Abstract
The prevalence of p16INK4A/CDKN2A genetic alterations in human oral cancers indicates that the p16 gene could be a potent and appropriate target for novel intervention. Chemically induced hamster cheek pouch (HCP) tumors are highly similar to human oral cancers in both histology and genetics and may be a good choice as a surrogate model for human oral cancers. Nevertheless, little is known about the genetic events in the p16 gene in the HCP tumor model. The purpose of this study was to evaluate chemically induced HCP tumor specimens for potential inactivating p16 alterations. HCP tumors were induced with 7, 12-dimethylbenz(a)anthracene (DMBA), and DNA extracted from 34 such specimens were analyzed for homozygous/hemizygous deletions, aberrant methylation of 5’ CpG islands, and point mutations using real-time multiplex PCR, methylation-specific PCR, and direct sequencing/cold single strand conformation polymorphism (SSCP), respectively. Homozygous deletions, hemizygous deletions, aberrant methylation of 5’-CpG islands, and point mutation were identified in 11, 4, 9, and 1 of 34 specimens, respectively. While the overall incidence of p16 alterations was 70.6% (24 of 34 specimens), the majority of inactivating events (67.6%) stemmed from deletion or methylation, which is consistent with the observations that in human oral SCC, the significant p16-inactivating events have been reported to be deletion and methylation. Our results strongly support the use of DMBA-induced HCP tumor model in evaluating novel p16-targeted therapy and prevention of human oral SCC.
INTRODUCTION
The P16/CDK4-cyclin D/RB/E2F pathway plays a key role in controlling cell growth by integrating multiple mitogenic and anti-mitogenic stimuli [1–3]. Through physical association with cyclin-dependent kinase 4 (CDK4), the protein product of p16INK4A/CDKN2A (hereafter, p16) specifically inhibits CDK4-mediated phosphorylation of the retinoblastoma susceptible gene product (RB). This association subsequently prevents the release of active transcription factors, E2Fs, from incompetent E2F/RB complexes, and negatively modulates E2F-mediated transcription of genes required for the G1-to-S transition [1]. Consequently, abrogation of the P16/CDK4/RB pathway contributes to unregulated cell proliferation and growth [2,3]. Additionally, P16 has been found to be implicated in senescence, apoptosis, DNA repair, aging, and multi-step oncogenesis through different mechanisms [4–6], such as promoting translocation of Rel A [4] and inducing an age-dependent decrease in the proliferative capacity of certain tissue-specific stem cells and unipotent progenitors [5]. Moreover, genetic alterations of p16 through homozygous deletion, hypermethylation, and point mutations have been found in a great number of human tumors [2,3], including human squamous cell carcinomas (SCCs) of the head and neck [7]. Together these findings indicate that genetic abnormality of the p16 gene contributes to, or may be, the primary cause of certain human tumors.
Squamous cell carcinomas represent the majority (about 90%) of oral cancers that are the sixth most prevalent cancer worldwide [8,9]. While current therapies for human oral SCC typically involve surgical resection in combination with radiation and chemotherapy, the clinical and quality of life consequences are suboptimal: First, the five-year survival rate is only about 50%, and the frequency of reoccurrence remains 20%–30%; Second, there are substantial side-effects relevant to surgical resection and radiotherapy, such as the disfiguring aesthetic effects associated with surgical procedures, tissue necrosis, fibrosis, atrophy and xerostomia [10,11]. Hence, alternative strategies as well as appropriate animal models in which these strategies can be tested need to be developed to effectively investigate the progression and ultimate prevention of oral SCCs. With regard to the prevalence of p16 genetic inactivation in human oral cancers, p16 represents a potent and appropriate target for novel intervention.
