Post-translational modification of many cellular proteins by covalent attachment of a 76 amino acid protein ubiquitin (Ub) or other Ub-like (UBL) modifiers controls a wide range of vital functions in eukaryotes, including cellular trafficking, DNA repair, ribosomal biogenesis, cell cycle progression, and antigen processing.[1] This modification involves a cascade of enzymatic reactions catalyzed by E1, E2, and E3 enzymes, and results in the formation of an isopeptide bond between the carboxylate group of the C-terminal glycine (G76 in Ub and Rub1) and the ε-amine of a lysine of the target protein.[2] Ubiquitination can result in the attachment of a single Ub or a polymeric chain of Ubs (polyUb) in which the same chemistry is involved in the formation of Ub-Ub linkages through any of the seven Ub lysine residues or via the α-amine of the N-terminal M1.[3] Rub1 (Nedd8 in mammalian cells) is the closest kin of Ub in the family of UBLs, with ~53% sequence identity and ~76% sequence similarity to Ub.[4] The key surface residues and the 3D structures of the two proteins are incredibly similar as well.[4] Despite these striking similarities, Ub, but not Rub1, is well established as a chain-forming protein.[5] The modification (rubylation) of cullin proteins with Rub1 and the subsequent activation of a multi-subunit SCF ligase is the most prevalent and clearly the best studied biological function of Rub1 reported so far.[6] Understanding of the outcomes of rubylation and the signaling properties of Rub1 requires the ability to attach it to other proteins at will.
We and others have recently shown that Rub1 and Ub can form heterologous (Rub1-Ub and Ub-Rub1) conjugates in vivo.[4, 7] Interestingly, mass spectrometry analyses of cellular proteins revealed that, similar to Ub, all lysines of Rub1 can be used to form isopeptide bonds (either with another Rub1 or with Ub).[8] Specifically, three types of heterologous conjugates are biologically evident under diverse stress conditions[4, 7]: Ub–48Rub1 (where G76 of Ub is linked to K48 of Rub1), Rub1–48Ub, and Rub1–29Ub, where G76 of Rub1 is linked to K48 or K29, respectively, of Ub (see ref [9] for chain/linkage nomenclature). These findings indicate that both rubylated Ub and ubiquitinated Rub1 (and possibly rubylated Rub1) are biologically relevant building blocks of heterologous polymeric chains. Despite their presence within cells, the physiological importance of these conjugates remains poorly understood. Recently, we showed that an enzymatically assembled Rub1–48Ub heterodimer can signal proteasomal targeting (i.e. is recognized by the proteasomal shuttles and Ub-receptors) and is disassembled by the proteasome essentially like the “canonical” Ub–48Ub.[4] All these results point to a possible role for Rub1 as part of the polyUb signaling system. However, investigation of the structural and functional properties of the conjugates of Rub1 and Ub of various linkages has been hindered by the unavailability of the corresponding linkage-specific conjugating E2 enzymes. Remarkably, the Ub-conjugation machinery allowed us to rubylate Ub but not to ubiquitinate Rub1.[4] Moreover, neither Rub1 E2 enzymes forming chains nor K29-selective Ub E2 enzymes are currently known. All this necessitates development of chemical methods for rubylation and ubiquitination that circumvent the need for E2 and E3 enzymes.
Toward this goal, we and others have recently developed various chemical methods for making Ub-Ub conjugates[10-14] (reviewed in [15]). Some of the methods use total chemical synthesis combined with native/isopeptide chemical ligation to generate dimers[10] and longer chains of Ub.[16] Although a remarkable achievement, the need to use total chemical synthesis has limited their utility in a biochemical laboratory setting. To circumvent this problem, we recently developed a nonenzymatic method for controlled assembly of unbranched or branched Ub chains from bacterially-expressed Ub monomers[12, 13] that (i) yields entirely natural polyUb chains of any desired length and linkage composition, (ii) can be used readily in any biochemical laboratory, and (iii) allows independent isotopic labeling of any monomer in the chain, making it amenable to atomic-resolution studies by NMR. While inspired by the GOPAL approach,[11] our method employs mutually orthogonal protecting groups, which provide the advantage of allowing assembly of chains longer than dimers with full control of chain length and linkage. However, the methodology has yet to be developed for conjugation of UBLs as well as for ubiquitination of proteins other than Ub (i.e. to bypass the E3 enzymes).
