Abstract
Here, we use a loss-of-function approach to demonstrate that the Arabidopsis (Arabidopsis thaliana) mitogen-activated protein kinase (MAPK) MPK6 plays a role in resistance to certain pathogens. MPK6-silenced Arabidopsis showed no apparent morphological phenotype or reduced fertility, indicating MPK6 is not required for development. However, resistances to an avirulent strain of Peronospora parasitica and avirulent and virulent strains of Pseudomonas syringae were compromised, suggesting that MPK6 plays a role in both resistance gene–mediated and basal resistance. Furthermore, this result demonstrates that MPK6's function cannot be fully complemented by other endogenous MAPKs. Although MPK6-silenced plants exhibited enhanced disease susceptibility, their ability to develop systemic acquired resistance or induced systemic resistance was unaffected. Expression of the pathogen-inducible gene VEGETATIVE STORAGE PROTEIN1 (VSP1) in MPK6-silenced plants was severalfold lower than in control plants, but the expression of other defense genes was comparable to the level observed in control plants. Taken together, these results provide direct evidence that a specific MAPK positively regulates VSP1 expression and resistance to a primary infection by certain pathogens, whereas systemic resistance and expression of several other defense genes appears to be mediated either by a functionally redundant MAPK(s) or independently from MPK6-dependent resistance.
INTRODUCTION
Over the past decade, our understanding of how plants activate defense responses after pathogen attack has grown substantially. A large number of resistance (R) genes, whose products are thought to interact directly or indirectly with pathogen avirulence proteins and thereby initiate the defense signaling pathway, have been isolated (Staskawicz et al., 1995; Bent, 1996; Baker et al., 1997; Cooley et al., 2000). Salicylic acid (SA) has been shown to play a critical signaling role in activating local and systemic defense gene expression (Durner et al., 1997; Dempsey et al., 1999). SA also activates the systemic, long-lasting, broad-spectrum resistance known as systemic acquired resistance (SAR) and may regulate the apoptotic-like cell death, known as a hypersensitive response (HR), which occurs at the site of pathogen entry. In addition, a variety of putative defense signaling components have been identified through analysis of mutants with altered defense responses.
With these discoveries has come the realization that the disease resistance signaling pathway in plants shares a number of common elements with those leading to innate immunity in animals and insects (Cohn et al., 2001; Menezes and Jared, 2002; Nurnberger and Brunner, 2002). For example, one class of plant R proteins containing Leu-rich repeats, as well as a critical downstream transducer of the SA signal (NPR1), share homology with components of the innate immunity pathways in Drosophila melanogaster and animals. The antimicrobial proteins activated by these pathways also share homology. Furthermore, protein kinases, including those associated with mitogen-activated protein kinase (MAPK) cascades, have been linked to the resistance signaling pathways of animals and plants (Sessa and Martin, 2000).
In yeast (Saccharomyces cerevisiae) and animals, MAPK cascades are an important part of the signaling machinery that transduces extracellular signals into a wide range of intracellular responses (Madhani and Fink, 1998; Chang and Karin, 2001). This cascade generally involves three functionally linked protein kinases, a MAP kinase kinase kinase (MAPKKK), a MAP kinase kinase (MAPKK), and a MAPK. In response to extracellular stimuli, MAPKKK activates MAPKK via phosphorylation of Ser and Ser/Thr residues within the SXXXS/T motif, where X denotes any amino acid. MAPKK, which is a dual-specificity protein kinase, then activates MAPK by phosphorylating the Thr and Tyr residues within the TXY motif. MAPK then phosphorylates specific effector proteins, which leads to activation of cellular responses. At least four MAPK cascades have been identified in mammals, and at least six have been detected in yeast (Davis, 1994; Herskowitz, 1995; Hirt, 1997; Chang and Karin, 2001).
A growing body of evidence suggests that MAPK cascades also operate in plants. Plant homologs for all three components of this cascade have been identified (Mizoguchi et al., 1993, 1996, 1998; Seo et al., 1995; Ligterink et al., 1997; Zhang and Klessig, 1997; Yang et al., 2001). Furthermore, MAPKs have been implicated in regulating certain aspects of plant growth and development, including cell division, hormone action, and pollen development (Hirt, 2000; Tena et al., 2001; Zhang and Klessig, 2001). Various biotic and/or abiotic stresses also activate plant MAPKs. For example, mechanical stress, drought, and/or cold activate MMK4 of Medicago sativa (alfalfa; Jonak et al., 1996; Bogre et al., 1997) and MPK4 and MPK6 of Arabidopsis (Arabidopsis thaliana; Ichimura et al., 2000). Wounding also activates the SA-induced protein kinase (SIPK) and the wounding-induced protein kinase (WIPK) of Nicotiana tabacum (tobacco; Seo et al., 1995; Zhang and Klessig, 1998b). In addition, SIPK and WIPK are activated by fungal elicitors, infection with avirulent pathogens, and/or SA (Zhang and Klessig, 1997, 1998a, 1998b; Zhang et al., 1998; Romeis et al., 1999). Elicitor treatment also activates MPK6, which is the Arabidopsis ortholog of SIPK (Nuhse et al., 2000; Asai et al., 2002).
Evidence that MAPKs regulate innate immunity in plants has come from several recent studies. In Arabidopsis, transpositional inactivation of the MPK4 gene conferred enhanced disease resistance and constitutive activation of defense responses (Petersen et al., 2000). Based on this phenotype, MPK4 was hypothesized to function as a negative regulator of SAR. Similarly, a portion of a MAPK cascade that positively regulates defense responses was identified in N. tabacum (Yang et al., 2001). Expression of a constitutively active NtMEK2 (a MAPKK) led to the activation of SIPK and WIPK and, subsequently, induced HR-like cell death and defense gene expression. Potato virus X–mediated silencing of all three of these components caused some attenuation of N gene–mediated resistance to Tobacco mosaic virus in N. benthamiana (Jin et al., 2003). Finally, a complete Arabidopsis MAPK cascade, consisting of MEKK1, MKK4/MKK5, and MPK3/ MPK6, that is activated in response to a 22–amino acid peptide derived from bacterial flagellin (flg22) was recently identified (Asai et al., 2002). Transient overexpression of constitutively activated MEKK1, MKK4, or MKK5 in Arabidopsis leaves enhanced resistance to bacterial and fungal pathogens, suggesting that this MAPK cascade plays an important role in signaling defense responses.
Although analyses of plants expressing constitutively active components of the MAPK cascade are highly informative, they must be interpreted with caution; sustained activation of MAPK cascade components may lead to pleiotrophic effects. For example, prolonged activation of MAPKs in Arabidopsis resulted in intracellular H2O2 formation, which preceded cell death (Ren et al., 2002). We therefore used a loss-of-function approach to investigate the role of MPK3 and MPK6 in disease resistance. A reliable virus-induced gene silencing system has not been established for Arabidopsis, and no other Arabidopsis MAPK knockout mutants have been identified (despite substantial efforts by many laboratories, including our own). Thus, we silenced MPK6 expression by generating an intron-containing hairpin loop RNA (ihpRNA); this strategy was previously shown to induce posttranscriptional gene silencing (PTGS) with almost 100% efficiency (Smith et al., 2000; Wesley et al., 2001). Analysis of lines in which MPK6 was silenced revealed that this MAPK is required to maintain basal resistance to a virulent bacterial pathogen and to activate full resistance to avirulent bacterial and oomycete pathogens. However, silencing MPK6 did not affect defense gene expression, SAR, or induced systemic resistance (ISR), although it did reduce the expression of VEGETATIVE STORAGE PROTEIN1 (VSP1). Based on these results, MPK6 appears to play a role in activating both local disease resistance regulated by specific R genes and basal resistance.