The choice of chemically-induced hamster cheek pouch (HCP) tumors as a surrogate model for human oral SCC is due to the striking resemblance in histology and genetics between these two types of tumors [11]. As with human SCCs, some HCP tumors may begin as verrucae or papillomas that become malignant and invasive, while other HCP tumors can evolve from relatively flat, dysplastic lesions [12]. Furthermore, there are genetic similarities between chemically-induced HCP tumors and human oral SCCs [13], including early H-ras mutations [14], amplification and over-expression of erbB2 [15] and EGFR [16], and alterations in TP53 [17]. With regard to the potential application of the chemically-induced HCP tumor model in p16-targeted intervention, there is little evidence concerning the genetic events in p16 in the HCP model. Our previous studies have shown that hamster P16 protein is nearly undistinguishable from its human homologue in both structure and function [18] and that genetic alterations of p16 in an N-nitroso-(2-propyl) amine (BOP)-induced hamster pancreatic tumor model are similar to those found in human pancreatic tumors [19]. These findings suggest that the chemically-induced HCP model may be appropriate for comparative studies evaluating oral SCC cancer intervention strategies targeting p16. To address this premise, we evaluated potential inactivating p16 alterations in chemically-induced HCP tumors. Our results show that, as in human oral SCC, p16 inactivation is a common event in chemically-induced HCP tumors, and the HCP model appears to be a suitable and relevant model to evaluate p16-targeted therapeutical and preventive strategies.
MATERIALS AND METHODS
Animals
Twenty-one male Syrian golden hamsters, 4–5 weeks of age, were obtained from Charles River Laboratories (Wilmington, MA). Three animals each were placed in plastic bottom cages with bedding and allowed to acclimate for one week. Food (AIN-76A, a modified semi-synthetic, high starch diet, formed into pellets (Dyets; Bethlehem, PA)) and water were given ad libitum. All experimental conditions were in accordance with NIH Guidelines and with approval from The Ohio State University Animal Care and Use Committee.
Chemicals
7,12-dimethylbenz(a)anthracene (DMBA) was obtained from Sigma-Aldrich (Milwaukee, WI) and dissolved at a concentration of 0.2% (w/v) in a solution of dimethylsulfoxide (DMSO; Fisher Scientific; Pittsburgh, PA). The carcinogen dosing solution was aliquoted (1.5 mL) into 13×100 mm glass tubes (Fisher Scientific), wrapped in aluminum foil, and stored at 4°C. At each dosing regimen, freshly-thawed carcinogen solutions were used (one tube will do approximately 6 cages).
Tumor Induction
At 5–6 weeks of age, animals were treated with carcinogen according to a modification of the initial methods previously described [20]. Hamsters were inhalation anesthetized with isoflurane and the opening of the cheek pouch was made accessible by inserting a small metal hook and gently pulling the hook laterally away from the hamster, so that the interior of the pouch was exposed. The medial surfaces of both pouches of all groups of animals were painted three times weekly for 6 weeks with a 0.2% DMBA in DMSO solution using a No. 4 camel hair brush. Animals were weighed bi-weekly during and after carcinogen treatment. Twelve weeks from the beginning of the carcinogen treatment, animals were sacrificed by a serial combination of CO2 asphyxiation and cervical dislocation. Both cheek pouches were everted using forceps and gross tumors (3–10 mm in length) were counted. Tumors were excised from the cheek pouches, divided into halves, and either placed in tissue cassettes for fixation in 10% neutral buffered formalin (NBF) at room temperature, or wrapped in aluminum foil and quick-frozen in liquid nitrogen. After 12 hrs formalin-fixed tissues were transferred from NBF into 1× phosphate buffer saline (PBS, pH 7.4) and stored at 4°C pending paraffin embedding and tissue sectioning. Following liquid nitrogen freezing, cheek pouch tissues were transferred and stored at –86°C pending subsequent nucleic acid isolation.
Nucleic Acid Isolation
Frozen tumor tissues were homogenized in appropriate lysis buffer using a Tissue-Tearor rotor/stator type dispersal tool (BioSpec Products; Bartlesville, OK). Genomic DNA was isolated from the cellular lysate using the DNeasy Mini Kit (Qiagen; Valencia, CA). DNA quantity and quality were assessed spectrophotometrically using a GeneQuant II RNA/DNA Calculator (Pharmacia Biotech/GE Healthcare; Pittsburgh, PA). Isolated DNA was stored at –20°C until used for assays.
Quantitative Real-Time PCR Deletion Assay
Homozygous or heterozygous deletion of hamster p16 exon 2 was analyzed by real-time PCR as previously described [19]. Each multiplex real-time PCR mixture contained 200 µM dNTPs, 3 mM MgSO4, 1.25 U Titanium Taq DNA polymerase (Takara Bio/Clontech; Mountainview, CA), 0.2 µM of each primer set (for both p16 and α-CMHC), 0.1 µM of each custom TaqMan probe, 1× PCR additive (1 mg/mL bovine serum albumin, 750 mM trehalose, and 1% Tween-20), 1× PCR Buffer, and 5–25 ng of genomic DNA (Table 1). The reaction was performed under the following PCR conditions: 96°C for 1 min, followed by 50 cycles of 95°C for 30 sec, 62°C for 30 sec, and 72°C for 30 sec.
