Abstract
Phototropin 1 (phot1) and phot2, which are blue light receptor kinases, function in blue light–induced hypocotyl phototropism, chloroplast relocation, and stomatal opening in Arabidopsis (Arabidopsis thaliana). Previous studies have shown that the proteins RPT2 (for ROOT PHOTOTROPISM2) and NPH3 (for NONPHOTOTROPIC HYPOCOTYL3) transduce signals downstream of phototropins to induce the phototropic response. However, the involvement of RPT2 and NPH3 in stomatal opening and in chloroplast relocation mediated by phot1 and phot2 was unknown. Genetic analysis of the rpt2 mutant and of a series of double mutants indicates that RPT2 is involved in the phot1-induced phototropic response and stomatal opening but not in chloroplast relocation or phot2-induced movements. Biochemical analyses indicate that RPT2 is purified in the crude microsomal fraction, as well as phot1 and NPH3, and that RPT2 makes a complex with phot1 in vivo. On the other hand, NPH3 is not necessary for stomatal opening or chloroplast relocation. Thus, these results suggest that phot1 and phot2 choose different signal transducers to induce three responses: phototropic response of hypocotyl, stomatal opening, and chloroplast relocation.
INTRODUCTION
Light is required to regulate plant growth and morphogenesis. Plants can respond to changes in light conditions, wavelength, intensity, and direction. In particular, blue light (390 to 500 nm) induces a wide range of physiological responses. Several responses, such as phototropism, stomatal opening, chloroplast relocation, and solar tracking by leaves, are thought to maximize photosynthetic light capture and control growth and development. Recent molecular genetic studies have shown that phototropin 1 (phot1) and phot2 function as photoreceptors for hypocotyl phototropism, chloroplast relocation, and stomatal opening in response to blue light (Briggs and Christie, 2002). phot1 was identified originally as a 120-kD plasma membrane protein showing blue light–dependent phosphorylation. The N-terminal region of the protein contains two LOV (light, oxygen, or voltage) domains, LOV1 and LOV2, which are kinds of PAS domain involved in protein–protein interaction and ligand binding (Taylor and Zhulin, 1999). The C-terminal region contains a Ser/Thr kinase domain (Huala et al., 1997). Biochemical and photochemical studies have demonstrated that each LOV domain binds to a blue light–absorbing chromophore, a flavin mononucleotide, and a recombinant protein of phot1 showed blue light–dependent autophosphorylation activity (Christie et al., 1998, 1999). phot2 is a phot1 homolog containing two LOV domains binding to a flavin mononucleotide at the N-terminal region and a kinase domain at the C-terminal region, which also shows blue light–dependent autophosphorylation activity (Sakai et al., 2001). Our previous genetic evidence showed that phot1 and phot2 function in a fluence-dependent manner to regulate hypocotyl phototropism (Sakai et al., 2001). phot1 functions at both low (0.01 to 1 μmol·m−2·s−1) and high (>1 μmol·m−2·s−1) fluence rates to mediate phototropic responses, but phot2 functions only at high light intensities. Furthermore, both phot1 and phot2 can mediate the accumulation response of chloroplasts to low-intensity blue light, although phot2 alone mediates the avoidance response to high-intensity light. Kinoshita et al. (2001) showed that phot1 and phot2 function redundantly in stomatal opening to blue light irradiation. Thus, phototropins are photoreceptors mediating a variety of photoinduced movement responses in Arabidopsis (Arabidopsis thaliana).
It is conceivable that RPT2 (for ROOT PHOTOTROPISM2) and NPH3 (for NONPHOTOTROPIC HYPOCOTYL3) function as signal transducers in phototropin signaling. RPT2 and NPH3 encode a novel family of plant-specific proteins, comprising 32 novel proteins in Arabidopsis (Motchoulski and Liscum, 1999; Sakai et al., 2000; see http://www.biosci.missouri.edu/liscum/research_page/LiscumLab_ResearchPage.html). Both RPT2 and NPH3 proteins possess a BTB/POZ (broad complex, tramtrack, bric à brac/pox virus and zinc finger) domain at the N-terminal region and a coiled-coil domain at the C-terminal region, which are thought to be protein–protein interaction domains. Members of the RPT2/NPH3 family have high similarity in primary sequence and secondary structure, but several differences have been reported. The nph3 mutant showed no phototropic response in hypocotyl or root (Okada and Shimura, 1992, 1994; Motchoulski and Liscum, 1999; Sakai et al., 2000). By contrast, the rpt2 mutant showed near normal phenotype at a low fluence rate of light, but its phototropic response decreased at a high fluence rate. RPT2 is induced by light in a manner dependent on light intensity, but NPH3 is highly expressed in dark-grown seedlings and is not light inducible (Sakai et al., 2000; Liscum, 2002). A yeast (Saccharomyces cerevisiae) two-hybrid study and a pull-down assay showed that NPH3 physically interacts with phot1 (Motchoulski and Liscum, 1999). From these results, it was speculated that NPH3 functions as a scaffold protein downstream of phot1 to transduce phototropic signals at low fluence rates, whereas RPT2 functions downstream of phot2 at high fluence rates (Liscum, 2002). Nevertheless, the interaction between RPT2 and phototropins remains unknown.
It is interesting that different movement responses to blue light—phototropic response, chloroplast relocation, and stomatal opening—in different tissues of a plant (hypocotyl, mesophyll cells, and guard cells) are mediated through the same photoreceptors, phot1 and/or phot2. How do phot1 and phot2 transfer the blue light signal for different blue light–induced responses? It is possible that phot1 and/or phot2 may choose signal transducers to suit each response. Indeed, phot1 and phot2 act in different manners to mediate the blue light–induced increase in cytosolic Ca2+ concentration (Harada et al., 2003), which has been shown to be a downstream component of phototropin signaling (Baum et al., 1999; Stoelzle et al., 2003).
Here, we show that phot1 signaling for stomatal opening follows an RPT2-dependent pathway and that RPT2 makes a complex with phot1 in vivo. Genetic analysis indicated that RPT2 and NPH3 are necessary for the phototropic response mediated by phot1 but not for chloroplast relocation. We discuss the roles of RPT2 and NPH3 in the phototropin signaling pathways of three blue light–induced responses.
RESULTS
Involvement of RPT2 and NPH3 in Hypocotyl Phototropism
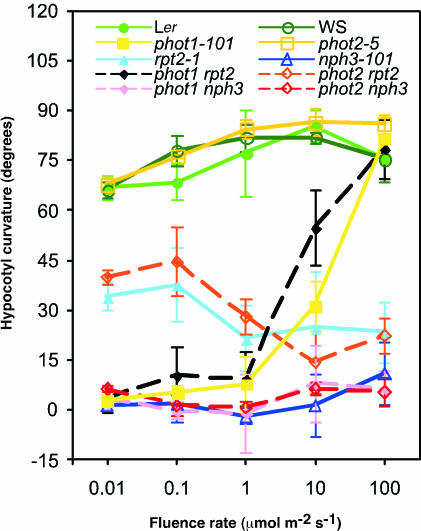
Our previous analysis of the rpt2-1 and nph3-101 mutants suggested that RPT2 and NPH3 transferred signals from blue light receptors for phototropic responses (Sakai et al., 2000). We also reported that both phot1 and phot2 functioned in a fluence rate–dependent manner to regulate hypocotyl phototropism; phot2 acted as a blue light receptor mediating phototropic response at high fluence rate, whereas phot1 regulated phototropism at both low and high fluence rates (Sakai et al., 2001). To examine the relationship between phototropin and signal transduction molecules, we analyzed the phototropic response of hypocotyls of double mutants phot1-101 rpt2-1, phot2-5 rpt2-1, phot1-101 nph3-101, and phot2-5 nph3-101 (see Methods). Previous analysis showed that the phot1-101, phot2-5/cav1-5/npl1-1, rpt2-1, and nph3-101 mutations were null alleles (Sakai et al., 2000, 2001).