RESULTS
Construction of MPK6-Silenced Arabidopsis Lines
Arabidopsis T-DNA knockout lines lacking MPK3 or MPK6 could not be identified in either the α or β T-DNA insertion populations available through the Wisconsin Biotechnology Center. Thus, MPK3- and MPK6-silenced Arabidopsis were constructed by inserting a 418-bp region of MPK3 (spanning a portion of the 5′ untranslated region and coding sequence) and a 393-bp region of MPK6 (encompassing the 5′ coding sequence) into the ihpRNA-forming vector pHannibal (Wesley et al., 2001). In four out of 12 independently transformed lines (ecotype Columbia [Col-0]) containing a single MPK6ihp insertion, MPK6 expression was almost completely silenced (Figure 1A; data not shown). Silencing of MPK3 expression by the MPK3ihp transgene was less effective; only a marginal reduction in mRNA levels was observed in some of the 12 lines tested (data not shown). The MPK6ihp-induced silencing appears to be specific, as MPK3 and MPK4 mRNA levels were not affected in these transgenic plants (Figure 1A). Three MPK6-silenced lines (L1, L4, and L7) were selected for further study. Two MPK3ihp lines that did not show any PTGS (MPK3ihp L4 and L5) were chosen as transgenic control lines and were used along with untransformed Col-0 plants.
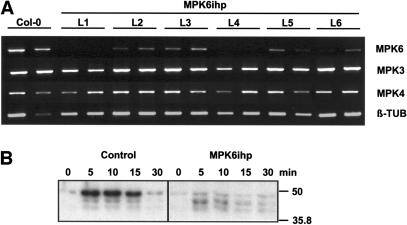
Figure 1.
Analysis of Arabidopsis Seedlings Harboring the MPK6ihp Construct.
(A) RT-PCR analysis of 2-week-old seedlings from Col-0 and MPK6ihp lines for expression of MPK3, MPK4, MPK6, and β-TUB. For each line (L1 to L6) and the Col-0 parent, RNA was extracted from two individual plants as described.
(B) Kinase activity in response to wounding. Leaves of 4-week-old Col-0 and MPK6ihp-expressing plants were wounded, and protein extracts prepared from samples harvested at the indicated times. Kinase activity was monitored with an in-gel kinase assay using MBP as the substrate. Molecular size markers are indicated at right. These experiments were independently performed twice with two control and two MPK6 silenced lines; similar results were observed.
Analysis of MPK6-silenced plants revealed no morphological differences as compared with wild-type Col-0 plants, except for a slight decrease in growth rate. To ensure that the reduced MPK6 mRNA levels corresponded with a loss or reduction in MPK6 activity, in-gel kinase activity assays were performed. In untransformed Col-0 plants, three myelin basic protein (MBP) kinases were activated within 5 min of wounding; their activity remained elevated for 15 min post-wounding (mpw) and then returned to nearly basal levels by 30 mpw. In the MPK6-silenced lines, wounding-induced activation of the highest molecular weight (MW) kinase activity was substantially reduced, arguing that this band corresponds to MPK6 (Figure 1B, right). A low level of kinase activity associated with this band was detected in MPK6-silenced plants, and this activity did not increase after wounding. Thus, the high MW kinase band observed in control plants probably represents more than one kinase activity. The lower MW MBP kinases displayed similar activation kinetics in MPK6-silenced and control plants. However, the amount of kinase activity exhibited by the middle MBP kinase at 5 mpw was twofold to threefold higher than that observed in control plants (Figure 1B). The relative sizes of these kinases (∼42 kD and 44 kD) suggest that they are MPK4 and MPK3, respectively.
Generation of Antibodies to MPK3, MPK4, and MPK6
To confirm that we had specifically silenced MPK6 but not other related MAPKs, polyclonal antibodies were raised against peptides derived from MPK3, MPK4, or MPK6. These peptides, which were selected based on antigenicity and their level of divergence between MAPK family members, correspond to the N-terminal 25 amino acids of MPK6, the C-terminal 14 amino acids of MPK4, and an internal stretch of 20 amino acids of MPK3 (residues 336 to 355). After affinity purification, the specificity and cross-reactivity of these antibodies were assessed using protein gel blot analysis. As shown in Figure 2A, all three antibodies specifically reacted with their cognate recombinant protein but not with the other recombinant MAPKs. The specificity of the different antibodies was further tested by performing immunoprecipitation experiments with in vitro expressed 35S-labeled MAPKs. Each antibody specifically immunoprecipitated its cognate MAPK (data not shown). Immunoprecipitations also were performed with 35S-labeled MPK11, which has an epitope very similar to that of MPK4. The α-C-4 antibody showed some affinity for MPK11, whereas α-C-3 and α-N-6 did not. Taken together, these results indicate that the α-C-3 and α-N-6 antibodies specifically bind their cognate MAPK, whereas α-C-4 binds MPK4 with high affinity and MPK11 with lower affinity.
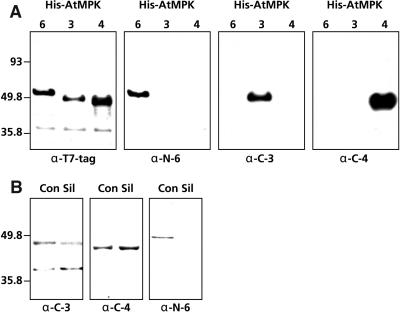
Figure 2.
Specificity of Arabidopsis MAPK Antibodies.
(A) Crude extracts (5 μg/lane) of E. coli-expressed His-tagged versions of MPK3, MPK4, and MPK6 were loaded onto duplicate blots and subjected to protein gel blot analysis with antibodies raised against MPK3 (α-C-3), MPK4 (α-C-4), and MPK6 (α-N-6). To ensure the recombinant proteins were loaded evenly on the gels, the blots were stripped and reprobed with a T7 antibody (α-T7-tag).
(B) Protein gel blot analysis of Arabidopsis MAPKs. Crude protein extracts (20 μg/lane) from Col-0 (Con) or MPK6-silenced (Sil) plants were prepared, loaded onto duplicate blots, and subjected to protein gel blot analysis with the α-C-3, α-C-4, or α-N-6 antibodies. Molecular size markers are indicated at left of the panels in (A) and (B).