Table 1.
PCR Primers and Probes.
| Real-time Genomic PCR Primers and Probes |
| p16: |
| 5’-ACTGCGATGTCGTACAGTATCTA-3’ (forward) |
| 5’-AGCCCTCAGATTTCAAACTCTT-3’ (reverse) |
| 5’-FAM-TAACTTCTGCCCAGACACCCCCA-TAMRA-3’ (probe) |
| α-CMHC: |
| 5’-ACCCAGGGCTTCATGCGTAT-3’ (forward) |
| 5’-CCGAGATAGGGAGCTAGGCA-3’ (reverse) |
| 5’-TET-CAAGCGTCTACCAGCGAGCTCCAGG-TAMRA-3’ (probe) |
| Methylation-specific PCR Primers |
| Modified/Methylated: |
| 5’-GCGGTTGTTTAGGGTCGC-3’ (forward) |
| 5’-CTACCTAAATCGAAATACGACCG-3’ (reverse) |
| Modified/Unmethylated: |
| 5’-GGAGTAGTATGGAGTTTTTTGTGGAT-3’ (forward) |
| 5’-TATACCTAAATCAAAATACAACCA-3’ (reverse) |
| Cold SSCP Amplification PCR Primers |
| Exon 1α |
| 5’-ATGGAGCCCTCTGCGGACG-3’ (forward) |
| 5’-GATGAGATACCCTGTGCCTAC-3’ (reverse) |
| Exon 2a |
| 5’-AGCGGCAGTCTGGCCTTGAT-3’ (forward) |
| 5’-GCCCCCGCCTGGTGTAGTAT-3’ (reverse) |
| Exon 2b |
| 5’-CGGGAGGGCTTCTTGGAAAC-3’ (forward) |
| 5’-TCAGGAGCCCTCAGATTTCA-3’ (reverse) |
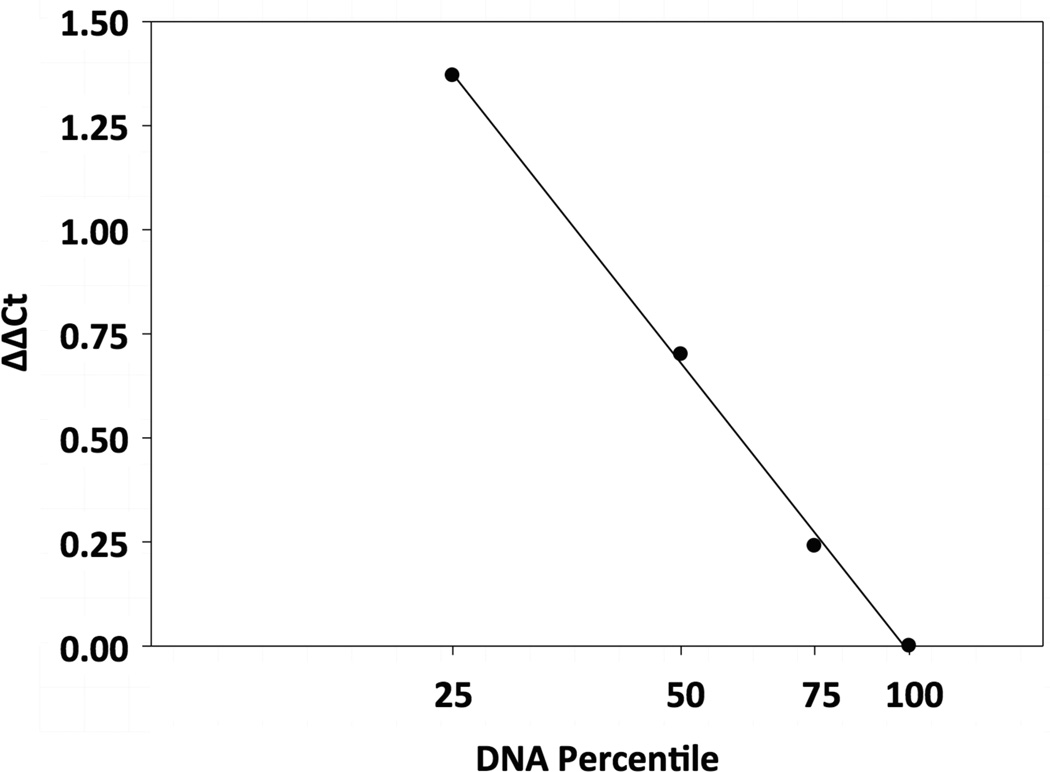
Deletion analyses were based on the assumption that the normalized cycle threshold (Ct) values resulting from multiplex PCRs are proportional to the dosage of p16 in the sample (in exponential form). The standard curve for defining p16 dosage (Figure 1) was acquired by titrating mixtures of hamster genomic DNA from POT2 cells (p16+/+) and H2T cells (p16−/−) [19]. The raw data were normalized using the relative approximation previously described [21] with the following equation:
ΔΔCt = (Ctp16 [sample] − Ctp16 [POT2]) − (Ctα-CMHC [sample] − Ctα-CMHC [POT2]),
where the housekeeping gene α-CMHC was used as an internal genomic amplification control. All experiments were performed in duplicate. The dosage of p16 in POT2 cells (p16+/+) was set as 100%, and the results were interpreted as follows [19]: dosage <25%, homozygous deletion (p16−/−); dosage 25%–75%, hemizygous deletion (p16+/−); and dosage >75%, wild type (p16+/+). The accuracy of the deletion assay was supported by the high correlation coefficient (r = 0.996).
