underlying the theory of crystal formation from a supersaturated macromolecular solution can be found in McPherson (1982). Here the authors present a method for macromolecular crystallization by the vapor-diffusion technique using hanging drops. It is the most convenient and commonly used technique, and often yields crystals from suitable macromolecular samples
Critical Parameters and Troubleshooting
The most critical parameter for obtaining macromolecular crystals that are suitable for structure determination using X-ray crystallography is the macromolecular sample itself. In general, homogeneous, compact, monodisperse macromolecular samples are straightforward to crystallize, while nonhomogeneous, polydisperse samples with flexible domains, or samples that tend to aggregate nonspecifically, are very difficult to crystallize (D’Arcy, 1994; Ferre-D’Amare and Burley, 1994). Generally, if crystals do not form with the appropriate sparse matrix crystallization screens at either room temperature or 4°C, it is likely that the macromolecule needs to be modified in some way to obtain well-ordered crystals. One modification that can be performed is to remove flexible domains of the protein. In general, flexible domains are most easily identified by limited proteolysis, followed by protein sequencing of the remaining globular product. Recombinant DNA technology can then be used to genetically engineer the desired protein fragment. Another modification is to prepare a macromolecular complex rather then a nascent protein. Formation of a complex sometimes serves to bury otherwise hydrophobic regions that can nucleate protein aggregation and thus hamper crystallization. In this regard, protein-antibody complexes sometimes crystallize more readily than free proteins.
Aside from the macromolecular sample, physiologically relevant cofactors often play major roles in crystallization. For example, enzymes may show entirely different crystallization behavior when they are bound to various nonhydrolyzable small-molecule or peptide substrates.
The most common problem associated with macromolecular crystallization is that the protein precipitates either upon concentration or in a large majority of crystallization conditions. This often indicates that the protein has a strong tendency to aggregate at high concentrations. If this is the case, one of the two modification strategies described above should be considered. Diluting the macromolecule 2- to 4-fold may also help. Alternatively, crystallization trays may show predominantly clear drops. This indicates that the protein concentration is too low and needs to be raised (between 2- and 4-fold), or that the protein is overly soluble. In the latter case, one might try crystallization screens at or very near the pI of the macromolecule in order to reduce its charge.
Another common problem in crystallizing macromolecules is that crystals of salt rather than of macromolecule are obtained. To determine if this is the case, set up a control drop containing everything except macromolecule. Salt crystals are commonly obtained with divalent ions (particularly with phosphate or sulfate counterions).
Anticipated Results
Crystallization trials generally show one of three types of results: clear drops, crystals, or precipitate or phase separation. Most often, one obtains a mixture of all of these results depending on the well conditions. If crystals are obtained under several different screen conditions, it is usually straightforward to proceed from small crystals to well-ordered, large crystals suitable for structure analysis by X-ray crystallography. Conditions that result in precipitate or phase separation generally are incompatible with crystal formation. If all, or nearly all, of the drops show precipitate, this is a bad sign and suggests that a modification of the macromolecule is required (see Critical Parameters and Troubleshooting).
Time Considerations
Once a purified macromolecule is on hand, it can usually be concentrated and set up in crystallization trials in a matter of a few days. Assessment and refinement of crystallization parameters can take between a week to several months, depending on the behavior of the macromolecular sample.
Contributor Information
David Friedmann, Wistar Institute.
Troy Messick, Vironika, LLC.
Ronen Marmorstein, Wistar Institute, 3601 Spruce St., Philadelphia, PA. 19104, Tel: 215-898-5006, FAX: 215-898- 0381, marmor@wistar.org.
Literature Cited
- Aggarwal AK. Crystallization of DNA binding proteins with oligonucleotides. Methods Companion. 1990;1:83–90. doi: 10.1016/S1046-2023(05)80150-1. [DOI] [PubMed] [Google Scholar]
- D’Arcy A. Crystallizing proteins: A rational approach? Acta Cryst. 1994;D50:469–471. doi: 10.1107/S0907444993014362. [DOI] [PubMed] [Google Scholar]
- Ferre-D’Amare A, Burley S. Use of dynamic light scattering to assess crystallizability of macromolecules and macromolecular assemblies. Structure. 1994;2:357–359. doi: 10.1016/s0969-2126(00)00037-x. [DOI] [PubMed] [Google Scholar]
- Jancarik J, Kim S-H. Sparse matrix sampling: A screening method for crystallization of proteins. J. Appl. Cryst. 1991;24:409–411. [Google Scholar]
- McPherson A. The Preparation and Analysis of Protein Crystals. New York: John Wiley & Sons; 1982. [Google Scholar]
- McPherson A. Crystalization of Biological Macromolecules. Cold Spring Harbor, New York: Cold Spring Harbor Press; 1999. [Google Scholar]
- Muecke M, Samuels M, Davey M, Jeruzalmi D. Preparation of Multimilligram Quantities of Large, Linear DNA Molecules for Structural Studies. Structure. 2008;16:837–841. doi: 10.1016/j.str.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Rupp B. Biomolecular Crystallography: Principles, Practice, and Application to Structural Biology. New York, New York: Garland Science; 2010. [Google Scholar]
- Scott WG, Finch JT, Grenfell R, Fogg J, Smith T, Gait MJ, Klug A. Rapid crystallization of chemically synthesized hammerhead RNAs using a double screening procedure. J. Mol. Biol. 1995;250:327–332. doi: 10.1006/jmbi.1995.0380. [DOI] [PubMed] [Google Scholar]
- Stura EA, Satterthwait AC, Calvo JC, Kaslow DC, Wilson IA. Reverse screening. Acta Cryst. 1994;50:448–455. doi: 10.1107/S0907444994001794. [DOI] [PubMed] [Google Scholar]
- Stura EA, Wilson IA. Analytical and production seeding techniques. Methods: A Companion to Methods in Enzymology. 1990;1:38–49. [Google Scholar]
- Stura EA, Wilson IA. Seeding Techniques. In: Ducruix A, Giegé G, editors. Crystallation of Nucleic Acids and Proteins: A Practical Approach. Oxford: IRL Press; 1991. pp. 241–254. [Google Scholar]
- Weber P. Overview of protein crystallization methods. Methods Enzymol. 1997;276:13–22. doi: 10.1016/S0076-6879(97)76048-8. [DOI] [PubMed] [Google Scholar]