The MAPK-specific antibodies were then reacted with protein gel blots containing crude Arabidopsis protein extracts from control and MPK6-silenced lines. The α-C-3 antibody detected two discrete bands in extracts from both control and MPK6-silenced plants (Figure 2B). The size of the upper band (∼44 kD) corresponds well with that predicted for MPK3. The identity of the lower band is undetermined. However, because the predicted sizes for Arabidopsis MAPKs range from 40 to 70 kD (Tena et al., 2001), this protein probably is too small to be a MAPK. The α-C-4 antibody reacted with a single band of ∼42 kD in extracts from control plants; this size is consistent with that predicted for MPK4 (Figure 2B). A band of the same size and intensity also was detected in lanes containing crude protein extracts from MPK6-silenced lines, indicating that the amount of MPK4 protein is unaffected by MPK6ihp transgene expression. The α-N-6 antibody reacted with a single band of ∼49 kD in lanes containing crude Arabidopsis protein extract, which correlates with the expected size for MPK6 (Figure 2B). In lanes containing crude protein extracts from MPK6-silenced plants, no such band was observed. These results show that, despite some cross reactivity of the α-C-3 and α-C-4 antibodies, ihp-induced silencing of MPK6 expression is highly effective and specific.
MPK6 Silenced Plants Are More Susceptible to Pathogen Infection
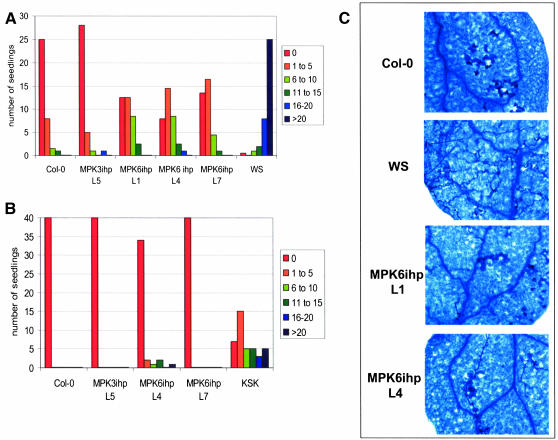
To assess whether disease resistance was affected in the MPK6-silenced plants, their response to inoculation with virulent and avirulent isolates of Peronospora parasitica was monitored. After inoculation with P. parasitica isolate Emwa1, which is avirulent on Col-0, the seedlings of nonsilenced control lines (MPK3ihp) and untransformed Col-0 plants displayed few to no sporangiophores (Figure 3A). By contrast, seedlings from the Wassilewskija ecotype (Ws-0), which are highly susceptible to this isolate, displayed large numbers of sporangiophores. The MPK6-silenced lines exhibited an intermediate level of resistance. Lactophenol trypan blue staining of infected seedlings confirmed that MPK6-silenced lines supported more hyphal growth than the Col-0 control lines but less than Ws-0 plants (Figure 3C). This analysis also revealed that MPK6-silenced plants develop an HR, although it was trailing, rather than the discrete HR exhibited by Col-0 plants. Because resistance to Emwa1 is regulated by RPP4 in Col-0 plants (van der Biezen et al., 2002), our results suggest that MPK6 is required for complete RPP4-mediated resistance. Analysis of MPK6-silenced and nonsilenced control seedlings inoculated with the avirulent Hiks1 isolate of P. parasitica revealed that MPK6 silencing did not affect resistance against this isolate (Figure 3B), which is mediated by RPP7 in Col-0 (Holub et al., 1994). Ecotype Keswick served as the susceptible control. Inoculation of MPK6-silenced seedlings with virulent Noco2 isolate of P. parasitica revealed only a slight increase in susceptibility as compared with the Col-0 control lines (data not shown). Thus, MPK6 appears to be required for full RPP4-mediated resistance but not for RPP7-mediated resistance.
Figure 3.
MPK6-Silenced Lines Exhibit Reduced Resistance to P. parasitica.
(A) Seven- to ten-day-old seedlings were inoculated with P. parasitica Emwa1 (105 spores/mL). At 6 to 8 dpi, single cotyledons from 35 to 40 seedlings were analyzed under the microscope and categorized into one of six categories (0, 1 to 5, 6 to 10, 11 to 15, 16 to 20, or >20) depending on the number of sporangiophores observed. Col-0 is mostly resistant to this isolate, whereas Ws-0 is highly susceptible. MPK3ihp L5 is the nonsilenced control. Experiments were repeated three times with similar results.
(B) Inoculation of Col-0, MPK3ihp L5, and MPK6 silenced lines with P. parasitica Hiks1, which is virulent on the Keswick (Ksk) accession. Inoculation and analysis of 40 to 44 seedlings was done as described in (A).
(C) Trypan blue staining of P. parasitica Emwa1-inoculated cotyledons at 96 hpi. Trypan blue stains dead leaf cells (including xylem cells) and oomycete hyphae.
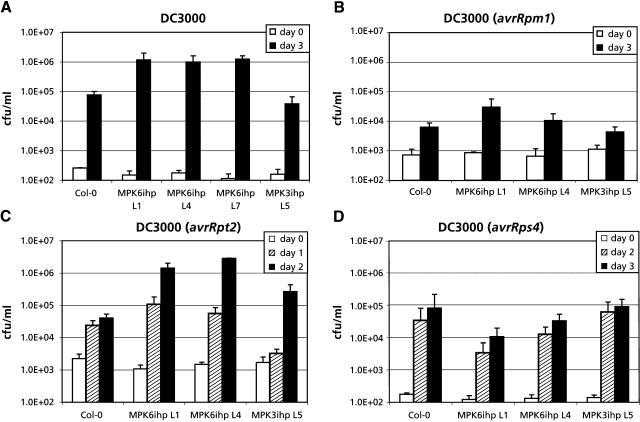
The response of MPK6-silenced plants to virulent and avirulent isolates of Pseudomonas syringae pv tomato was then monitored. By 3 d postinoculation (dpi) with virulent P. s. tomato, MPK6-silenced plants contained at least 10-fold greater titer of bacteria than Col-0 or MPK3ihp control plants (Figure 4A). This result suggests that MPK6 plays a role in maintaining basal resistance against a virulent isolate of P. s. tomato. By comparison, MPK6-silenced and control plants inoculated with avirulent P. s. tomato expressing avrRpm1 contained similar (<10-fold difference) levels of bacteria at 3 dpi (Figure 4B), suggesting that MPK6 is probably not required for the RPM1-mediated resistance signaling pathway. However, inoculation with avirulent P. s. tomato expressing avrRpt2 resulted in significantly higher bacterial titers in the leaves of MPK6-silenced plants as compared with Col-0 or the nonsilenced control lines (Figure 4C). The RPS2-mediated resistance response was not completely compromised because infected leaves of both control and MPK6-silenced lines exhibited tissue collapse and necrosis, indicative of an HR. Inoculation with avirulent P. s. tomato expressing avrRps4 resulted in similar levels of bacteria at 3 dpi in MPK6-silenced and control plants (Figure 4D). Note that the slightly lower levels of P. s. tomato growth in MPK6ihp L4 and particularly in MPK6ihp L1 compared with controls in Figure 4D were not observed in a duplicate experiment. Taken together, these results indicate that MPK6 is required for maintaining basal resistance to this bacterial pathogen and plays a role in RPS2-mediated resistance.
Figure 4.
MPK6-Silenced Lines Display Enhanced Susceptibility to P. s. tomato.
(A) Four-week-old plants were inoculated with P. s. tomato, and four leaf disks per sample were harvested at 0 dpi and 3 dpi. The number of colony forming units (cfu) per sample was calculated and expressed as cfu/mL. Col-0 and MPK3ihp L5 were used as controls, along with the MPK6ihp L1, L4, and L7 lines. Experiments were done twice with similar results.