Figure 1. Standard Curve of the Real-Time PCR Deletion Assay.
Genomic DNA from hamster POT2 and H2T cells was mixed at ratios of 100:0, 75:25, 50:50, 25:75, and 0:100, respectively, and was analyzed by multiplex real-time PCR for both p16 and α-CMHC loci. The resulting cycle threshold (Ct) values were normalized against POT2 (p16+/+, dosage = 100%). The normalized Ct values, ΔΔCt, were plotted against the p16 dosage in the POT2/H2T DNA mixtures (in exponential form) to yield a standard curve. For each tumor sample, the p16 dosage was determined from the standard curve based on its normalized Ct values (Materials and Methods Section).
Methylation-specific PCR in p16 exon 1α
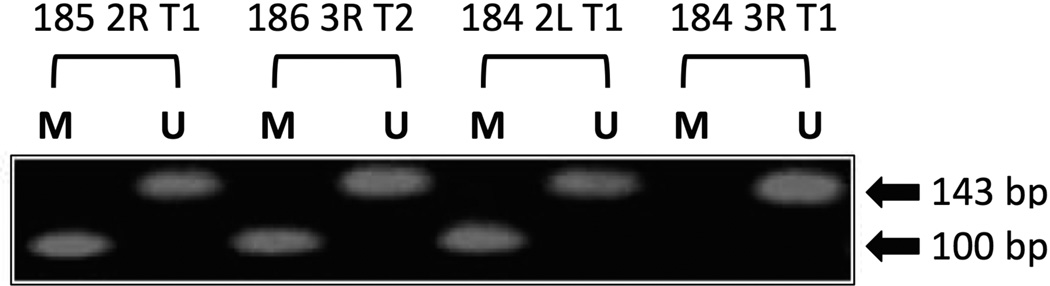
Since specific information about the promoter region of hamster p16 is not currently available, aberrant 5’-CpG methylation in p16 exon 1α was evaluated by a methylation-specific PCR assay [19]. Genomic DNA was bisufite-modified using the CpGenome DNA modification kit (Millipore; Temecula, CA) according to the manufacturer’s recommendation. Genomic targets were amplified using the PCRx Enhancer Kit (Invitrogen; Carlsbad, CA) in the presence of 1× Enhancer Solution, Titanium Taq DNA polymerase, and 0.2 µM of each specific forward and reverse primer (Table 1). The samples were subjected to 95°C for 2 min, followed by 50 cycles of 95°C for 30 sec, 54°C for 30 sec,and 72°C for 45 sec. After purification with a PCR Product Purification Kit (Qiagen; Valencia, CA), PCR products were electrophoresed through a 2.0% agarose gel. The sizes of PCR products from methylated and unmethylated p16 were 100 bp and 143 bp, respectively.