The phot1 rpt2 mutant showed a positive phototropic response at 10 μmol·m−2·s−1 and 100 μmol·m−2·s−1 but no response at 0.01 to 1 μmol·m−2·s−1 (Figure 1), which was phot1-like, as described previously (Sakai et al., 2000). The phot2 rpt2 double mutant showed a response similar to that of the rpt2-1 single mutant; phototropic curvature was induced by weak blue light, but the degree of curvature decreased as the fluence rate increased. These results indicated that phot1 and RPT2 act in the same genetic pathway. However, RPT2 does not act in the phot2-mediated pathway.
Figure 1.
Hypocotyl Phototropism in Wild-Type (Ler and WS) and Mutant Seedlings.
Hypocotyl curvatures were measured after 12-h irradiation with blue light at the fluence rates indicated. Each symbol represents an average of three experiments (13 to 20 measurements each) ±sd.
The phot1 nph3 and phot2 nph3 double mutants and the nph3-101 mutant showed impaired phototropic response. This result supports previous observations suggesting that NPH3 is necessary for hypocotyl phototropism. Thus, NPH3 participates in the common pathway to induce the phototropic responses of hypocotyl by phot1 and phot2.
phot1 Signaling Leading to Stomatal Opening Depends on RPT2
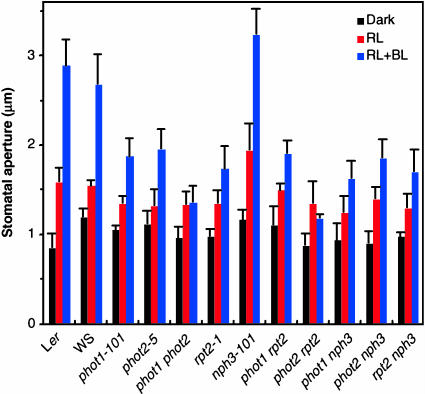
Recently, Kinoshita et al. (2001) reported that phot1 and phot2 are photoreceptors that induce stomatal opening under blue light irradiation. Therefore, we used rpt2-1, nph3-101, and their mutants to examine the involvement of RPT2 and NPH3 in stomatal responses. Red light induced stomatal opening slightly in epidermal strips from wild-type or mutant leaves (Figure 2), as described previously (Kinoshita et al., 2001). The addition of blue light at 10 μmol·m−2·s−1 induced stronger stomatal opening under background red light, although no significant increase in stomatal aperture was obtained at 1 μmol·m−2·s−1 (data not shown). The stomatal aperture of the phot1-101 or phot2-5 mutants was smaller than that of the wild-type plants after blue light irradiation at 10 μmol·m−2·s−1. Although a slight increase was observed under red light irradiation in the phot1 phot2 double mutant, the stomata did not respond to blue light, as described previously (Kinoshita et al., 2001). The nph3-101 mutant opened normally. Furthermore, additive phenotypes were not found in the phot1 nph3 and phot2 nph3 double mutants. These results strongly suggest that NPH3 does not mediate stomatal opening under blue light. However, the stomatal opening of the rpt2-1 mutant was small, as in phot1-101 and phot2-5. The phenotype of the rpt2 nph3 double mutant was similar to that of the rpt2-1 mutant. Interestingly, the stomata did not open in the phot2 rpt2 double mutant, and the phot1 rpt2 double mutant did not exhibit an additive phenotype. Because the stomatal opening under blue light irradiation is induced by two blue light receptors, phot1 and phot2, these results indicate that RPT2 is involved in the signaling process from phot1 for stomatal opening, but the signaling pathway activated by phot2 does not depend on RPT2.
Figure 2.
Stomatal Opening under Blue Light in Wild-Type Plants (Ler and WS) and Mutants.
Stomatal apertures were measured after 1-h incubation in the dark (Dark) and then after 2-h incubation under red light (50 μmol·m−2·s−1; RL) or under blue light (10 μmol·m−2·s−1) in a red light background (RL + BL). Each bar represents an average of three experiments (20 measurements each) ±sd.
Neither RPT2 nor NPH3 Is Involved in Phototropin Signaling for Chloroplast Relocation
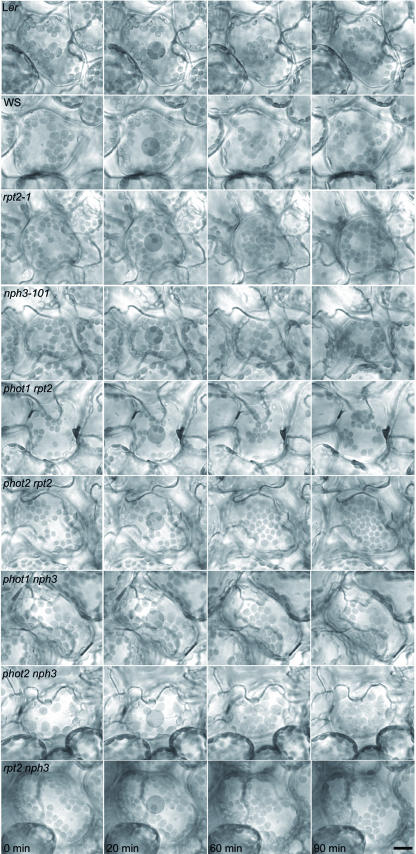
Our recent studies showed that phot2 is a blue light receptor that regulates the avoidance response of chloroplasts to high-intensity blue light, and both phot1 and phot2 can mediate the accumulation response of chloroplasts under low-light conditions (Kagawa et al., 2001; Sakai et al., 2001). To investigate the downstream molecular mechanism of chloroplast relocation, we analyzed the responses of rpt2-1, nph3-101, and their mutants by microbeam irradiation. Chloroplasts in wild-type cells moved toward the area irradiated with low-intensity blue light (2 μmol·m−2·s−1; Figure 3) but moved away from that area at 40 μmol·m−2·s−1. Both responses of the phot1 mutant were normal at 2 μmol·m−2·s−1 and 40 μmol·m−2·s−1, but chloroplasts in the phot2 mutant entered the irradiated area under both low- and high-intensity light, as described previously (Kagawa et al., 2001; Sakai et al., 2001). The rpt2-1 and nph3-101 mutants exhibited normal accumulation and avoidance responses. To examine the possibility that RPT2 and NPH3 function in a redundant manner, we observed chloroplast relocation in the rpt2 nph3 double mutant. However, both responses were similar to that of the wild type. Furthermore, the phot1 rpt2 and phot1 nph3 double mutants exhibited a normal phenotype, like the phot1 single mutant, whereas the response of phot2 rpt2 and phot2 nph3 was phot2-like. These results were confirmed by a slit assay (data not shown), which indicates the blue light–activated chloroplast relocation in a whole leaf (Kagawa et al., 2001; Sakai et al., 2001). Thus, the defects in RPT2 and NPH3 showed no effect on chloroplast relocation in phot1 and phot2. These findings suggest that signaling pathways from phototropins for chloroplast relocation do not depend on the presence of either RPT2 or NPH3.