(B) Four-week-old plants were inoculated with P. s. tomato expressing avrRpm1, and four leaf disks per sample were harvested at 0 dpi and 3 dpi.
(C) Four-week-old plants were inoculated with P. s. tomato expressing avrRpt2, and four leaf disks per sample were harvested at 0, 1, and 2 dpi.
(D) Four-week-old plants were inoculated with P. s. tomato expressing avrRps4, and four leaf disks per sample were harvested at 0, 2, and 3 dpi.
MPK6 Silencing Alters VSP1 Gene Expression
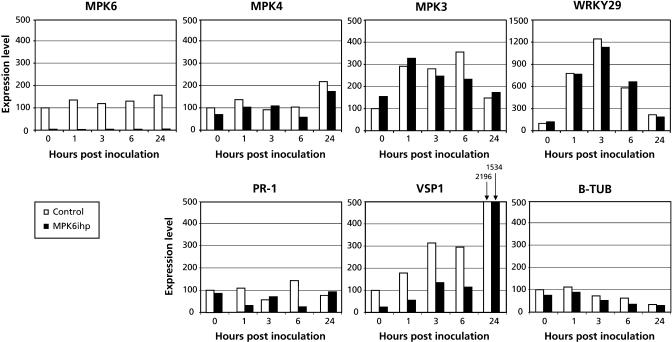
To determine whether MPK6 plays a role in regulating defense gene expression, transcript levels for PR-1, VSP1, Eli3-2, and the putative regulatory factor WRKY29 were monitored by quantitative PCR (Q-PCR) in MPK6-silenced and control plants inoculated with virulent P. s. tomato. In the control lines, MPK3 and WRKY29 were rapidly induced after P. s. tomato inoculation, whereas transcripts for VSP1 and Eli3-2 accumulated more gradually (Figure 5; data not shown). There was little difference in steady state mRNA levels between P. s. tomato–inoculated MPK6-silenced lines and control lines for all of these genes, except VSP1 and possibly PR-1. VSP1 expression levels in the MPK6-silenced lines were approximately threefold lower than in control lines before infection. Upon inoculation the expression of VSP1 was induced similarly in control and MPK6-silenced lines, but mRNA levels remained approximately threefold lower up to 6 h postinoculation (hpi). Expression of MPK4 and PR-1 exhibited little induction by P. s. tomato inoculation above their basal levels in both sets of plants (Figure 5). By contrast, PR-1 expression in MPK6-silenced plants appears to be reduced after infection with virulent P. s. tomato. However, because this reduction was quite variable among experiments and even time points, its significance is unclear. These results suggest that MPK6 activity affects the basal level of VSP1 transcripts but is not required or is functionally redundant for inducing this gene after pathogen challenge.
Figure 5.
Pathogen-Induced Gene Expression in MPK6-Silenced Plants.
Four-week-old MPK6-silenced and control plants were dipped in virulent P. s. tomato suspension (108 cfu/mL), and samples were taken at selected time points between 0 hpi and 24 hpi. Samples were snap frozen in liquid nitrogen, and RNA extraction and Q-PCR were performed as described in Methods. Equal amounts of RT reaction products were used as templates to amplify selected regions of the indicated genes with specific primer pairs. Expression levels are represented by arbitrary units, which were set at 100 for control line at t = 0. The expression level of all other samples were related to the control sample t = 0. These experiments were done with two controls (Col-0 and MPK3ihp L5) and two MPK6-silenced lines (MPK6ihp L1 and L4), and averages of both lines are shown.
SAR and ISR Are Not Affected by MPK6 Silencing
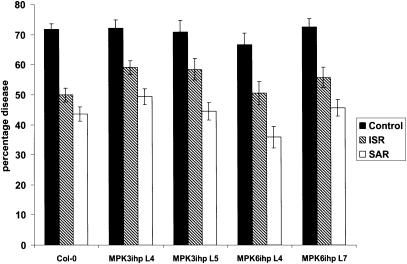
Because MPK6-silenced plants exhibited enhanced susceptibility to certain virulent and avirulent pathogens, we tested their ability to develop SAR or an SA-independent ISR. SAR was induced by inoculating three lower leaves with avirulent P. s. tomato carrying avrRpt2 3 d before challenge inoculation with virulent P. s. tomato, whereas ISR was induced by growing plants in soil containing P. fluorescens WCS 417r before P. s. tomato inoculation. Control plants received no treatment before P. s. tomato inoculation. As expected, wild-type Col-0 plants that received an SAR- or ISR-inducing treatment developed substantially fewer disease symptoms after inoculation with virulent P. s. tomato than untreated control plants (Figure 6). A similar reduction in disease symptom levels was observed in SAR- or ISR-induced nonsilenced MPK3ihp and MPK6-silenced lines. Thus, MPK6 either is not required for SAR or ISR development, or a functionally redundant MAPK participates in these responses in the silenced plants.
Figure 6.
Induced/Acquired Resistance Is Intact in MPK6-Silenced Lines.
Percentage diseased leaves at 3 dpi with virulent P. s. tomato in ISR- and SAR-expressing plants and control plants. The effectiveness of ISR and SAR expression was tested in MPK6-silenced lines (MPK6ihp4 and MPK6ihp7) and compared with nonsilenced transgenic lines (MPK3ihp4 and MPK3ihp5) and the parent Columbia (Col-0). Error bars indicated SE (n = 18).
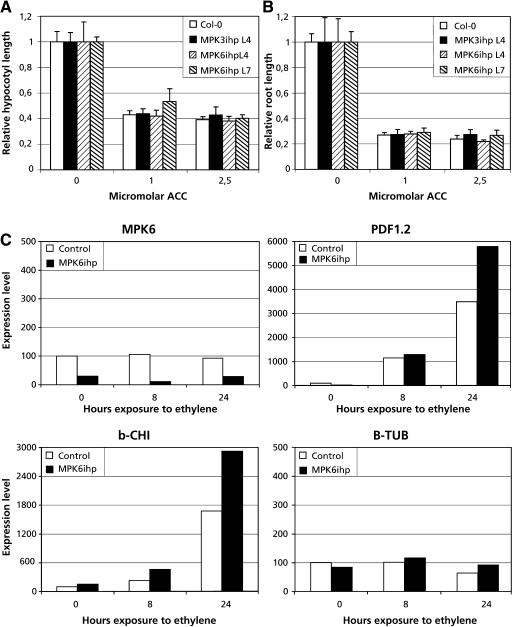
Because development of ISR requires a functional ethylene signaling pathway (Pieterse et al., 1998) and ISR developed normally in MPK6-silenced plants, it appears that transduction of the ethylene signal does not require MPK6. However, recently MPK6 has been implicated in ethylene signaling because overexpression of an MAPK kinase led to constitutive MPK6 activation and elevated expression of several ethylene-inducible genes (Ouaked et al., 2003). We therefore tested ethylene-inducible responses in MPK6-silenced lines. Consistent with our observation on ISR in MPK6-silenced lines, ethylene-induced expression of the defense genes PDF1.2 and basic Chitinase (b-CHI) was similar in MPK6-silenced lines as compared with control lines (Figure 7C). Although induction appeared to be slightly higher in the MPK6-silenced plants, the very modest difference of less than twofold is within the standard error for this type of experiment and, thus, not considered significant. Furthermore, etiolated seedlings of MPK6-silenced and control lines also displayed similar shortening of the hypocotyl and roots in response to the ethylene precursor 1-aminocyclopropane-1-carboxylate (Figures 7A and 7B). Our data suggest that MPK6 is not required for these ethylene responses and together with the results of Ouaked et al. (2003) suggest that MPK6 may be functionally redundant in ethylene signaling.