Mutational Analysis using Sequencing and Cold SSCP
p16 exon 1α and exon 2 were PCR amplified from hamster tumor DNA using Titanium Taq DNA Polymerase and the PCRx Enhancer Kit (Table 1). Exon 2 was amplified as two overlapping fragments, designated exon 2a and exon 2b, while exon 3 was not analyzed since it contributes only four amino acid residues at the C-terminus of P16, and these four residues play minor roles in the structure and function of P16 [22,23]. Reactions were performed in 1× PCR Buffer containing 1.5 mM MgSO4 and supplemented with 1.5× Enhancer Solution, 1.25 mM each dNTP, 1.0 U of Platinum Taq DNA polymerase, 0.2 µM of each primer, and 20–50 ng of genomic DNA. Thermocycling was performed as follows: 96°C for 1 min, followed by 50 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 45 sec. After purification with a PCR Product Purification Kit (Qiagen; Valencia, CA), PCR products were sequenced at the Plant-Microbe Genomics Facility at The Ohio State University. Since the sensitivity of direct sequencing mutation detection is relatively low, an enhanced sensitivity method, cold single-stranded conformational polymorphism (SSCP) analysis was used to detect nucleotide changes. PCR fragments were electrophoresed through Novex (Invitrogen) non-denaturing 10% TBE-PAGE gels for 4 hrs at 10°C using a Novex ThermoFlow electrophoresis apparatus according to methods previously described [7]. Following electrophoresis, gels were stained with 1× SYBR Gold (Invitrogen/Molecular Probes) and digitally analyzed using a ChemiImager 4400 cooled CCD camera system (Alpha Innotech; San Leandro, CA). If initial sequence alterations were detected by direct sequencing or cold SSCP analysis, both DNA strands were sequenced to confirm the results. The multiple sequence alignment software, T-Coffee (www.Expasy.com), was used to analyze the sequence data for possible nucleotide alterations.
RESULTS AND DISCUSSION
Table 2 summarizes the genetic alterations of p16 in 34 HCP tumor specimens. Homozygous (p16−/−) and hemizygous (p16−/+) deletions were detected in 11 (32.4%) and 4 (11.8%) of 34 specimens, respectively, while aberrant 5’-CpG methylation in p16 exon 1α was identified in 26.5% of specimens (9/34) (Figure 2), indicating that deletion and methylation of p16 are common events in chemically-induced HCP tumors. Interestingly, only one point mutation was identified in these HCP tumor specimens and this mutation was a one-base insertion after the codon encoding amino acid residue D76 in hamster P16 protein, which led to a frame shift after D76. While mutations at the corresponding residue, D84 in human P16, have been found in human SCCs of the head and neck, lung, prostate and esophagus cancers [23], this frame shift resulted in grossly a truncated P16 containing only the first two ankyrin repeats [22]. As revealed in the crystal structure of the P16/CDK6 complex, the ankyrin repeat before D76 and the ankyrin repeat after D76 form the core structure of P16 and provide most of contacts for CDK4 interactions [23]. Therefore, the D76FS mutation disrupts the tertiary structure of hamster P16 and abolishes its ability to inhibit CDK4-mediated phosphorylation of RB.
Table 2.
Summary of p16 alterations in chemically-induced HCP tumors.
| Sample |
p160 Status |
Methylation Exon 1α |
Point Mutations Exons 1α and Exon 2 |
|---|---|---|---|
| 184 2L T1 | +/+ | Yes | No |
| 184 2R T1 | +/+ | No | No |
| 184 2R T2 | +/+ | No | No |
| 184 3L T1 | −/− | No | No |
| 184 3L T2 | −/− | No | No |
| 184 3R T1 | +/+ | No | No |
| 185 1R T1 | −/− | No | No |
| 185 1R T2 | +/+ | No | No |
| 185 2R T1 | +/+ | Yes | No |
| 185 2R T2 | −/− | No | No |
| 185 3L T1 | −/− | No | No |
| 186 1R T1 | +/+ | No | No |
| 186 1R T3 | +/− | Yes | No |
| 186 2L T1 | +/+ | No | No |
| 186 3R T1 | −/− | No | No |
| 186 3R T2 | +/+ | Yes | No |
| 187 1R T1 | −/− | No | No |
| 187 3R T1 | +/− | No | No |
| 188 1L T1 | +/− | No | No |
| 188 1L T2 | +/+ | Yes | No |
| 188 1R T1 | +/+ | Yes | No |
| 188 2L T1 | −/− | No | No |
| 188 2L T2 | +/+ | No | D74FS* |
| 188 2R T3 | +/+ | No | No |
| 188 3L T1 | +/+ | Yes | No |
| 188 3L T2 | −/− | No | No |
| 188 3L T4 | +/+ | No | No |
| 189 1L T1 | −/− | No | No |
| 189 2L T1 | +/+ | Yes | No |
| 190 1R T1 | +/+ | No | No |
| 190 3L T1 | +/+ | No | No |
| 190 3L T2 | +/+ | Yes | No |
| 190 3R T1 | +/− | No | No |
| 190 3R T2 | −/− | No | No |
FS, frame shift.