Figure 3.
Chloroplast Relocation in Wild-Type Plants (Ler and WS) and Mutants.
Mesophyll cells were irradiated with red light for 0 to 20 min, with a microbeam of low-intensity blue light (2 μmol·m−2·s−1) for 20 to 60 min, and with high-intensity blue light (40 μmol·m−2·s−1) for 60 to 90 min. Shaded circles (20 μm in diameter) in the cells represent the irradiated area. Bar = 20 μm.
RPT2 Interacts with phot1 and NPH3 in Yeast
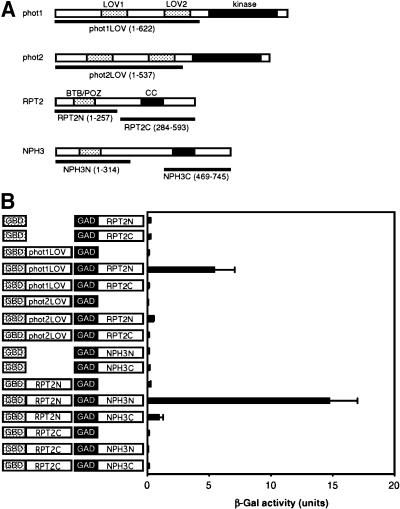
A previous study showed that NPH3 interacts with phot1 and that its interaction may contribute to the phototropic response mediated by phot1 (Motchoulski and Liscum, 1999). This genetic study indicated that RPT2 is necessary for the contribution of phot1 to stomatal opening and that its contribution is independent of NPH3. Our results suggest that RPT2 also interacts with phot1 directly, not mediated by NPH3, so we examined its interaction by yeast two-hybrid assay. We tested interactions between the N-terminal regions containing the two LOV domains of phot1 or phot2 and the N- or C-terminal regions of RPT2 (Figure 4A), similar to a study of the interaction between phot1 and NPH3 by Motchoulski and Liscum (1999). The N- and C-terminal regions included BTB/POZ and coiled-coil domains, respectively, thought to be protein–protein interaction domains. As shown in Figure 4B, strong interactions occurred between phot1 and the N-terminal portion of RPT2. Although the binding of NPH3 to phot1 is strongly mediated by its C-terminal region (Motchoulski and Liscum, 1999), the C-terminal portion of RPT2 did not show binding activity to phot1 in yeast. On the other hand, the N-terminal region of phot2 did not show a significant interaction with either region of RPT2 in yeast.
Figure 4.
Interaction between Phototropins and RPT2 or between RPT2 and NPH3.
(A) Schematic of phot1, phot2, RPT2, and NPH3 structures. The protein kinase and LOV domains of phot1 and phot2 proteins are shown as solid and dotted blocks, respectively. The BTB/POZ and coiled-coil (CC) domains of RPT2 and NPH3 are shown as dotted and solid blocks, respectively. Amino acid residues used for each underlined construct are shown in parentheses.
(B) Yeast two-hybrid assay in phot1–RPT2 or RPT2–NPH3 interactions. Solution assays of β-galactosidase (β-Gal) activity were performed for the combinations shown at left. One unit of β-Gal activity was defined as the amount of enzyme that converted 1 μmol of o-nitrophenyl-β-d-galactopyranoside to o-nitrophenol and d-galactose in 1 min at 30°C. Each bar represents an average of 3 to 10 measurements ±sd.
RPT2 shows genetic interaction with NPH3 in the phototropic response; both signal transducers are involved in the phototropic response induced by phot1. We examined the physical interaction between RPT2 and NPH3 in the same assay. The N-terminal region of RPT2 bound strongly to the N-terminal region of NPH3. It also bound to the C-terminal region of NPH3 but very weakly, but the C-terminal region of RPT2 did not show binding activity to either region of NPH3 (Figure 4B).
RPT2 Makes a Complex with phot1 in Vivo
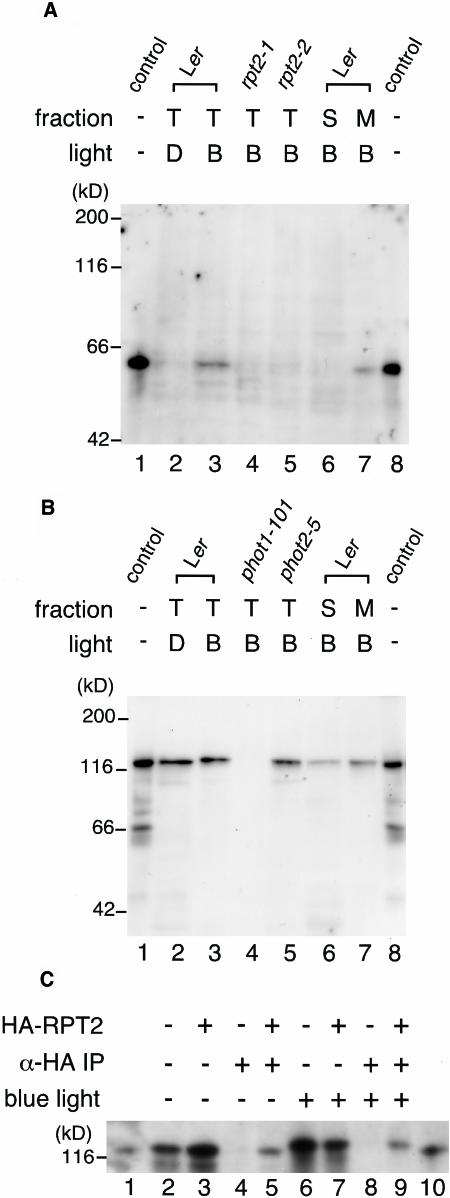
If RPT2 interacts with phot1 and/or NPH3 in vivo, it should localize at the plasma membrane with these proteins because previous studies indicated that these proteins localize at the plasma membrane (Motchoulski and Liscum, 1999; Sakamoto and Briggs, 2002). First, we examined whether RPT2 is fractionated in the crude microsomal fraction, including the plasma membrane, using anti-RPT2 antiserum. We detected the RPT2 protein in the total protein fraction from wild-type seedlings irradiated by blue light but not from rpt2-1 and rpt2-2 (Figure 5A). The RPT2 protein was hardly detected in the seedlings held in the dark. This result agrees with the results of the previous study showing that mRNA of RPT2 is induced by blue light irradiation (Sakai et al., 2000). When we prepared the crude microsomal membrane and the supernatant after high-speed centrifugation at 100,000g from wild-type seedlings, RPT2 was fractionated in the microsomal membrane rather than in the supernatant. This result shows the possibility that RPT2 interacts with phot1 and/or NPH3 at the plasma membrane.
Figure 5.
Immunological Analysis of the Association of RPT2 with phot1.