Here we describe a nonenzymatic method that allowed us to achieve, for the first time, rubylation of both Ub and Rub1 and ubiquitination of Rub1 at desired lysine positions. We made all the above-mentioned naturally occurring heterodimers of Rub1 and Ub. This allowed us to obtain previously unavailable structural insights into interdomain interactions and receptor recognition of these chains. Moreover, since we recently attributed a derubylase function to the 26S proteasome,[4] here we use the heterologous conjugates of Rub1 and Ub to examine the derubylase/deubiquitinase activity of several deubiquitinating enzymes (DUBs). Surprisingly, we found that some of the DUBs (USP5, OTUB1) can act as derubylases.
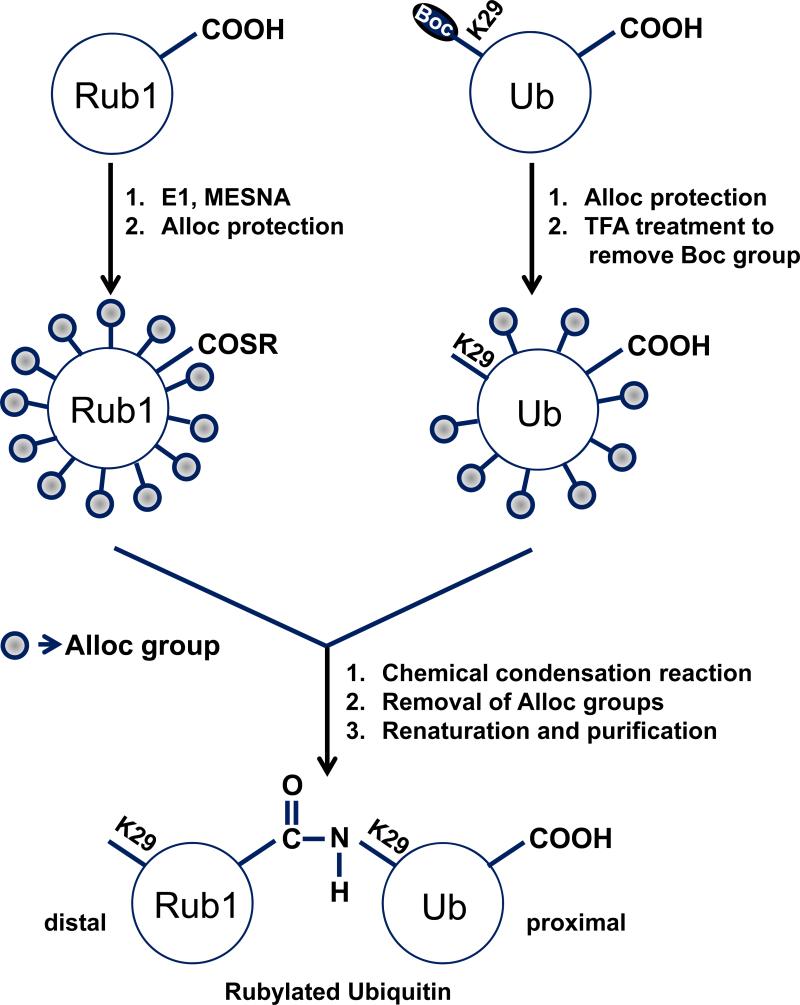
Our method for isopeptide bond formation uses chemical condensation reaction between the thioesterified C-terminus of one monomer (distal-to-be unit, e.g., Rub1 in Rub1–29Ub) and the ε-amine of a specific lysine residue of the other monomer (proximal-to-be unit, e.g., Ub in Rub1–29Ub) (see Scheme 1 for details). Critically, in order to direct the chemical reaction to a specific lysine on the proximal unit, we use a combination of two mutually orthogonal removable protecting groups, Boc and Alloc. The former is introduced as a Lys(Boc) at the desired conjugation site in the proximal-to-be unit using genetic incorporation of unnatural amino acids and is removed prior to the conjugation reaction, while the latter group is introduced as a chemical modification of each unit (reactant) post-purification, to protect all remaining amines. The mutual orthogonality of the two protecting groups and of the thioester allows their attachment and removal at desired steps without affecting the other groups. [12, 13]
Scheme 1.
Nonenzymatic rubylation of Ub. Similar schemes are used for ubiquitination or rubylation of Rub1. The basic steps involved in this method are as follows: (1) the C-terminus of either Rub1 or Ub (distal-to-be unit) is thioesterified (SR) by incubating the protein with its respective cognate E1 activating enzyme; (2) to direct the reaction to a specific lysine (e.g., K29) on the proximal-to-be unit (Ub or Rub1), the ε-amine of that lysine is protected with a Boc group (incorporated in the form of Lys(Boc) into the proximal-to-be unit as a genetically encoded unnatural amino acid (UAA); (3) all remaining free amines of both distal-to-be and proximal-to-be monomers are blocked with Alloc. Note that there are 12 such groups in distal-to-be Rub1 (8 lysines, 3 histidines, and the N-terminus) and 8 groups in proximal-to-be Ub (6 lysines, 1 histidine, and the N-terminus) which need to be protected with Alloc; (4) the Boc-group is removed to make the ε-amine of the lysine of interest the sole site for the ligation reaction; (5) ligation of the two monomers by Ag-mediated condensation reaction; (6) removal of all the Alloc groups from the product; followed by (7) renaturaton and separation of the dimer from the unreacted monomers, to yield a fully natural product, in this case Rub1–29Ub. Boc=tert-butoxycarbonyl, Alloc=allyloxycarbonyl, MESNA=sodium 2-mercaptoethanesulfonate.