Figure 2. Analysis of Aberrant 5’-CpG Methylation in Hamster p16 Exon 1α.
Results from four representative examples are presented here. PCR products in lanes “M” and “U” are from reactions using modified/methylated primers and modified/unmethylated primers, respectively. The presence of a PCR product of 100 bp in lane “M” indicates the presence of aberrant CpG methylation of p16 in the specimen.
In the current study, genetic alterations of the p16 gene were detected in 70.6% (24/34) of DMBA-induced HCP tumor specimens. Except for one tumor specimen, 186 1R T3 which harbored both a hemizygous deletion and p16 exon 1α methylation, only a single inactivating event was found in the other 23 specimens, suggesting that only one form of genetic inactivation may be sufficient to silence p16 [19]. Moreover, the majority of genetic alterations of p16 in HCP tumors (67.6%) stemmed from deletion or methylation, which is consistent with the observations in human oral SCCs; i.e., significant p16-inactivating events are deletions and methylation [24,25]. The incidence of somatic p16 point mutation in DMBA-induced HCP tumor specimens is minor (about 3%). In comparison, the reported frequencies of p16 point mutation in human head and neck SCC varied from 3.4% [26], 5.4% [27] to 27% [7], which could be attributed to the differences in the development stage of human head and neck SCC as well as in a heterogeneous patient population that is most likely exposed to a diversity of pro-carcinogenic compounds (e.g., cigarette smoke) and risk-enhancing behaviors. Additionally, our previous studies [19] have showed that a BOP-induced hamster pancreatic tumor model is a good genetic proxy for human pancreatic tumors with respect to p16 modifications, and as in DMBA-induced HCP tumors, most of the genetic alterations in p16 (83.3%) were secondary to mechanisms involving methylation or deletion. Last, a chemically induced hamster SCC model has the advantages of ease in tumor induction, a high rate of tumor incidence following chemical carcinogen exposure, and a short latency period preceding tumor development [7,13,19]. Taken together, these observations support the premise that the Syrian hamster is a useful model to evaluate p16-targeted intervention. Interestingly, 4-nitroquinoline-1-oxide (4N-O) induced rat oral tumors are an animal model commonly used for human head and neck SCC, tongue, and esophagus tumors [28], and the incidence of p16 inactivation in this model is about 62.2%, including deletion (36%), methylation (22.2%), and point mutation (4.4%) [29]. Hence, the DMBA-induced HCP tumor model is as good as the widely used 4N-O-induced rat oral tumor model.
It is worthwhile to note that, like its human counterpart, the hamster p16 locus also harbors an ortholog to the human p16Arf gene [30,31]. Since information about the hamster p16Arf gene is not currently available, it is difficult to evaluate the genetic status of p16Arf in the current HCP tumor specimens. While aberrant 5v-CpG methylation in p16 exon 1α should not affect p16Arf exon 1β, deletions and point mutations in p16 exon 2 may also bring about alterations in p16Arf, which consequently down-regulates another tumor suppressive pathway, the CDKN2A-ARF/P53/MDM2 pathway [30,31]. Moreover, while inactivation of p16 has been regarded as an early genotypic event in the development of human oral cancers [32], it remains to be elucidated whether inactivation of p16 occurs at the early developmental stage of DMBA-induced HCP tumors.
In summary, the incidence and mechanisms of inactivating genetic alterations of the p16 gene in DMBA-induced HCP tumors appear to be consistent with those found in human oral SCC. Furthermore, our data strongly support the use of a DMBA-induced HCP tumor model in evaluating novel p16-targeted therapies and cancer prevention strategies for human oral squamous cell carcinogenesis.