(A) Immunoblot of RPT2. Etiolated wild-type seedlings (Ler; lanes 2, 3, 6, and 7), rpt2-1 seedlings (lane 4), and rpt2-2 seedlings (lane 5) were irradiated by blue light at 100 μmol·m−2·s−1 for 1 h (B; lanes 3 to 7) or were mock-irradiated (D; lane 2), and then fractions of total proteins (T; lanes 2 to 5), soluble proteins (S; lane 6), and crude microsomal proteins (M; lane 7) were prepared. Proteins (10 μg) in each fraction were separated by 8% SDS-PAGE, followed by immunoblotting with anti-RPT2 antiserum. Lanes 1 and 8 show the molecular size marker of RPT2.
(B) Immunoblot of phot1. Etiolated wild-type seedlings (Ler; lanes 2, 3, 6, and 7), phot1-101 seedlings (lane 4), and phot2-5 seedlings (lane 5) were irradiated by blue light at 100 μmol·m−2·s−1 for 1 h (B; lanes 3 to 7) or mock-irradiated (D; lane 2), and then fractions of total proteins (T; lanes 2 to 5), soluble proteins (S; lane 6), and crude microsomal proteins (M; lane 7) were prepared. Proteins (10 μg) in each fraction were separated by 8% SDS-PAGE, followed by immunoblotting with anti-phot1 antibody. Lanes 1 and 8 show the molecular size marker of phot1.
(C) Immunoprecipitation of the phot1–RPT2 complex. Etiolated wild-type seedlings (Ler; lanes 2, 4, 6, and 8) and transgenic seedlings expressing HA-RPT2 (lanes 3, 5, 7, and 9) were irradiated by blue light at 10 μmol·m−2·s−1 for 1 h (lanes 6 to 9) or mock-irradiated (lanes 2 to 5), and then fractions of crude microsomal proteins were prepared. These were immunoprecipitated with anti-HA antibody–conjugated agarose and then were separated by 6% SDS-PAGE, followed by immunoblotting with anti-phot1 antibody (lanes 4, 5, 8, and 9). Altogether 0.005 volumes of crude microsomal proteins used for immunoprecipitation were also electrophoresed without immunoprecipitation as a phot1 protein control (lanes 2, 3, 6, and 7). Lanes 1 and 10 show the molecular size marker of phot1.
Next, we tried to immunoprecipitate RPT2 from the crude microsomal fraction. Because anti-RPT2 antiserum was not suited to the assay, we used transgenic plants expressing hemagglutinin (HA) epitope–tagged RPT2 (HA-RPT2). The transgene complemented the phototropic response in the rpt2-1 mutant, and it was functional in vivo (data not shown). To detect the phot1 protein, anti-phot1 antibody was used. Anti-NPH3 antibody could not be obtained, and the interaction with NPH3 in vivo was not observed in this study.
Anti-phot1 antibody detected the phot1 protein in wild-type seedlings but not in the phot1-101 mutants (Figure 5B). Its molecular size in seedlings irradiated by blue light was slightly larger than that produced in vitro or in the seedlings held in the dark. This result is consistent with the previous result, indicating that phot1 is phosphorylated by blue light irradiation (Short et al., 1993; Christie et al., 1998). Its expression level and mobility in SDS-PAGE did not change in the phot2-5 mutant. phot1 was fractionated in the crude microsomal membrane and the supernatant; this result is consistent with the previous study indicating that phot1 localizes at the plasma membrane and in the cytosol under blue light irradiation (Sakamoto and Briggs, 2002). phot1 was immunoprecipitated with anti-HA antibody–conjugated agarose to detect the HA-RPT2 protein (Figure 5C). This result suggests that RPT2 makes a complex with phot1 in vivo. Because the HA-RPT2 was expressed ectopically by the 35S promoter of Cauliflower mosaic virus even in the dark, phot1 from seedlings in the dark was also immunoprecipitated with HA-RPT2. The molecular mass of the phot1 protein isolated from the seedlings under blue light condition was larger than that of phot1 protein isolated from the seedlings under the dark condition. The result suggests that the phosphorylation state of phot1 has no effect on the association with RPT2.
Transcriptional Expressions of RPT2 and NPH3 in Various Tissues
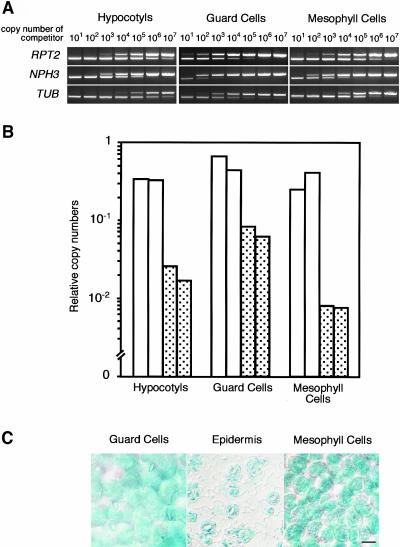
Our genetic study indicated that the rpt2-1 mutant shows defects in the phototropic response and stomatal opening and that the nph3-101 mutant shows a defect in the phototropic response when induced by blue light irradiation. We suppose that both RPT2 and NPH3 are expressed in hypocotyls and RPT2 in guard cells. We confirmed the expression of RPT2 and NPH3 mRNA in hypocotyls of etiolated seedlings irradiated by blue light for 12 h and in guard cells using the competitive RT-PCR method. We collected hypocotyls 1-cm long under cotyledons. To check the transcriptional level in guard cells of mature leaves under long-day conditions (16 h/8 h, day/night), we prepared the cells as protoplasts and extracted total RNA from them.
Our data show that RPT2 mRNA was expressed at almost the same level in hypocotyls and guard cells (Figures 6A and 6B). RPT2 mRNA was also detected in mesophyll cells, although RPT2 does not contribute to the chloroplast relocation induced by blue light irradiation. In agreement with the results of competitive RT-PCR analysis, transgenic plants expressing the β-glucuronidase (GUS) reporter gene driven by the RPT2 promoter also expressed RPT2 in guard cells and mesophyll cells (Figure 6C). NPH3 was also expressed in hypocotyls, guard cells, and mesophyll cells. Calculations of copy numbers of mRNA by comparison with competitors suggested that copy numbers of NPH3 mRNA were ∼0.0625, 0.125, and 0.023 of those of RPT2 mRNA in hypocotyls, guard cells, and mesophyll cells, respectively. Thus, both RPT2 and NPH3 were expressed in all tissues examined, regardless of their contribution to phototropin signaling for blue light responses.
Figure 6.
Expression Patterns of RPT2 and NPH3.
(A) RT-PCR with total RNA extracted from hypocotyls, highly purified guard cell protoplasts, and mesophyll cell protoplasts. The expression level of the tubulin gene (TUB) was used as an internal control.
(B) Copy numbers of RPT2 and NPH3 cDNAs in hypocotyls, guard cells, and mesophyll cells. Numbers were calculated against each competitor DNA and normalized to TUB. The graph shows relative copy numbers of RPT2 and NPH3 cDNAs (open and dotted bars, respectively) when the copy number of TUB was assigned a value of 1.0. The experiments were performed two times.
(C) The RPT2 promoter drives GUS activity (indicated by blue staining) in guard cells and mesophyll cells of wild-type plants expressing the RPT2-GUS fusion construct. To remove the mesophyll cell background, epidermal strips were peeled off the leaves. Bar = 20 μm.