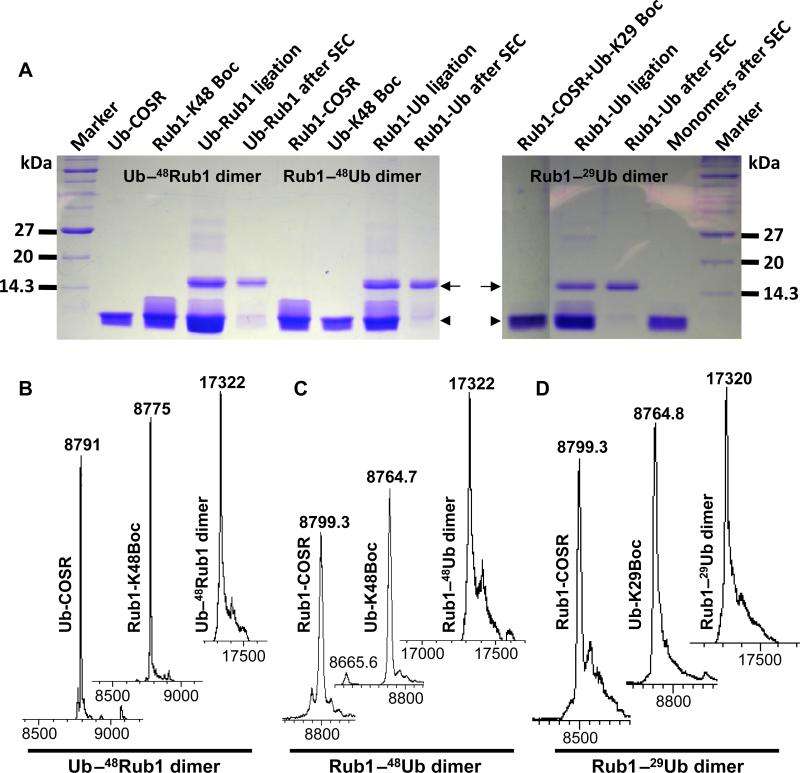
Using this method, we successfully assembled uniformly 15N-enriched Rub1–29Ub, Rub1–48Ub, and Ub–48Rub1 heterodimers (Figure 1A). Note that as each monomer is bacterially expressed separately, our method allows unit-specific isotopic labeling, which is critical for NMR studies of polyUb chains.[12, 17] However, because most of the NMR signals of Ub and Rub1 do not overlap, it was possible to perform NMR characterization of the heterodimers containing both units isotopically enriched at the same time. All the steps in the assembly of the heterodimers were closely monitored by ESI-MS. Consistent with the expected mass, the ESI-MS results confirmed the complete attachment of SR and Boc groups to their respective monomers (see Figure 1B-D) prior to the condensation reaction. The observed mass (≈17,322 Da) of the assembled Ub–48Rub1, Rub1–48Ub, and Rub1–29Ub heterodimers clearly indicates that the resulting products are fully natural (Figure 1B-D). It should be emphasized that by successfully rubylating Ub and Rub1 (see below) we demonstrated here, for the first time, that a protein other than Ub can be conjugated to other proteins without E2/E3 enzymes. Also, by achieving the successful ubiquitination of Rub1, we showed that Ub can be attached to a protein other than Ub without E2/E3 enzymes using our method.
Figure 1.
Chemical assembly of Ub–48Rub1, Rub1–48Ub, and Rub1– 29Ub dimers. (A) 2 mg of each reacting monomers, as indicated, were incubated in a condensation reaction for 36 hr. A 15% SDS-PAGE was performed with 2 μL of each reacting monomers, ligated product mix (non-purified), and the SEC-purified dimers/monomers. The running positions of monomers and dimers are indicated by the arrowhead and the arrow, respectively. (B-D) ESI-MS spectra of various intermediate and final steps during the assembly of Ub–48Rub1 (B), Rub1–48Ub (C), and Rub1–29Ub (D) heterodimers. The molecular masses of the 15N-enriched Ub and Rub1 are 8,665 Da and 8,675 Da, respectively. The protection with the Boc group increases the mass by 100 Da, while the attachment of the SR group increases the mass by 125 Da.