Acknowledgments
This work was supported by Research Grants from NIH R01 CA69472 to Ming-Daw Tsai (Department of Chemistry and Biochemistry), and R01 DE011943 and R21 DE016361 to C. M. W at The Ohio State University.
Abbreviations
- α-CMHC
hamster alpha-cardiac myosin heavy chain, also referred to as Myh6, myosin heavy polypeptide 6, cardiac muscle, alpha
- Arf
alternative reading frame of p16, also called p16beta, p16β
- BOP
N-nitroso-bis-(2-oxopropyl) amine
- CDK4
cyclin-dependent kinase 4
- CDKN2A
human cyclin-dependent kinase inhibitor 2A gene, also referred to as human p16INK4A/MTS1 gene
- Cdkn2a
hamster cyclin-dependent kinase inhibitor 2A gene, also referred to as hamster p16INK4A gene
- Ct
threshold cycle
- DMBA
7,12-dimethylbenz(a)anthracene
- DMSO
dimethylsulfoxide
- FS
frame shift
- HCP
hamster cheek pouch
- INK4
inhibitors of CDK4, including P15INK4B, P16INK4A, P18INK4C, and P19INK4D
- PBS
phosphate buffer saline
- PCR
polymerase chain reaction
- NBF
neutral buffered formalin
- RB
retinoblastoma susceptible gene
- SCC
squamous cell carcinoma
- SSCP
single-strand conformational polymorphism
REFERENCES
- 1.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 2.Ortega S, Malumbres M, Barbacid M. Cyclin-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta. 2002;1602:73–87. doi: 10.1016/s0304-419x(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Byeon I-JL, Poi MJ, et al. The structure-function relationship of p16 tumor suppressor. In: Ehrlich M, editor. DNA alterations in cancer: Genetic and epigenetic changes. BioTechniques books. Massachusetts: Eaton Publishing; 2000. pp. 71–83. [Google Scholar]
- 4.Thomas HC, Dunlop MG, Stark LA. CDK4 inhibitors and apoptosis: A novel mechanism requiring nuclear targeting of Rel A. Cell Cycle. 2007;6:1293–1297. doi: 10.4161/cc.6.11.4312. [DOI] [PubMed] [Google Scholar]
- 5.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Canepa ET, Scassa ME, Ceruti JM, et al. INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. IUBMB Life. 2007;59:419–426. doi: 10.1080/15216540701488358. [DOI] [PubMed] [Google Scholar]
- 7.Poi MJ, Yen T, Li J, et al. INK4a-ARF locus mutations: A significant mechanism of gene inactivation in squamous cell carcinomas of the head and neck. Mol Carcinogenesis. 2001;30:26–36. doi: 10.1002/1098-2744(200101)30:1<26::aid-mc1010>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 8.Parkin DM, Pisani P, Ferlay J. Global cancer statistics. CA A Cancer J Clin. 1999;49:33–64. doi: 10.3322/canjclin.49.1.33. [DOI] [PubMed] [Google Scholar]
- 9.Lippman SM, Hong WK. Chemoprevention of aerodigestive epithelial cancers. Adv Exp Med Biol. 1992;320:151–161. doi: 10.1007/978-1-4615-3468-6_20. [DOI] [PubMed] [Google Scholar]
- 10.Larson D, Lindberg RD, Lane E, Goepfert H. Major complications of radiotherapy in cancer of the oral cavity and oropharynx. Am J Surg. 1983;146:531–536. doi: 10.1016/0002-9610(83)90247-7. [DOI] [PubMed] [Google Scholar]
- 11.Casto BC, Kresty LA, Kraly CL, et al. Chemoprevention of oral cancer by black raspberries. Anticancer Res. 2002;22:4005–4016. [PubMed] [Google Scholar]
- 12.Gimenez-Conti IB, Shin DM, Bianchi AB, et al. Changes in keratin expression during, 7, 12-dimethylbenz(a)anthracene induced hamster cheek pouch carcinogensis. Cancer Res. 1990;50:4441–4445. [PubMed] [Google Scholar]
- 13.Gimenez-Conti IB, Slaga TJ. The hamster cheek pouch carcinogenesis model. J Cell Biochem Suppl. 1993;17F:83–90. doi: 10.1002/jcb.240531012. [DOI] [PubMed] [Google Scholar]
- 14.Kwong YY, Husain A, Biswas DK. c-Ha-ras gene mutation and activation precede pathological changes in DMBA-induced in vivo carcinogenesis. Oncogene. 1992;7:1481–1489. [PubMed] [Google Scholar]