DISCUSSION
On the basis of genetic studies in Arabidopsis, it has become clear that the phototropin family of blue light receptors regulates three movements—hypocotyl phototropism, chloroplast relocation, and stomatal opening—in response to blue light irradiation. In addition, RPT2 and NPH3 appear to function in the signaling pathway of phototropism. We investigated the possibilities that the signal transducers of RPT2 and NPH3 can induce the other phototropin-mediated responses, stomatal opening, and chloroplast relocation and that RPT2 functions downstream of phot2 to induce phototropic response of hypocotyls (Table 1).
Table 1.
Contributions of RPT2 and NPH3 to Signaling Pathways Mediated by Phototropins
| Phototropic Response in Hypocotyls
|
Stomatal Opening
|
Chloroplast Relocation
|
||||
|---|---|---|---|---|---|---|
| phot1-induced | phot2-induced | phot1-induced | phot2-induced | phot1-induced | phot2-induced | |
| RPT2 | + | − | + | − | − | − |
| NPH3 | + | + | − | − | − | − |
RPT2 contributes to the phot1-induced phototropic response, but it is not essential at low fluence rates. +, Necessary; −, unnecessary. See text for details.
Our previous studies using the rpt2-1 or phot1 phot2 mutants indicated that phot2 acts redundantly with phot1 in mediating hypocotyl phototropism at high fluence rates and that RPT2 and phot2 function at higher light levels (Sakai et al., 2000, 2001). The PHOT2 and RPT2 genes are induced by light irradiation in a manner depending on fluence rate (Sakai et al., 2000, 2001). Furthermore, RPT2 has two protein–protein interaction domains, BTB/POZ, and coiled-coil domains. These findings suggest that RPT2 acts as a scaffold to bring phot2 together with its downstream signaling partner(s) at high fluence rates because NPH3 was thought to be a scaffold allowing phot1 and other proteins to induce the phototropic response under a low fluence rate of blue light (Liscum, 2002). If so, the phot1 rpt2 double mutant would show a hypocotyl curvature like that of the phot1 phot2 double mutant, and the phot2 rpt2 mutant would show a phenotype like the phot2 single mutant. However, this study indicates that the defect in RPT2 influenced the phototropic curvature in phot2 but not in phot1. Thus, the defect in RPT2 influenced the phot1-induced curvature but not the phot2-induced curvature. This result suggests that RPT2 is involved in the phot1-induced phototropic response and that phot2 and RPT2 do not act in the same genetic pathway. It should be noted that partial bending of rpt2 at low fluence rate of blue light indicated that RPT2 is not an essential signaling partner for the phot1 in the phototropic response of hypocotyls, as described previously (Sakai et al., 2000). On the other hand, previous studies and this study indicate that NPH3 is a common and essential regulator in the induction of the phototropic response by phot1 and phot2 (Liscum and Briggs, 1996; Sakai et al., 2000). For this reason, RPT2 may contribute to the phot1-induced phototropic response by association with NPH3.
Although RPT2 is not necessary for the phot2-induced phototropic response, why doesn't the rpt2-1 mutant show the phot1-like phenotype at high fluence rates? There is a large difference in responses between rpt2-1 and phot1 or phot1 rpt2 at high fluence rates. We examined the phototropic response of the hypocotyl in an RPT2 knockout mutant, rpt2-2, which is mutated by the insertion of a transposon in the RPT2 locus, and it also showed a phenotype similar to that of rpt2-1 (data not shown). Thus, the requirement of RPT2 to induce the phototropic response at high fluence rates is not specific to the rpt2-1 allele. These results suggest that the presence of phot1 acts negatively in the phototropic response of hypocotyls at high fluence rates in the rpt2-1 mutant for an unknown reason, as described previously (Sakai et al., 2000).
The signaling pathways to induce the phototropic response of roots should be considered separately from the pathways in hypocotyls. The phot1 and rpt2 mutants showed barely any phototropic response in roots (Sakai et al., 2000). So, we do not know the involvement of phot2 or the relationship between phot1 and RPT2 in the phototropic response of roots.
Recently, it was shown that phot1 and phot2 act in a functionally redundant manner to mediate stomatal opening induced by blue light; normal opening response was observed in the phot1 or phot2 single mutant, but no response was detected in the phot1 phot2 double mutant (Kinoshita et al., 2001). However, we could detect a significant difference in stomatal aperture between the wild-type plant and the phot1-101 or phot2-5 single mutant; the phot1 phot2 double mutant exhibited no response. In addition, the opening response in the phot1-101 mutant was similar to that in the phot2-5 mutant, and no significant difference in stomatal aperture was detected between middle- and high-intensity irradiation (10 μmol·m−2·s−1 and 100 μmol·m−2·s−1; data not shown). We do not know why phot1 and phot2 showed a more severe phenotype in stomatal opening in our study, although some experimental conditions may have been different. Almost the same results were confirmed by observation of stomatal opening in intact leaves (data not shown), in addition to epidermal strips. Thus, the sole presence of phot1 or phot2 induced a sufficient phototropic response in hypocotyl like that in the wild type but not in the stomatal opening by blue light irradiation at 10 μmol·m−2·s−1 in our study. If so, both signaling pathways from phot1 and phot2 are necessary to induce a sufficient response of stomata, and there is a possibility of different contributions of phot1 and phot2 between the phototropic response of hypocotyl and the stomatal opening.
Genetic and physiological analyses indicate that RPT2 is a possible partner of phot1 in stomatal opening, although NPH3 does not function in phototropin signaling. Thus, phot1 and phot2 may function in two different pathways, RPT2 dependent and RPT2/NPH3 independent, respectively, for signaling stomatal opening. Blue light causes phosphorylation and activation of the plasma membrane H+-ATPase in guard cells (Kinoshita and Shimazaki, 1999). Activation of H+-ATPase was lacking in the phot1 phot2 double mutant (Kinoshita et al., 2001). The electrical potential gradient created by H+-ATPase drives a voltage-gated, inward-rectifying K+ channel, which is involved in stomatal opening (Schroeder et al., 1987). Although the involvement of RPT2 in phosphorylation and activation of the plasma membrane H+-ATPase must be elucidated, the phot1–RPT2 complex may play an important role at the plasma membrane.
Our previous findings also suggested that phot1 and phot2 act redundantly in mediating chloroplast accumulation at low fluence rates, whereas phot2 mediates avoidance by chloroplasts at high fluence rates (Kagawa et al., 2001; Sakai et al., 2001). Therefore, it is reasonable that RPT2 and NPH3 function as signal transducers, leading to chloroplast relocation. The rpt2-1, nph3-101, and rpt2 nph3 mutants showed no defects in chloroplast relocation. We suggest that phototropin signaling for chloroplast relocation may follow RPT2/NPH3-independent pathways; other signal transducers may act in the pathway instead of RPT2 and NPH3. In addition, although the leaves of the phot1 phot2 double mutant are curled (Kinoshita et al., 2001; Sakai et al., 2001; Sakamoto and Briggs, 2002), abnormality in that morphogenesis was not observed in rpt2-1, nph3-101, and a series of double mutants (data not shown). The RPT2/NPH3 family has at least 32 genes in the Arabidopsis genome (Liscum, 2002). Several members of the RPT2/NPH3 family have two protein–protein interaction domains, a BTB/POZ domain at the N-terminal region and a coiled-coil domain at the C-terminal region. It is conceivable that some members of the RPT2/NPH3 family may be involved in the signaling pathways for chloroplast relocation and the leaf morphogenesis and in phot2 signaling mediating hypocotyl phototropism and stomatal opening. This possibility is a subject for future research.