Our success with the assembly of the abovementioned heterodimers (and previously with various Ub–Ub homodimers and longer chains[12]) has motivated us to extend our method to the assembly of a fully natural Rub1–48Rub1 homodimer. A polymeric chain of Rub1 has been reported to modify some of the cullin proteins,[18] thus suggesting that a Rub1–48Rub1 conjugate may be a biologically relevant signal. However, such a conjugate has not been made so far. Importantly, we succeeded in the assembly of Rub–48Rub1 homodimer (Figure S1) by following the above-described procedure (Scheme 1) up to step 6, and confirmed the product by SDS-PAGE. However, when we attempted to renature the product (step 7), the dimer precipitated. Currently we are testing various renaturing approaches to solubilize and study Rub1–48Rub1.
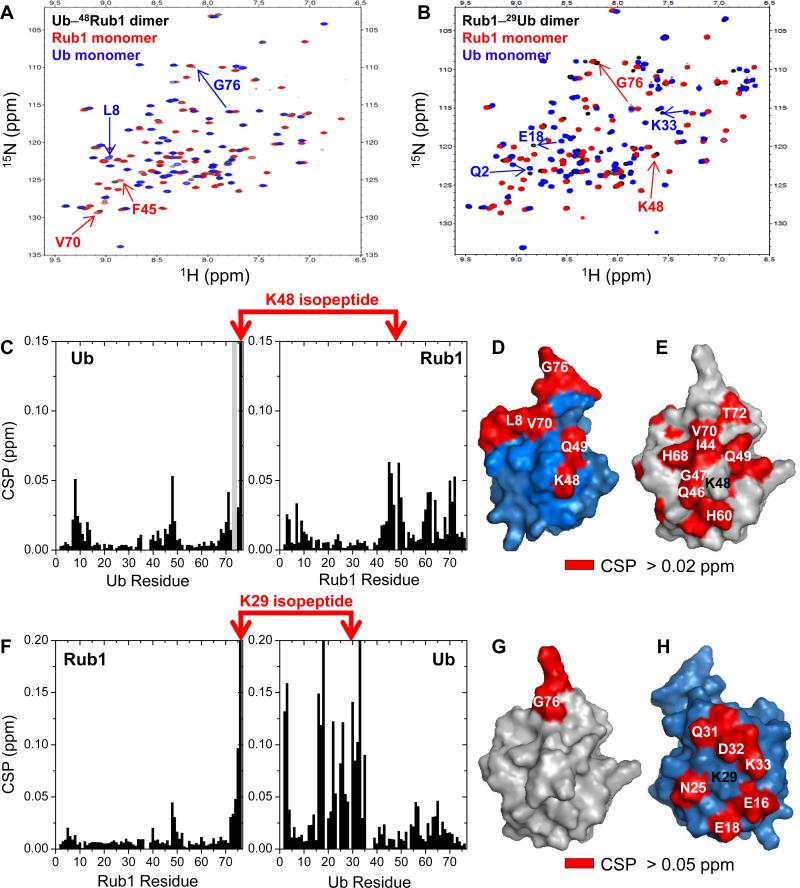
We recently showed that the enzymatically assembled Rub1–48Ub adopts a conformation where Rub1 and Ub form an interface mediated by the hydrophobic surface patches (comprising L8, I44, and V70) of both monomers.[4] To determine whether the chemically assembled Rub1–48Ub adopts a similar conformation, we recorded its 1H-15N NMR spectrum (at pH 6.8) and quantified the chemical shift perturbations (CSP) for each residue in Rub1–48Ub versus the corresponding monomer (Figure S2A-B). The strong similarity between the CSPs observed in the chemically (Figure S2B) and in the enzymatically[4] assembled Rub1–48Ub indicates that the heterodimers made by the two different methods are structurally identical. Moreover, mapping the significant CSPs and signal attenuations on the 3D structures of Rub1 and Ub (Figure S2C-D) revealed that the surfaces involved in the interdomain interface are essentially same as in the enzymatically assembled heterodimer.[4]
Next, to examine whether Rub1 and Ub in the other two heterodimers, Ub–48Rub1 and Rub1–29Ub, form an interface, we recorded 1H-15N NMR spectra of 15N-enriched dimers (Figure 2A-B) and quantified the CSPs (versus the corresponding monomers) for each residue (Figure 2C,F). The NMR signals are well dispersed, and the close similarity with the spectra of monomeric Rub1 and Ub indicates fully folded heterodimers (Figure 2A-B). Significant CSPs clustered around residues L8, I44, and V70 of Ub clearly indicate the formation of an interdomain interface in Ub–48Rub1 (Figure 2C). Mapping the CSPs and signal attenuations on the surface of both Ub and Rub1 revealed that the largest perturbations are located on one side of each unit (Figure 2D-E), thus suggesting a specific interdomain contact in Ub–48Rub1, mediated by the hydrophobic surface patches of both Ub and Rub1. By contrast, no significant CSPs (except for the C-terminus, conjugated to Ub) were detected for Rub1 in Rub1–29Ub (Figure 2F-G), indicating absence of a specific noncovalent contact between Rub1 and Ub in this heterodimer. Consistent with this observation, residues perturbed in Ub are located primarily on the opposite side of its surface from the hydrophobic patch and clustered around residue K29 (Figure 2H), suggesting that the observed CSPs are mainly due to the isopeptide bond formation. These results indicate that there is no defined interdomain interface in Rub1–29Ub.