Figure 7.
Ethylene Responses in MPK6-Silenced Lines.
(A) and (B) Seeds for MPK6-silenced lines and control lines were germinated in the dark on MS medium containing the indicated amount of 1-aminocyclopropane-1-carboxylic acid (ACC). Four days after germination, the hypocotyl and root lengths of the etiolated seedlings was measured.
(C) Q-PCR analysis of ethylene-inducible gene expression in control and MPK6-silenced lines. Two-week-old plants were exposed to 5 ppm ethylene, and samples harvested and snap frozen in liquid nitrogen at indicated times. Q-PCR was performed as described in Figure 5. b-CHI, basic Chitinase; B-TUB, β-Tubulin.
DISCUSSION
Here, we report the successful use of ihpRNA-induced PTGS to silence the Arabidopsis MAPK MPK6. All 12 independent transgenic Arabidopsis lines expressing the MPK6ihp construct showed moderate to high levels of silencing; in four lines, MPK6 expression was almost completely suppressed. Despite the loss of MPK6 expression, these lines did not show any major alteration in morphological phenotype. This finding could indicate that MPK6 does not play a major role in regulating developmental processes. Alternatively, because the Arabidopsis genome contains 23 genes encoding MAPK-like proteins (Tena et al., 2001), some of the effect(s) of silencing MPK6 could be masked by other functionally redundant MAPKs.
Despite the potential for functional redundancy, the fact that MPK6-silenced plants exhibited several differences from wild-type plants indicates that at least certain functions cannot be complemented by other MAPKs. For example, the growth rate of MPK6-silenced plants was slightly slower than that of wild-type plants. In addition, the MAPK activation kinetics after wounding differed between MPK6-silenced and wild-type plants. In wild-type plants, a 49-kD kinase, subsequently identified as MPK6, was rapidly activated by wounding, whereas a 44-kD and a 42-kD kinase, which likely represent MPK3 and MPK4, were activated to much lower levels. Similar to our results, Ichimura et al. (2000) demonstrated that a variety of abiotic stresses, including wounding, activate MPK6, MPK4, and an MBP kinase of 44 kD. In the MPK6-silenced lines, no MPK6 activity was detected before or after wounding, and the level of 42 kD activity was comparable to that observed in control plants. However, the level of 44 kD activity was twofold to threefold higher. One possible explanation for this result is that MPK6 is required to attenuate the 44 kD activity after wounding. In N. tabacum, silencing expression of SIPK (orthologous to MPK6) led to increased and sustained activation of WIPK (orthologous to MPK3) in response to prolonged ozone treatment (Samuel and Ellis, 2002). Additional evidence that SIPK may regulate WIPK protein was reported by Liu et al. (2003), who showed that SIPK-dependent phosphorylation positively regulates WIPK transcription and protein accumulation. The lack of sustained 44-kD MAPK activation in wounded MPK6-silenced plants, however, suggests that the Arabidopsis MAPKs are regulated differently from their N. tabacum orthologs.
An alternative explanation for the elevated levels of 44 kD activity detected in wounded MPK6-silenced plants is that MPK6 and the 44-kD MAPK are partially redundant for mediating wounding responses. The 44 kD activity might therefore be elevated in the absence of MPK6 to take over some of MPK6's functions. Support for this possibility comes from the observation that MPK3 and MPK6, which are members of the same MAPK subfamily, are activated with similar kinetics by diverse stimuli, including wounding and flg22 (F.L.H. Menke and D.F. Klessig, unpublished data). Furthermore, MPK3 and MPK6 are both activated by the MAPKKs MKK4 and MKK5, which suggests that they belong to the same flg22-induced MAPK cascade (Asai et al., 2002). Although the 42-kD kinase also is activated by wounding, it does not appear to share functional redundancy with MPK6; its activity level in wounded plants is unaffected by the loss of MPK6. Because activation of MPK4, which is predicted to be 42 kD, has been linked to the MAPKK MKK1 (Mizoguchi et al., 1998; Huang et al., 2000; Matsuoka et al., 2002), wounding may activate MPK4 via a MAPK cascade unrelated to that associated with MPK3 and MPK6.
The most striking difference between MPK6-silenced and wild-type plants is that silenced plants displayed increased susceptibility to certain virulent and avirulent pathogens. Silencing of MPK6 resulted in enhanced susceptibility to a virulent and an avirulent isolate of P. s. tomato and reduced resistance to the avirulent Emwa1 isolate of P. parasitica. However, loss of MPK6 did not completely abolish resistance to Emwa1 or to P. s. tomato expressing avrRpt2. Growth of P. parasitica Emwa1 still triggered a trailing necrosis in MPK6-silenced plants, and these plants supported less sporulation than susceptible Ws-0 plants, whereas P. s. tomato carrying avrRpt2 still induced an HR in the MPK6-silenced plants. Because double silenced lines lacking MPK3 as well as MPK6 might exhibit a greater reduction in resistance to P. parasitica, we cannot exclude the possibility that MPK6's function is at least partially redundant. However, our results argue that MPK6 is required for full RPP4-mediated resistance to Emwa1 and plays a role in RPS2-mediated resistance against P. s. tomato expressing avrRpt2. Consistent with these results, the N. tabacum MPK6 ortholog SIPK was recently linked to N gene–mediated resistance to Tobacco mosaic virus (Jin et al., 2002). By contrast, resistance to avirulent P. syringae carrying AvrRpm1 or avrRps4 was affected very little in MPK6-silenced plants, and RPP7-mediated resistance to P. parasitica Hiks1 was not compromised in these silenced lines. Thus, MPK6 may not be required for RPM1-, RPS4-, or RPP7-mediated resistance, or it may be functionally redundant with another MAPK for activating these responses. Supporting the possibility that MPK6 is required for specific resistance pathways (such as RPS2 and RPP4 but not RPM1, RPS4, or RPP7) is the demonstration that NPK1-silenced N. tabacum exhibit reduced resistance to pathogens recognized by the N, Bs2, and Rx resistance genes but not the Pto or Cf4 resistance genes (Jin et al., 2002). Genetic evidence suggests that there are two major pathways for resistance signaling. Resistance mediated by the Toll-interleukin-1 receptor-nucleotide-binding site-Leu-rich repeat (TIR-NBS-LRR) class of R genes involves the pathway containing EDS1, whereas the coiled coil-nucleotide-binding site-Leu-rich repeat (CC-NBS-LRR) class of R genes generally used the pathway containing NDR1 (Aarts et al., 1998). Silencing of MPK6 affects resistance mediated by some members of both the TIR-NBS-LRR (RPP4) and CC-NBS-LRR (RPS2) classes but not other members (i.e., RPS4 of the TIR-NBS-LRR class, RPM1 of the CC-NBS-LRR class, and RPP7, a non-TIR type R gene [Tor et al., 2002; Muskett and Parker, 2003]). Therefore, R gene class is not the critical factor determining whether MPK6 silencing compromises resistance mediated by a particular R gene. Rather, it may be the strength of the defense signal(s) initiated by the R gene. Both RPS2 and RPP4 provide relatively weak resistance to P. s. tomato and P. parasitica, respectively (Van der Biezen et al., 2002; Tao et al., 2003), and this resistance was compromised in MPK6-silenced plants. Taken together, our results confirm the findings of Asai et al. (2002) and establish a direct link between MPK6 and the positive regulation of both basal and R gene–mediated resistance.