Analyses by RT-PCR and GUS reporter gene showed that RPT2 is expressed in hypocotyls and guard cells, as expected by its function. On the other hand, it was expressed also in mesophyll cells, at least as its transcripts. NPH3 is also expressed in guard cells and mesophyll cells in addition to hypocotyls. We do not know whether these expressions are important to their functions, but there is a possibility that RPT2 and NPH3 have further functions besides those we identified in the signaling pathways of blue light responses.
From our observation and analysis, we suggest that phototropins use signal transducers for different photoinduced movement responses in different tissue. In the phototropic response of hypocotyls, NPH3 is a common regulator in the phot1- and phot2-signaling pathways, but RPT2 is not necessary for the phot2-mediated pathway. Neither RPT2 nor NPH3 is a signal component of stomatal opening mediated by phot2, but RPT2 is a phot1-signaling partner. Neither RPT2 nor NPH3 is a signal component of chloroplast relocation induced by phot1 and phot2. Our recent study suggested that phot1 and phot2 mediate the blue light–dependent increase in cytoplasmic Ca2+ in different manners (Harada et al., 2003). We demonstrated that phot1 and phot2 have separate partners and pathways for transducing the signal, although they have high similarity in primary sequence and secondary structure and regulate the same movement responses induced by blue light irradiation. Functional analysis of RPT2 and NPH3 will be necessary to understand the phototropin-signaling pathways in detail in the phototropic response and stomatal opening by blue light irradiation.
METHODS
Plant Materials and Growth Conditions
Arabidopsis seedlings were grown from seeds of the wild types (ecotypes Landsberg erecta [Ler] and Wassilewskija [WS]), phot1-101 (a nonsense mutation at residue 120; Ler background), phot2-5/npl1-1/cav1-5 (a mutation by T-DNA insertion; WS background), nph3-101 (a nonsense mutation at residue 461; Ler background), rpt2-1 (a frameshift mutation at the splice acceptor site of the first intron; Ler background), phot1-101 phot2-5, phot1-101 rpt2-1, phot2-5 rpt2-1, phot1-101 nph3-101, phot2-5 nph3-101, and rpt2-1 nph3-101 mutants. Strains of these mutants were isolated as described previously (Okada and Shimura, 1992, 1994; Sakai et al., 2001). rpt2-2, which shows a mutation by dSpm (a nonautonomous defective Spm element) insertion in the third exon, was isolated from the SLAT collection (Tissier et al., 1999). Transgenic plants expressing HA epitope–tagged RPT2 were made as follows. The RPT2 cDNA gene was cloned into the NotI site on the pHA-NotI plasmid (Harada et al., 2003). Then, the HA-RPT2 gene was cloned in the pBI121 binary vector and transformed into rpt2-1 by the vacuum infiltration method mediated by Agrobacterium tumefaciens (Bechtold et al., 1993).
Measurement of Phototropic Curvature
Hypocotyl curvatures were assayed as described previously (Sakai et al., 2000). Seeds were surface-sterilized and planted in square Petri dishes containing 1.5% agar medium (Okada and Shimura, 1992). Three-day-old etiolated seedlings were irradiated for 12 h with unilateral blue light from blue light–emitting diode (LED) lamps (maximum wavelength of 470 nm with a half-bandwidth of 30 nm; Eyela, Tokyo, Japan). The fluence rate of the light source was controlled by the use of blue filters (film number 72; Tokyo Butai Shoumei, Tokyo, Japan). Seedling images were obtained with a 3D Digital Fine Scope (VC4500-PC; Omron, Tokyo, Japan) after irradiation, and the curvatures were measured with an Omron Image-Ana LITE.
Measurement of Stomatal Aperture
Stomatal opening was analyzed as described (Kinoshita et al., 2001). Wild-type plants and mutants were grown at 21°C to 22°C under fluorescent lamps (16 h/8 h, day/night; 50 μmol·m−2·s−1) on soil. Epidermal strips from 3- to 5-week-old plants were peeled off from the abaxial surface of a leaf and incubated in 5 mM Bis-Tris/propane, pH 6.5, 50 mM KCl, and 0.1 mM CaCl2 in the dark for 1 h. They were then irradiated with 10 μmol·m−2·s−1 blue light under background 50 μmol·m−2·s−1 red light with blue and red light LED lamps (maximum wavelength of 660 nm with a half-bandwidth of 20 nm; Eyela) for 2 h. Images of stomatal apertures were obtained with an Olympus BX51 microscope connected to a CCD camera (CS330; Olympus, Tokyo, Japan) after incubation.
Microbeam Assay of Chloroplast Relocation
Chloroplast relocation in a single mesophyll cell was observed using a microbeam irradiation system (Kagawa and Wada, 2000). Wild-type plants and mutants were grown under long-day conditions (16 h/8 h, day/night). Monochromatic blue light was transmitted through an interference filter (OPTOS224; Olympus), which had a transmission peak at 450 nm and a spectral full width at half-maximum of 13 nm. Monochromatic red light was transmitted through an interference filter (OPTOS223; Olympus), which had a relatively flat plateau (∼85% over 640 to 653 nm) and a half-bandwidth of 27 nm. Background red light irradiation activated chloroplast movement in mesophyll cells (Kagawa and Wada, 2000). Therefore, leaves from 3- to 5-week-old plants were irradiated by a blue light microbeam (20-μm diameter) at 2 μmol·m−2·s−1 for 40 min and then at 40 μmol·m−2·s−1 for 30 min in combination with red light background at 120 μmol·m−2·s−1. Images of mesophyll cells were obtained with an Olympus BX51 microscope connected to a CCD camera (CS330) after irradiation. An Advantest TQ8210 optical power meter and Q82017A optical sensor (Advantest, Tokyo, Japan) were used to measure the fluence rate of the microbeam.
cDNA Isolation
cDNAs of PHOT1, PHOT2, and NPH3 were isolated from Arabidopsis cDNA by a method described previously (Sakai et al., 2000).
Yeast Two-Hybrid Assay
PCR was used to generate the coding sequences of PHOT1, PHOT2, and RPT2 with NdeI-PstI sites incorporated into the 5′ and 3′ primers. Products were subcloned into the NdeI-PstI sites of the GAL4 DNA binding domain vector pGBDKT7 (Clontech, Palo Alto, CA). The coding sequences of RPT2 and NPH3 were also generated by PCR with NdeI-XhoI sites incorporated into the 5′ and 3′ primers, respectively. Products were subcloned into the NdeI-XhoI sites of the GAL4 activation domain vector pGADT7 (Clontech). All constructions were checked by DNA sequencing.
Pairwise combinations of vectors were cotransformed into yeast strain Y187 (Clontech) and were plated on the same selective medium. Quantitative β-galactosidase assays were performed in liquid cultures of yeast using o-nitrophenyl-β-d-galactopyranoside (Ausubel et al., 2001).