Figure 2.
Analysis of the interdomain interface in the heterodimers of Ub and Rub1. (A-B), 1H-15N SOFAST-HMQC spectra (black) of Ub–48Rub1 (A) and Rub1–29Ub (B) overlaid with the spectra of Rub1 (red) and Ub (blue) monomers. Selected residues showing significant signal shifts are marked with numbers and indicated by arrows. Note that residue K48 in the proximal units was not 15N-labeled. (C, F), Chemical shift perturbations (CSP, black bars) and significant signal attenuations (>75%; grey bars) of backbone amides in Ub–48Rub1 (C) and Rub1–29Ub (F) plotted as a function of the residue number. Isopeptide- forming residues are connected by red arrows. (D-E, G-H), The perturbed residues (painted red) in Ub–48Rub1 (D-E) and Rub1–29Ub (G-H) are mapped on the 3D structures of Ub (blue) and Rub1 (grey), oriented such that the hydrophobic-patch surfaces face the reader except for (H) where the opposite side of Ub surface is shown.
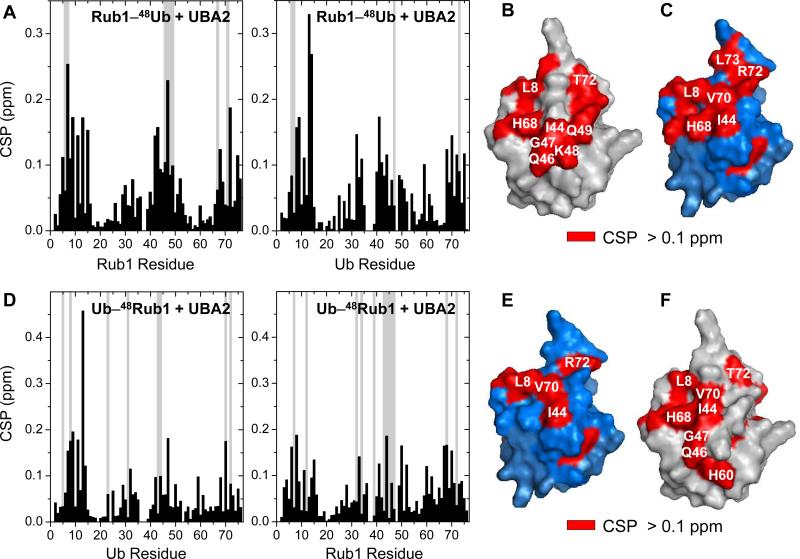
To verify that chemically assembled Rub1–48Ub is not only structurally but also functionally similar to the enzymatically conjugated heterodimer, we examined its interactions with the UBA2 domain of the proteasomal shuttle protein hHR23a using NMR titration assays. Interestingly, the CSP patterns (Figure 3A) and the mapped UBA2-interacting surfaces (Figure 3B-C) are essentially same as observed for the enzymatically conjugated Rub1–48Ub,[4] thus confirming that our chemically assembled rubylated Ub is both structurally and functionally indistinguishable from the heterodimer synthesized by the enzymatic machinery. Next, to examine whether Ub–48Rub1 heterodimer is recognized by the UBA2 domain of hHR23a, unlabeled UBA2 was titrated into 15N-labeled Ub–48Rub1. The observed strong CSPs and signal attenuations (Figure 3D) clearly indicate that ubiquitinated Rub1 binds to UBA2. As in Rub1–48Ub (and Ub–48Ub[19]), this interaction is residue specific and mediated by the hydrophobic-patch surfaces of both Ub and Rub1 (Figure 3E-F), suggesting that Ub–48Rub1 can be a signal for proteasomal targeting.
Figure 3.