Two Arabidopsis mutants, dth9 and phx3, that display enhanced susceptibility to P. syringae and P. parasitica but normal induction of defense-related genes were previously identified (Morel and Dangl, 1999; Mayda et al., 2000). The similarities to the MPK6-silenced phenotype suggested that these mutations might reside within MPK6. However, although dth9 and MPK6 both map to chromosome 2, dth9 is linked to the SSLP marker Nga1126, which is ∼33 centimorgans from MPK6; phx3 was mapped to chromosome 5. Van der Biezen et al. (2002) recently reported that RPP4-mediated resistance requires 12 downstream defense components, including DTH9. Based on our results, it is possible that MPK6 also belongs on this list. But because MPK6-silenced plants expressed normal SAR and ISR, we can only conclude that these plants are compromised in local disease resistance.
How might MPK6 affect disease resistance? One clue may be provided by the observation that expression of VSP1 was approximately threefold lower in MPK6-silenced versus control lines. Ellis and Turner (2001) recently showed that the cev1 mutant, which exhibits activated ethylene- and jasmonate-dependent defense responses, has both enhanced VSP1 expression and fungal resistance. This correlation between VSP1 expression and resistance is similar to that seen in MPK6-silenced lines and suggests that VSP1 may play a role in resistance. Presumably MPK6 also regulates expression of other, yet to be identified genes that play a role in resistance.
In summary, we have used a loss-of-function approach to demonstrate that MPK6 is required for resistance to some virulent and avirulent strains of P. s. tomato and is partially required for RPP4-mediated resistance to P. parasitica Emwa1. Because SAR and ISR were unaffected in pathogen-infected MPK6-silenced plants, our results suggest that these responses are regulated by a pathway(s) that either is independent of MPK6 or is complemented by a functionally redundant MAPK in MPK6-suppressed plants. Future analysis with plants in which more than one MAPK has been silenced should provide greater insight into how MPK6, and potentially other MAPKs, regulate disease resistance in plants.
METHODS
Arabidopsis Growth and Transformation
Seeds for soil-grown plants were sown on autoclaved moist soil and vernalized overnight at 4°C. Plants were germinated and grown under a 16-h-light/8-h-dark regime at 22°C and 70% relative humidity.
Construction of ihp Constructs
A 418-bp region of MPK3 (spanning a portion of the 5′ untranslated region and coding sequence) and a 393-bp region of MPK6 (encompassing the 5′ coding sequence) were inserted into the pHannibal vector and mobilized into the corresponding binary vector pART27 (Wesley et al., 2001). Four-week-old Arabidopsis plants (Col-0) were transformed by the floral dip method as described before (Cooley et al., 2000). Seeds were surface-sterilized with 1% hypochlorite and 0.05% Tween 20 and washed three times with sterile water. Seeds were spread on solid MS medium with 1% sucrose and vernalized 2 d at 4°C. Germination and the first 10 d of growth occurred under the 16-h light condition at 22°C in Petri dishes containing kanamycin (100 mg/L), after which resistant seedlings were transferred to soil. The kanamycin-resistant lines were assessed at 2 weeks old for silencing of MPK3 or MPK6 by RT-PCR.
RNA Isolation and RT-PCR and Q-PCR
Tissue samples were frozen in liquid nitrogen and stored at −80°C. RNA was extracted using Trizol (Invitrogen, Carlsbad, CA) according to manufacturer's instructions. RNA samples were incubated with 1 unit of DNase (Promega, Madison, WI)/50μL at 37°C for 30 min. DNase-treated RNA was extracted with Trizol as before. RT reactions were done with 1 μg of RNA and 0.5 μg of oligo(dT)18 primer at 42°C with 0.1 unit of RT (Promega) and 2 units of RNasin (Promega) for 1 h in 25-μL reactions. Aliquots of the resulting RT reaction product were used as template for RT-PCR and Q-PCR analysis. For RT-PCR, amplification of cDNA was performed with 0.5 μL of RT reaction product in 20 μL with 10 pmol of each primer, 200 pmol deoxynucleotide triphosphate, 0.5 unit of Taq polymerase in a thermal cycler (Peltier; MJ Research, Watertown, MA) for 1 cycle at 94°C for 2 min 30 s, followed by 30 s at 94°C, 30 s at 60°C, and 1 min 30 s at 72°C for 22 to 30 cycles, ending with 2 min 30 s at 72°C (Kang and Singh, 2000). Primers for amplification reactions for β-Tubulin (β-TUB) were described previously (Kang and Singh, 2000), while the following were used: for MPK3 forward, 5′-atacggagacgaacgagctag-3′; MPK3 reverse, 5′-cgcatttgatgaacatggtct-3′; MPK4 forward, 5′-atgtcggcggagagttgtttcg-3′; MPK4 reverse, 5′-tcacactgagtcttgaggattg-3′; MPK6 forward, 5′-atggacggtggttcaggtcaa-3′; and MPK6 reverse, 5′-ttgaaagcaagcgcctcgcgg-3′. Q-PCR was performed using the SYBR Green protocol (Applied Biosystems, Foster City, CA) with 100 nM primers and 0.5-μL aliquot of RT reaction product in a total of 50 μL per reaction. Reactions were run and analyzed on ABI Prism 7700 (Applied Biosystems) according to the manufacturer's instructions. For each reaction, the threshold cycle value (Ct) was determined by setting the threshold within the logarithmic amplification phase. A standard curve was made by determining the Ct values for a dilution series of the RT reaction product for each primer pair. Using this standard curve, the relative quantification for each reaction was calculated from its Ct value because there is a linear relation between Ct value and log (amount RT reaction product; User Bulletin 2, ABI Prism 7700 sequence detection system). The relative quantity for the control sample at t = 0 was arbitrarily set to 100. All other samples were expressed in relation to this control sample by a conversion with the same factor used for the control sample at t = 0. Q-PCR reactions were done in triplicate and averaged for each line individually. Primers were for MPK6 (At2g43790) forward, 5′-accaccaccaacctcaaaag-3′; MPK6 reverse, 5′-cctccaggagcttctgtcat-3′; MPK4 (At4g01370) forward, 5′-cgttgtgccacccatattt-3′; MPK4 reverse, 5′-aaaattgaacggcctcacac-3′; MPK3 (At3g45640) forward, 5′-ccaagaagccatagcactca-3′; MPK3 reverse, 5′-agccattcggatggttattg-3′; WRKY29 (At4g23550) forward, 5′-cccaaaccaaccttaaccaa-3′; WRKY29 reverse, 5′-atcagcggatgggatcatag-3′; PR-1 (At2g19990) forward, 5′-aagaaacgctcgtggttcac-3′; PR-1 reverse, 5′-tctgtgcataggtcgcaaga-3′; VSP1(At5g24780) forward, 5′-cgtggttagagtccggagaa-3′; VSP1 reverse, 5′-atcccgagttccaagaggtt-3′; ELI3.2 (At4g37990) forward, 5′-gacagctgcaccgaaggtat-3′; ELI3.2 reverse, 5′-tacgggaatccgtacgtttg-3′; PDF1.2 (At5g44420) forward, 5′-gcacagaagttgtgcgagaa-3′; PDF1.2 reverse, 5′-tgtttggctccttcaaggtt-3′; b-CHI (At3g12500) forward, 5′-ggtctatgctgcagcgagtt-3′; b-CHI reverse, 5′-ggctgcttacagtatggttcg-3′; β-TUB (At2g29550) forward, 5′-ggaagaagctgagtacgagca-3′; and β-TUB reverse, 5′-gcaactggaagttgaggtgtt-3′.