In Vitro Transcription and Translation
To prepare proteins of RPT2 and phot1 as molecular size controls, in vitro transcription and translation was performed using the TNT T7 Quick for PCR DNA kit (Promega, Madison, WI). DNA templates for in vitro transcription and translation were prepared by PCR with primers RPT2invitroFW (5′-GGATCCTAATACGACTCACTATAGGGAACAGCCACCATGGCAACAGAAGGAAAAAACCCC-3′) and RPT2invitroRV (5′-T32AAGAGATTGAGAATCTTCGTCT-3′) for cDNA of RPT2 and primers PHOT1invitroFW (5′-GGATCCTAATACGACTCACTATAGGGAACAGCCACCATGGAACCAACAGAAAAACCATCG-3′) and PHOT1invitroRV (5′-T31CAAAAAACATTTGTTTGCAGATCTTC-3′) for cDNA of PHOT1. Reaction solution was loaded for SDS-PAGE.
Protein Extraction and Fractionation of Crude Microsome Membrane
Etiolated seedlings 3.5 d old were irradiated by 10 μmol·m−2·s−1 blue light for 1 h or were mock-irradiated, and were harvested from agar medium with forceps. The seedlings were frozen in liquid nitrogen, ground with a mortar and pestle, and homogenized in extraction buffer (50 mM Tris-Mes, pH 7.5, 300 mM sucrose, 150 mM NaCl, 10 mM potassium acetate, 5 mM EDTA, and a protease inhibitor mixture [Complete EDTA-free; Roche Diagnostics, Mannheim, Germany]) with 0.5% Triton X-100. The extract was mixed with an equal volume of 2× SDS gel loading buffer. After centrifugation at 10,000g for 10 min to exclude cell debris, the supernatant was used as the total protein fraction.
To prepare a crude extract of microsomal membranes, seedlings were homogenized with extraction buffer without Triton X-100. Cell debris and macro-organelles, such as nuclei and chloroplasts, were excluded by centrifugation two times at 10,000g, 4°C, for 10 min, and then crude microsomal membranes were isolated by centrifugation at 100,000g, 4°C, for 75 min. The microsomal pellet from the high-speed centrifugation was then resuspended in extraction buffer with 0.5% Triton X-100 by pipetting. The supernatant after the high-speed centrifugation was used as the soluble protein fraction.
Immunoprecipitation
The crude microsomal proteins were incubated at 4°C for 2 h with 10 μL of anti-HA antibody–conjugated agarose (Santa Cruz Biotechnology, Santa Cruz, CA). Agarose was collected by centrifugation at 800g, 4°C, for 20 s and washed five times in extraction buffer with 0.5% Triton X-100. Proteins were then eluted from the agarose in 1× SDS gel loading buffer, and they were separated by SDS-PAGE followed by immunoblotting with anti-phot1 antibody. Samples from plants held in the dark were prepared in a darkroom under dim red light.
Antibodies and Immunoblot Analysis
Anti-RPT2 antiserum was produced from a rabbit using 6× His-tagged RPT2 protein as an antigen. Horseradish peroxidase (HRP)–conjugated anti-rabbit IgG antibody was obtained from Amersham Biosciences (Piscataway, NJ). Anti-phot1 antibody, which is an affinity-purified goat polyclonal antibody, and HRP-conjugated anti-goat IgG antibody were obtained from Santa Cruz Biotechnology. HRP-conjugated anti-HA antibody (rat monoclonal antibody, clone 3F10) was obtained from Roche Diagnostics. HRP activity was detected by the ECL Advance protein gel blotting detection kit (Amersham Biosciences) and imaged on the x-ray film (Hyperfilm ECL; Amersham Biosciences). All immunoblot analyses were done twice on independent samples.
RT-PCR Analysis
Total RNA was extracted from the frozen hypocotyl segments of seedlings or mesophyll cell protoplasts or guard cell protoplasts using an RNeasy plant mini kit (Qiagen, Valencia, CA). Four-day-old etiolated seedlings were irradiated for 12 h with unilateral blue light by blue LED lamps at 100 μmol·m−2·s−1. After irradiation, 1-cm apical segments excluding cotyledons were excised from the hypocotyls. Mesophyll cell protoplasts and guard cell protoplasts were isolated enzymatically from 4- to 6-week-old leaves according to previous methods with modifications (Pei et al., 1997; Kinoshita and Shimazaki, 1999). A hundred Arabidopsis rosette leaves were homogenized in a Waring blender in cold (4°C) deionized water twice for 20 s each time at 15,000 rpm and were collected between blendings on a 300-μm nylon mesh. The homogenate was incubated in first-step enzyme solution containing 10 mM Mes-KOH, pH 5.5, 0.1% pectolyase Y-23 (Kikkoman, Tokyo, Japan), 3% cellulase Onozuka R10 (Yakult Pharmaceutical, Tokyo, Japan), 0.05% (w/v) BSA (BSA), 1 mM CaCl2, and 250 mM mannitol for 1 h at room temperature on an orbital shaker (30 rpm). Precipitating mesophyll cell protoplasts were collected and then washed twice in wash buffer containing 1 mM CaCl2 and 500 mM mannitol and suspended in suspension medium containing 5 mM Mes-KOH, pH 6.0, 10 mM KCl, 1 mM CaCl2, and 500 mM mannitol at 4°C. Floating epidermal strips were collected and then incubated in second-step enzyme solution containing 0.05% pectolyase Y-23, 3% cellulase Onozuka R10, 0.05% (w/v) BSA, 1 mM CaCl2, 500 mM mannitol, and 10 mM Mes-KOH, pH 5.5, for 2 h at room temperature on an orbital shaker (15 rpm). Isolated guard cell protoplasts were collected through a 20-μm nylon mesh, washed twice, and suspended in suspension medium at 4°C. The purity of the guard cell protoplast and mesophyll cell protoplast preparations was 83% and 99%, respectively, on a cell number basis.
DNA competitors were designed in sequences derived from GUS genes in pBI121 (Clontech), in which each set of gene-specific primers could be annealed. Three primer sets were used for PCR synthesis of each competitor: 5′-GAAACTTGGGAAGTTAAATCCGGAATCCATCGCAGCGTAATG-3′ and 5′-GCATCAAGAAGGGAACAATAAGCGTGACGCACAGTTCATAGAG-3′ for RPT2 competitor; 5′-TCTTGAGATGACAGAGGATCTGGTGATTACCGACGAAAACG-3′ and 5′-CGTTCCATCTCGGACTAGAAGTGACGCACAGTTCATAGAGA-3′ for NPH3 competitor; and 5′-TCCTACTTTGTGGAATGGATGGAATCCATCGCAGCGTAATG-3′ and 5′-GCTTCAGTGAACTCCATCTCGTGACGCACAGTTCATAGAG-3′ for tubulin gene (TUB) competitor. A graded series from 101 to 107 copies of competitors was used for the analysis. Each total RNA was pretreated with DNase I (TaKaRa Bio, Otsu, Japan), and then cDNA was synthesized by SuperScriptIII RT (Invitrogen, Carlsbad, CA) with three gene-specific primers. PCR was performed with the following primer sets: 5′-GAAACTTGGGAAGTTAAATCC-3′ and 5′-GCATCAAGAAGGGAACAATAAGC-3′ for RPT2; 5′-TCTTGAGATGACAGAGGATCT-3′ and 5′-CGTTCCATCTCGGACTAGAA-3′ for NPH3; and 5′-TCCTACTTTGTGGAATGGAT-3′ and 5′-GCTTCAGTGAACTCCATCTC-3′ for TUB. Sequences of PCR primers for TUB were highly conserved in TUB4, TUB6, and TUB7 genes. The PCR was performed for 40 cycles of 15 s at 94°C, 30 s at 63°C, and 30 s at 72°C. Equal volumes of PCR products were separated in 3% (w/v) agarose gels. We confirmed that each gene was not amplified from genomic DNA by a control experiment without reverse transcriptase. The expected lengths of amplified fragments derived either from RPT2 cDNAs or from each competitor were 0.20 kb and 0.30 kb, respectively. Those of NPH3 were 0.28 kb and 0.36 kb, and those of TUB were 0.22 kb and 0.30 kb, respectively. The gel images were captured and analyzed using an ImageMaster TotalLab (Amersham Biosciences). Copy numbers of cDNAs of RPT2 and NPH3 were calculated against each competitor DNA and normalized to TUB.