K48-linked heterodimers of Rub1 and Ub form residue-specific interactions with the UBA2 domain of hHR23a. CSPs (black bars) and significant signal attenuations (>75%; grey bars) of backbone amides in Rub1–48Ub (A) and Ub–48Rub1 (D) at saturation with UBA2 are plotted as a function of the residue number. Residues with significant CSPs (>0.1 ppm) and signal attenuations are mapped (red) on the surface of the distal and proximal units of Rub1–48Ub (B-C) and Ub–48Rub1 (E-F). Ub and Rub1 are shown in blue and grey, respectively.
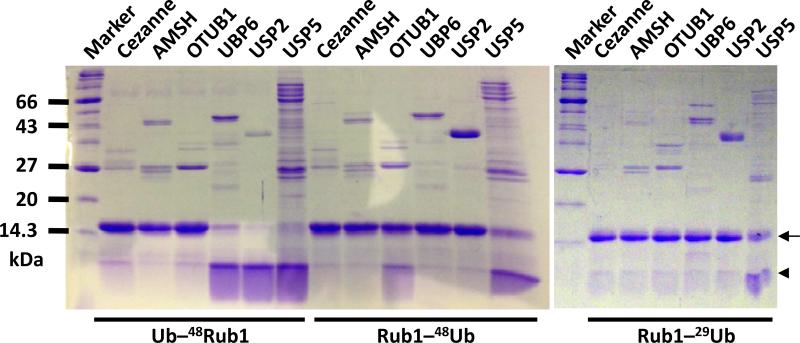
Disassembly of Ub conjugates and subsequent recycling of Ub is critical for maintaining the Ub homeostasis in the cell.[20] A variety of DUBs play a central role in disassembly of different types of Ub conjugates. Among them, OTUB1,[21] UBP6[22] (USP14 in humans), and USP2[23] are mainly K48-linkage specific DUBs found in the cells, while Cezanne and AMSH have the specificity for K11 and K63 linkages, respectively. USP5 (Isopeptidase T) has been shown to recycle Ub by hydrolyzing the isopeptide bond in a variety of unanchored polyUb linkages.[24] The remarkable similarity between Rub1 and Ub[4] raises the question whether DUBs can recognize and treat Rub1 as Ub. To study the recognition pattern of these DUBs as well as their enzymatic activity towards the heterodimers of Ub and Rub1, we performed an in-vitro deubiquitination assay (Figure 4; see controls in Figure S3). Interestingly, UBP6 and USP2 completely disassembled ubiquitinated Rub1 but did not cleave rubylated Ub (K48- or K29-linked). This indicates that Ub at the position distal to the isopeptide bond is required for the deubiquitinase function of UBP6 and USP2, thus a distal Rub1 blocks this activity. By contrast, OTUB1 partially cleaved Rub1–48Ub, but not Ub–48Rub1 or Rub1–29Ub, indicating that the enzyme is particularly selective for the proximal Ub and for K48-linkage. The results also suggest that Ubs on both sides of the K48-isopeptide bond are needed for optimal activity of OTUB1-mediated cleavage. This agrees with the role of UBP6 and USP2 in removal of a (poly)Ub tag from a substrate destined for degradation[22] and with OTUB1 recognizing both the distal and proximal Ubs simultaneously.[21] Interestingly, USP5 cleaved all three heterodimers tested here, confirming its ability to cleave different types of linkages. These results clearly show that USP5 and OTUB1 have both deubiquitinase and derubylase activity. Neither Cezanne nor AMSH disassembled any of the dimers, which is consistent with their specificity towards K11 and K63 linkages, respectively.
Figure 4.
Cleavage of the synthesized heterodimers of Rub1 and Ub by deubiquitinases. The heterodimers were incubated for 16 hr in a cleavage reaction with indicated deubiquitinases in a 10:1 molar ratio. The reactions were stopped by adding SDS loading buffer and loaded onto a 15% SDS PAGE. The running positions of monomers and dimers are shown by the arrowhead and the arrow, respectively.