Antibody Production and Protein Gel Blot Analysis
MPK3, MPK4, and MPK6 were subcloned into pET28a (Novagen, Madison, WI) downstream of the EcoRI restriction site to produce N-terminal fusion proteins containing both a His tag and T7 tag. Fusion proteins were expressed in Escherichia coli strain BL21 (DE3; Novagen) as instructed by the manufacturer. 35S-Met–labeled His- and T7-tag fusions of MPK3, MPK4, and MPK6 were made by coupled in vitro transcription translation using TNT coupled rabbit reticulocyte lysate system (Promega) according to the manufacturer's instructions, using pET28a constructs as a DNA template. Polyclonal member specific antibodies were raised in rabbits against peptides of MPK3 (NH2-QKPFSFEFEQQPLDEEQIK-COOH), MPK4 (NH2-ELIYRETVKFNPQDSV-COOH), and MPK6 (NH2-MDGGSGQPAADTEMTEAPGGFPAAAPS-COOH) by Zymed (San Francisco, CA) and affinity purified. Protein gel blot analysis was performed using duplicate blots, in which equal amounts of crude extract from E. coli expressing the different recombinant T7-tagged AtMPKs were separated on 10% SDS-PAGE and transferred to nitrocellulose (Schleicher and Schuell, Keene, NH) by wet transfer. Blots were blocked in Tris-buffered saline with Tween 20 (TTBS; 25 mM Tris, pH 7.5, 150 mM NaCl, and 0.05% Tween 20) and 5% (w/v) nonfat dry milk. Antibodies were diluted in TTBS with 3% (w/v) BSA (Sigma, St. Louis, MO) to 1 μg/mL of α-C-4 and α-N-6 or 5 μg/mL of α-C-3. Protein gel blots were incubated with diluted antibody for 1 h at room temperature. Blots were washed four times for 10 min in TTBS and subsequently incubated with anti-rabbit horseradish peroxidase conjugate diluted 1:50,000 in TTBS with 3% BSA. Blots were washed twice with TTBS and twice with Tris-buffered saline. Immunocomplexes were visualized with the Western lightning chemiluminescence kit (DuPont–New England Nuclear Life Science Products, Boston, MA) according to manufacturer's instructions.
In-Gel Kinase Assay and Immunocomplex Kinase Assay
In-gel kinase assays and immunocomplex kinase assays were performed as described previously (Zhang et al., 1998; Zhang and Klessig, 1998b).
Wounding and Ethylene Treatment
For wounding, leaves were cut into 0.5-cm strips and floated on water. At the indicated times, samples were collected and snap frozen in liquid nitrogen. Ethylene was applied to 2-week-old plants in a controlled chamber constantly flushed with air containing 5 ppm ethylene. At the indicated times, samples were collected and snap frozen in liquid nitrogen.
Pathogen Inoculation and Analysis of Resistance Status
P. parasitica isolates were maintained as described before (Cooley et al., 2000). Inoculation of P. parasitica was done on 7- to 10-d-old seedlings by spraying with 105 spores/mL. Sporangiophore formation was evaluated on single cotelydons of 35 to 40 individual seedlings at 6 to 8 dpi with a dissecting microscope. Lactophenol trypan blue staining was described before (Cooley et al., 2000). For resistance tests, P. s. tomato was inoculated at a density of 104 colony-forming units (cfu)/mL by infiltrating the abaxial leaf surface with a 1-mL syringe. P. s. tomato carrying avrRpm1, avrRpt2, or avrRps4 were inoculated at a density of 105 cfu/mL. Four leaf disks of 0.6 cm were randomly taken per sample from four individual plants, and four samples were taken from each line at indicated times. Leaf disks were ground in 10 mM MgCl2, and dilutions were plated in Kings B medium with 40 μg/mL of rifampicin and 50 μg/mL of kanamycin and incubated at 27°C for 48 h, after which the number of colonies formed were counted. At least two independent experiments were done for each strain of P. syringae. For P. s. tomato–induced gene expression, plants were incubated for 48 h under high humidity and then dipped in a suspension of virulent P. s. tomato (108 cfu/mL) containing 0.015% swilet. At indicated times, leaf tissue was frozen in liquid nitrogen. Expression of ISR and SAR were tested as described before (Pieterse et al., 1996).
Acknowledgments
We thank Joseph Kuhl and Pradeep Kachroo for helpful discussions and useful comments and D'Maris Dempsey for critical reading of the manuscript. We also thank Eric Holub for providing us with the P. parasitica Hiks1 isolate and Jonathan Jones for providing us with P. s. tomato expressing avrRps4. F.L.H.M. was supported by a long-term fellowship from the Human Frontier Science Program Organization. This work was supported in part by Grant MCB-0110404 from the National Science Foundation to D.F.K.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Daniel F. Klessig (dfk8@cornell.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.015552.