Analysis of Expression Pattern Using the GUS Reporter Gene
A 2.5-kb fragment of the RPT2 promoter region in front of the start codon of RPT2 was cloned by PCR amplification with primers 5′-AAGCTTGGTGTGTAGAAATTGCA-3′ and 5′-GGATCCTTTTTTTGGTTCTCTATTGAAG-3′ from Columbia genomic DNA and inserted between the HindIII and BamHI sites of the pBI121 binary vector (Clontech). A transgenic Arabidopsis plant was obtained by the vacuum infiltration method using A. tumefaciens (Bechtold et al., 1993). GUS activity was assayed in leaves from 21-d-old T3 homozygous plants. To observe guard cells without the mesophyll cell background, we peeled epidermal strips off the abaxial side (Figure 6C, Epidermis). Evacuated leaf or epidermal strips were stained in X-Gluc solution containing 1 mM X-Gluc (5-bromo-4-chloro-3-indolyl-β-glucuronide), 100 mM sodium phosphate buffer, pH 7.0, and 0.1% Triton X-100 for 6 h at 37°C. Stained samples were incubated in fixation solution containing 5% formaldehyde, 5% acetic acid, and 20% ethanol for 30 min and then dehydrated in 70% ethanol overnight at room temperature.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AF030864, AF053941, AF181683, and AF180390.
Acknowledgments
We thank the Sainsbury Laboratory at the John Innes Centre for providing the rpt2-2 mutant in the SLAT collection. We also thank Noriaki Kondo and Naoko Asai for valuable advice on measurement of stomatal aperture.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Tatsuya Sakai (tsakai@psc.riken.go.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.019901.
References
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (2001). Current Protocols in Molecular Biology. (New York, NY: John Wiley & Sons).
- Baum, G., Long, J.C., Jenkins, G.I., and Trewavas, A.J. (1999). Stimulation of the blue light phototropic receptor NPH1 causes a transient increase in cytosolic Ca2+. Proc. Natl. Acad. Sci. USA 96, 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis plants. C. R. Acad. Sci. III. Sci. Vie 316, 1194–1199. [Google Scholar]
- Briggs, W.R., and Christie, J.M. (2002). Phototropins 1 and 2: Versatile plant blue-light receptors. Trends Plant Sci. 7, 204–210. [DOI] [PubMed] [Google Scholar]
- Christie, J.M., Reymond, P., Powell, G.K., Bernasconi, P., Raibekas, A.A., Liscum, E., and Briggs, W.R. (1998). Arabidopsis NPH1: A flavoprotein with the properties of a photoreceptor for phototropism. Science 282, 1698–1701. [DOI] [PubMed] [Google Scholar]
- Christie, J.M., Salomon, M., Nozue, K., Wada, M., and Briggs, W.R. (1999). LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): Binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA 96, 8779–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada, A., Sakai, T., and Okada, K. (2003). phot1 and phot2 mediate blue light-induced transient increases in cytosolic Ca2+ differently in Arabidopsis leaves. Proc. Natl. Acad. Sci. USA 100, 8583–8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala, E., Oeller, P.W., Liscum, E., Han, I.-S., Larsen, E., and Briggs, W.R. (1997). Arabidopsis NPH1: A protein kinase with a putative redox-sensing domain. Science 278, 2120–2123. [DOI] [PubMed] [Google Scholar]
- Kagawa, T., Sakai, T., Suetsugu, N., Oikawa, K., Ishiguro, S., Kato, T., Tabata, S., Okada, K., and Wada, M. (2001). Arabidopsis NPL1: A phototropin homolog controlling the chloroplast high-light avoidance response. Science 291, 2138–2141. [DOI] [PubMed] [Google Scholar]
- Kagawa, T., and Wada, M. (2000). Blue light-induced chloroplast relocation in Arabidopsis thaliana as analyzed by microbeam irradiation. Plant Cell Physiol. 41, 84–93. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Doi, M., Suetsugu, N., Kagawa, T., Wada, M., and Shimazaki, K. (2001). phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414, 656–660. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., and Shimazaki, K. (1999). Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J. 18, 5548–5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, E. (2002). Phototropism: Mechanism and outcomes. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0042, http://www.aspb.org/publications/arabidopsis/.
- Liscum, E., and Briggs, W.R. (1996). Mutations of Arabidopsis in potential transduction and response components of the phototropic signaling pathway. Plant Physiol. 112, 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motchoulski, A., and Liscum, E. (1999). Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science 286, 961–964. [DOI] [PubMed] [Google Scholar]
- Okada, K., and Shimura, Y. (1992). Mutational analysis of root gravitropism and phototropism of Arabidopsis thaliana seedlings. Aust. J. Plant Physiol. 19, 439–448. [Google Scholar]
- Okada, K., and Shimura, Y. (1994). Modulation of root growth by physical stimuli. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 665–684.
- Pei, Z.-M., Kuchitsu, K., Ward, J.M., Schwarz, M., and Schroeder, J.I. (1997). Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9, 409–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, T., Kagawa, T., Kasahara, M., Swartz, T.E., Christie, J.M., Briggs, W.R., Wada, M., and Okada, K. (2001). Arabidopsis nph1 and npl1: Blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. USA 98, 6969–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, T., Wada, T., Ishiguro, S., and Okada, K. (2000). RPT2: A signal transducer of the phototropic response in Arabidopsis. Plant Cell 12, 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, K., and Briggs, W.R. (2002). Cellular and subcellular localization of phototropin1. Plant Cell 14, 1723–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, J.I., Raschke, K., and Neher, E. (1987). Voltage dependence of K+ channels in guard-cell protoplasts. Proc. Natl. Acad. Sci. USA 84, 4108–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short, T.W., Reymond, P., and Briggs, W.R. (1993). A pea plasma membrane protein exhibiting blue light–induced phosphorylation retains photosensitivity following triton solubilization. Plant Physiol. 101, 647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoelzle, S., Kagawa, T., Wada, M., Hedrich, R., and Dietrich, P. (2003). Blue light activates calcium-permeable channels in Arabidopsis mesophyll cells via the phototropin signaling pathway. Proc. Natl. Acad. Sci. USA 100, 1456–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, B.L., and Zhulin, I.B. (1999). PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63, 479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier, A.F., Marillonnet, S., Klimyuk, V., Patel, K., Torres, M.A., Murphy, G., and Jones, J.D.G. (1999). Multiple independent defective Suppressor-mutator transposon insertions in Arabidopsis: A tool for functional genomics. Plant Cell 11, 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]