In vitro synthesis and subsequent functional studies of various types of chains composed of Ub and/or UBL proteins remained challenging for many years mainly due to the lack of proper enzymes to form such conjugates. Here we presented a chemical ligation method by which heterologous (Ub–Rub1 and Rub1–Ub) and homologous (Rub1–Rub1) conjugates can be made of recombinant monomers in a controlled manner, with fully natural connectivity, and in sufficient amounts for structural and biophysical studies. Moreover, our method can be readily used in any biochemical laboratory and yields fully natural products devoid of any chain-terminating mutations. By taking advantage of bacterial expression of recombinant monomers, our method allows cost-effective unit-specific isotopic labeling of the chains, which is required for high-resolution NMR studies. The NMR characterization of the two novel heterodimers assembled with our method revealed that in Ub–48Rub1, but not in Rub1–29Ub, Ub and Rub1 form an interface which is very similar to the interface in Rub1–48Ub and the corresponding Ub–48Ub homodimer,[17] and provided site-specific information on the binding interactions with the UBA2 domain of a proteasomal shuttle protein hHR23a. Furthermore, the availability of heterodimers allowed us to examine, for the first time, requirements for Ub-units recognition and cleavage by several DUBs. Moreover, we discovered an unexpected derubylase activity of some of the deubiquitinases. All these results demonstrate that the method presented here expands the repertoire of chemical-biology tools by opening previously unavailable opportunities to make polymeric chains containing various UBL modifiers and to study their structural and functional properties. We foresee a straightforward extension of our methodology to other UBLs, for example, SUMO, which forms both homologous (SUMO 2/3)[25] chains and heterologous (with Ub) chains.[26] It would be interesting to examine whether the structural properties of such chains correlate with their specific functional roles. Our ultimate goal is to be able to attach Ub or UBL, as a monomer or as a chain of any desired length and linkage composition, to a substrate protein at will, with no need for E2 or E3 enzymes. This would open endless possibilities to study the structural and functional roles of the attachment of Ub/UBL monomers or chains to their physiological substrate proteins. By developing our method for rubylation of Ub and Rub1 and ubiquitination of Rub1, we definitely moved a few steps, if not many, closer to achieving this goal.
Supplementary Material
Footnotes
This work was supported by the NIH grants GM065334 and GM095755 to D.F. We thank M.H.Glickman for plasmids of USP2 and UBP6, R.E.Cohen for AMSH plasmid, B. Nicholson for the Cezanne plasmid, C.A. Castañeda for Ub(K29Boc), and M.A.Nakasone and A.Chaturvedi for some of the deubiquitinases used here.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- 1.a Marmor MD, Yarden Y. Oncogene. 2004;23:2057–2070. doi: 10.1038/sj.onc.1207390. [DOI] [PubMed] [Google Scholar]; b Jackson SP, Durocher D. Mol Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]; c Ulrich HD, Walden H. Nat Rev Mol Cell Biol. 2010;11:479–489. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]; d Finley D, Bartel B, Varshavsky A. Nature. 1989;338:394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]; e Teixeira LK, Reed SI. Annu Rev Biochem. 2013 doi: 10.1146/annurev-biochem-060410-105307. [DOI] [PubMed] [Google Scholar]; f Loureiro J, Ploegh HL. Adv Immunol. 2006;92:225–305. doi: 10.1016/S0065-2776(06)92006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a Hochstrasser M. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]; b Pickart CM. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 3.Fushman D, Wilkinson KD. F1000 Biol Rep. 2011;3:26. doi: 10.3410/B3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh RK, Zerath S, Kleifeld O, Scheffner M, Glickman MH, Fushman D. Mol Cell Proteomics. 2012;11:1595–1611. doi: 10.1074/mcp.M112.022467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hori T, Osaka F, Chiba T, Miyamoto C, Okabayashi K, Shimbara N, Kato S, Tanaka K. Oncogene. 1999;18:6829–6834. doi: 10.1038/sj.onc.1203093. [DOI] [PubMed] [Google Scholar]
- 6.a Rabut G, Peter M. EMBO Rep. 2008;9:969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Merlet J, Burger J, Gomes JE, Pintard L. Cell Mol Life Sci. 2009;66:1924–1938. doi: 10.1007/s00018-009-8712-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leidecker O, Matic I, Mahata B, Pion E, Xirodimas DP. Cell Cycle. 2012;11:1142–1150. doi: 10.4161/cc.11.6.19559. [DOI] [PubMed] [Google Scholar]