- 15.Wong DTW, Biswas DK. Expression of c-erbB proto-oncogene during dimethylbenzathracene induced tumorigenesis in hamster cheek pouch. Oncogene. 1987;2:67–72. [PubMed] [Google Scholar]
- 16.Shin DM, Gimenez IB, Lee JS, et al. Expression of epidermal growth factor receptor, polyamine levels, ornitthine decarboxylase activity, micronuclei, and tansglutaaminase I in 7,12-dimethylbenz(a)anthracene-induced hamster buccal pouch carcinogenesis model. Cancer Res. 1990;50:2505–2510. [PubMed] [Google Scholar]
- 17.Gimenez-Conti IB, LaBate M, Liu F, Osterndorff E. p53 alterations in chemically induced hamster cheek-pouch lesions. Mol Carcinogenesis. 1996;16:197–202. doi: 10.1002/(SICI)1098-2744(199608)16:4<197::AID-MC3>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Qin D, Knobloch TJ, et al. Expression and characterization of Syrian golden hamster p16, a homologue of human tumor suppressor p16 INK4A. Biochem Biophys Res Commun. 2003;304:241–247. doi: 10.1016/s0006-291x(03)00577-1. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Weghorst CM, Tsutsumi M, et al. Frequent p16INK4A/CDKN2A alterations in chemically induced Syrian golden hamster pancreatic tumors. Carcinogenesis. 2004;25:263–268. doi: 10.1093/carcin/bgh007. [DOI] [PubMed] [Google Scholar]
- 20.Morris AL. Factors influencing experimental carcinogenesis in the hamster cheek pouch. J Dent Res. 1961;40:3–15. doi: 10.1177/00220345610400012001. [DOI] [PubMed] [Google Scholar]
- 21.Pfaffl MW. A new mathematical model for relative quantification in the real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byeon IL, Li J, Ericson K, et al. Tumor suppressor p16INK4A: Determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4. Mol Cell. 1998;1:421–431. doi: 10.1016/s1097-2765(00)80042-8. [DOI] [PubMed] [Google Scholar]
- 23.Russo AA, Tong L, Lee J, Jeggrey PD, Pavletich NP. Structural basis for inhibition of the cyclin-dependent kinase Cdk 6 by the tumor suppressor p16INK4a. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- 24.Olshan AF, Weissler MC, Pei H, et al. Alterations of the p16 gene in head and neck cancer: Frequency and association with p53, PRAD-1 and HPV. Oncogene. 1997;14:811–818. doi: 10.1038/sj.onc.1200892. [DOI] [PubMed] [Google Scholar]
- 25.Puri SK, Si L, Fan CY, Hanna F. Aberrant promoter hypermethylation of multiple genes in head and neck squamous cell carcinomas. Am J Otolaryngol. 2005;26:12–17. doi: 10.1016/j.amjoto.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Reed AL, Caligano J, Cairns P, et al. High frquency of p16(CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res. 1996;56:3630–3633. [PubMed] [Google Scholar]
- 27.Olshan AF, Weissler MC, Pei H, et al. Alterations of the p16 gene in head and neck cancer: Frequency and association with p53, PRAD-1 and HPV. Oncogene. 1997;14:811–818. doi: 10.1038/sj.onc.1200892. [DOI] [PubMed] [Google Scholar]
- 28.Kanojia D, Vaidya MM. 4-Nitroquinoline-1-oxide induced experimental oral carcinogenesis. Oral Oncol. 2006;42:655–667. doi: 10.1016/j.oraloncology.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Ogawa K, Tanuma J, Hirano MY, Semba I, Shisa H, Kitano M. Selective loss of resistant alleles at p15INK4B and p16INK4A genes in chemically-induced rat tongue cancers. Oral Oncol. 2006;42:710–717. doi: 10.1016/j.oraloncology.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 30.Sharpless NE, Depinho RA. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 31.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumor suppressor locus: All for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 32.Kresty LA, Mallery SR, Knobloch TJ, et al. Alterations of p16(INK4a) and p14(ARF) in patients with severe oral epithelial dysplasia. Cancer Res. 2002;62:5295–5300. [PubMed] [Google Scholar]