- 8.a Jeram SM, Srikumar T, Zhang XD, Eisenhauer H. Anne, Rogers R, Pedrioli PG, Matunis M, Raught B. Proteomics. 2010;10:254–265. doi: 10.1002/pmic.200900648. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Jones J, Wu K, Yang Y, Guerrero C, Nillegoda N, Pan ZQ, Huang L. J Proteome Res. 2008;7:1274–1287. doi: 10.1021/pr700749v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakasone MA, Livnat-Levanon N, Glickman MH, Cohen RE, Fushman D. Structure. 2013;21:727–740. doi: 10.1016/j.str.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a Kumar KS, Spasser L, Erlich LA, Bavikar SN, Brik A. Angew Chem Int Ed Engl. 2010;49:9126–9131. doi: 10.1002/anie.201003763. [DOI] [PubMed] [Google Scholar]; b El Oualid F, Merkx R, Ekkebus R, Hameed DS, Smit JJ, de Jong A, Hilkmann H, Sixma TK, Ovaa H. Angew Chem Int Ed Engl. 2010;49:10149–10153. doi: 10.1002/anie.201005995. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Yang R, Pasunooti KK, Li F, Liu XW, Liu CF. Chem Commun (Camb) 2010;46:7199–7201. doi: 10.1039/c0cc01382j. [DOI] [PubMed] [Google Scholar]
- 11.a Virdee S, Ye Y, Nguyen DP, Komander D, Chin JW. Nat Chem Biol. 2010;6:750–757. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]; b Virdee S, Kapadnis PB, Elliott T, Lang K, Madrzak J, Nguyen DP, Riechmann L, Chin JW. J Am Chem Soc. 2011;133:10708–10711. doi: 10.1021/ja202799r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castañeda C, Liu J, Chaturvedi A, Nowicka U, Cropp TA, Fushman D. J Am Chem Soc. 2011;133:17855–17868. doi: 10.1021/ja207220g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon EK, Castaneda CA, Kashyap TR, Wang Y, Fushman D. Bioorg Med Chem. 2013;21:3421–3429. doi: 10.1016/j.bmc.2013.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a Eger S, Scheffner M, Marx A, Rubini M. J Am Chem Soc. 2010;132:16337–16339. doi: 10.1021/ja1072838. [DOI] [PubMed] [Google Scholar]; b Valkevich EM, Guenette RG, Sanchez NA, Chen YC, Ge Y, Strieter ER. J Am Chem Soc. 2012;134:6916–6919. doi: 10.1021/ja300500a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a Spasser L, Brik A. Angew Chem Int Ed Engl. 2012;51:6840–6862. doi: 10.1002/anie.201200020. [DOI] [PubMed] [Google Scholar]; b Brik A. Bioorg Med Chem. 2013;21:3398–3399. doi: 10.1016/j.bmc.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 16.a Kumar KS, Bavikar SN, Spasser L, Moyal T, Ohayon S, Brik A. Angew Chem Int Ed Engl. 2011;50:6137–6141. doi: 10.1002/anie.201101920. [DOI] [PubMed] [Google Scholar]; b Moyal T, Bavikar SN, Karthikeyan SV, Hemantha HP, Brik A. J Am Chem Soc. 2012;134:16085–16092. doi: 10.1021/ja3078736. [DOI] [PubMed] [Google Scholar]
- 17.Varadan R, Walker O, Pickart C, Fushman D. J Mol Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 18.Ohki Y, Funatsu N, Konishi N, Chiba T. Biochem Biophys Res Commun. 2009;381:443–447. doi: 10.1016/j.bbrc.2009.02.090. [DOI] [PubMed] [Google Scholar]
- 19.Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 20.a Clague MJ, Coulson JM, Urbé S. J Cell Sci. 2012;125:277–286. doi: 10.1242/jcs.090985. [DOI] [PubMed] [Google Scholar]; b Komander D, Clague MJ, Urbé S. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]; c Komander D. Subcell Biochem. 2010;54:69–87. doi: 10.1007/978-1-4419-6676-6_6. [DOI] [PubMed] [Google Scholar]
- 21.Wang T, Yin L, Cooper EM, Lai MY, Dickey S, Pickart CM, Fushman D, Wilkinson KD, Cohen RE, Wolberger C. J Mol Biol. 2009;386:1011–1023. doi: 10.1016/j.jmb.2008.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.a Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]; b Guterman A, Glickman MH. J Biol Chem. 2004;279:1729–1738. doi: 10.1074/jbc.M307050200. [DOI] [PubMed] [Google Scholar]; c Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. EMBO J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.a Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK. EMBO J. 2007;26:976–986. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Oberfeld B, Ruffieux-Daidie D, Vitagliano JJ, Pos KM, Verrey F, Staub O. Am J Physiol Renal Physiol. 2011;301:F189–196. doi: 10.1152/ajprenal.00487.2010. [DOI] [PubMed] [Google Scholar]
- 24.Amerik A, Swaminathan S, Krantz BA, Wilkinson KD, Hochstrasser M. EMBO J. 1997;16:4826–4838. doi: 10.1093/emboj/16.16.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.a Ulrich HD. Mol Cell. 2008;32:301–305. doi: 10.1016/j.molcel.2008.10.010. [DOI] [PubMed] [Google Scholar]; b Vertegaal A. Biochem Soc Trans. 2007;35:1422–1423. doi: 10.1042/BST0351422. [DOI] [PubMed] [Google Scholar]
- 26.Hunter T, Sun H. Ernst Schering Found Symp Proc. 2008:1–16. doi: 10.1007/2789_2008_098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.