References
- Aarts, N., Metz, M., Holub, E.B., Staskawicz, B.J., Daniels, M.J., and Parker, J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene mediated pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Baker, B., Zambryski, P., Staskawicz, B., and Dinesh-Kumar, S.P. (1997). Signaling in plant-microbe interactions. Science 276, 726–733. [DOI] [PubMed] [Google Scholar]
- Bent, A.F. (1996). Plant disease resistance genes: Function meets structure. Plant Cell 8, 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogre, L., Ligterink, W., Meskiene, I., Barker, P.J., Heberle-Bors, E., Huskisson, N.S., and Hirt, H. (1997). Wounding induces the rapid and transient activation of a specific MAP kinase pathway. Plant Cell 9, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, L.F., and Karin, M. (2001). Mammalian MAP kinase signalling cascades. Nature 410, 37–40. [DOI] [PubMed] [Google Scholar]
- Cohn, J., Sessa, G., and Martin, G.B. (2001). Innate immunity in plants. Curr. Opin. Immunol. 13, 55–62. [DOI] [PubMed] [Google Scholar]
- Cooley, M.B., Pathirana, S., Wu, H.-J., Kachroo, P., and Klessig, D.F. (2000). Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12, 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, R.J. (1994). MAPKs: New JNK expands the group. Trends Biochem. Sci. 19, 470–473. [DOI] [PubMed] [Google Scholar]
- Dempsey, D.A., Shah, J., and Klessig, D.F. (1999). Salicylic acid and disease resistance in plants. CRC Crit. Rev. Plant Sci. 18, 547–575. [Google Scholar]
- Durner, J., Shah, J., and Klessig, D.F. (1997). Salicylic acid and disease resistance in plants. Trends Plant Sci. 2, 266–274. [Google Scholar]
- Ellis, C., and Turner, J.G. (2001). The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13, 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz, I. (1995). Map kinase pathways in yeast: For mating and more. Cell 80, 187–197. [DOI] [PubMed] [Google Scholar]
- Hirt, H. (1997). Multiple roles of MAP kinases in plant signal transduction. Trends Plant Sci. 2, 11–15. [Google Scholar]
- Hirt, H. (2000). Connecting oxidative stress, auxin, and cell cycle regulation through a plant mitogen-activated protein kinase pathway. Proc. Natl. Acad. Sci. USA 97, 2405–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holub, E.B., Beynon, J.L., and Crute, I.R. (1994). Phenotypic and genotypic characterization of interactions between isolates of Peronospora parasitica and accessions of Arabidopsis thaliana. Mol. Plant Microbe Interact. 7, 223–239. [Google Scholar]
- Huang, Y.F., Li, H., Gupta, R., Morris, P.C., Luan, S., and Kieber, J.J. (2000). ATMPK4, an arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiol. 122, 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura, K., Mizoguchi, T., Yoshida, R., Yuasa, T., and Shinozaki, K. (2000). Various abiotic stresses vapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 24, 655–665. [DOI] [PubMed] [Google Scholar]
- Jin, H., Axtell, M.J., Dahlbeck, D., Ekwenna, O., Zhang, S., Staskawicz, B., and Baker, B. (2002). NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev. Cell 3, 291–297. [DOI] [PubMed] [Google Scholar]
- Jonak, C., Kiegerl, S., Ligterink, W., Barker, P.J., Huskisson, N.S., and Hirt, H. (1996). Stress signaling in plants: A mitogen-activated protein kinase pathway is activated by cold and drought. Proc. Natl. Acad. Sci. USA 93, 11274–11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H.G., and Singh, K.B. (2000). Characterization of salicylic acid-responsive, Arabidopsis Dof domain proteins: Overexpression of OBP3 leads to growth defects. Plant J. 21, 329–339. [DOI] [PubMed] [Google Scholar]
- Ligterink, W., Kroj, T., zurNieden, U., Hirt, H., and Scheel, D. (1997). Receptor-mediated activation of a MAP kinase in pathogen defense of plants. Science 276, 2054–2057. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Jin, H., Yang, K., Kim, C., Baker, B., and Zhang, S. (2003). Interaction between two mitogen-activated protein kinases during tobacco defense signaling. Plant J. 34, 149–160. [DOI] [PubMed] [Google Scholar]
- Madhani, H.D., and Fink, G.R. (1998). The riddle of MAP kinase signaling specificity. Trends Genet. 14, 151–155. [DOI] [PubMed] [Google Scholar]
- Matsuoka, D., Nanmori, T., Sato, K., Fukami, Y., Kikkawa, U., and Yasuda, T. (2002). Activation of AtMEK1, an Arabidopsis mitogen-activated protein kinase kinase, in vitro and in vivo: Analysis of active mutants expressed in E. coli and generation of the active form in stress response in seedlings. Plant J. 29, 637–647. [DOI] [PubMed] [Google Scholar]
- Mayda, E., Mauch-Mani, B., and Vera, P. (2000). Arabidopsis dth9 mutation identifies a gene involved in regulating disease susceptibility without affecting salicylic acid-dependent responses. Plant Cell 12, 2119–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes, H., and Jared, C. (2002). Immunity in plants and animals: Common ends through different means using similar tools. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 132, 1–7. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Hayashida, N., Yamaguchishinozaki, K., Kamada, H., and Shinozaki, K. (1993). ATMPKs: A gene family of plant MAP kinases in Arabidopsis thaliana. FEBS Lett. 336, 440–444. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Ichimura, K., Irie, K., Morris, P., Giraudat, J., Matsumoto, K., and Shinozaki, K. (1998). Identification of a possible MAP kinase cascade in Arabidopsis thaliana based on pairwise yeast two-hybrid analysis and functional complementation tests of yeast mutants. FEBS Lett. 437, 56–60. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Irie, K., Hirayama, T., Hayashida, N., Yamaguchi-Shinozaki, K., Matsumoto, K., and Shinozaki, K. (1996). A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93, 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, J.B., and Dangl, J.L. (1999). Suppressors of the Arabidopsis lsd5 cell death mutation identify genes involved in regulating disease resistance responses. Genetics 151, 305–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskett, P., and Parker, J. (2003). Role of SGT1 in the regulation of plant R gene signalling. Microbes Infect. 5, 969–976. [DOI] [PubMed] [Google Scholar]
- Nuhse, T.S., Peck, S.C., Hirt, H., and Boller, T. (2000). Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J. Biol. Chem. 275, 7521–7526. [DOI] [PubMed] [Google Scholar]
- Nurnberger, T., and Brunner, F. (2002). Innate immunity in plants and animals: Emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 5, 318–324. [DOI] [PubMed] [Google Scholar]
- Ouaked, F., Rozhon, W., Lecourieux, D., and Hirt, H. (2003). A MAPK pathway mediates ethylene signaling in plants. EMBO J. 22, 1282–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., et al. (2000). Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J., van Wees, S.C.M., Hoffland, E., van Pelt, J.A., and van Loon, L.C. (1996). Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8, 1225–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J., van Wees, S.C.M., van Pelt, J.A., Knoester, M., Laan, R., Gerrits, N., Weisbeek, P.J., and van Loon, L.C. (1998). A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10, 1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, D.T., Yang, H.P., and Zhang, S.Q. (2002). Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 277, 559–565. [DOI] [PubMed] [Google Scholar]
- Romeis, T., Piedras, P., Zhang, S.Q., Klessig, D.F., Hirt, H., and Jones, J.D.G. (1999). Rapid Avr9- and Cf-9-dependent activation of MAP kinases in tobacco cell cultures and leaves: Convergence of resistance gene, elicitor, wound, and salicylate responses. Plant Cell 11, 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel, M.A., and Ellis, B.E. (2002). Double jeopardy: Both overexpression and suppression of a redox-activated plant mitogen-activated protein kinase render tobacco plants ozone sensitive. Plant Cell 14, 2059–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, S., Okamoto, N., Seto, H., Ishizuka, K., Sano, H., and Ohashi, Y. (1995). Tobacco MAP kinase: A possible mediator in wound signal-transduction pathways. Science 270, 1988–1992. [DOI] [PubMed] [Google Scholar]
- Sessa, G., and Martin, G.B. (2000). Signal recognition and transduction mediated by the tomato Pto kinase: A paradigm of innate immunity in plants. Microbes Infect. 2, 1591–1597. [DOI] [PubMed] [Google Scholar]
- Smith, N.A., Singh, S.P., Wang, M.B., Stoutjesdijk, P.A., Green, A.G., and Waterhouse, P.M. (2000). Gene expression: Total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320. [DOI] [PubMed] [Google Scholar]
- Staskawicz, B.J., Ausubel, F.M., Baker, B.J., Ellis, J.G., and Jones, J.D.G. (1995). Molecular-genetics of plant-disease resistance. Science 268, 661–667. [DOI] [PubMed] [Google Scholar]
- Tao, Y., Xie, Z., Chen, W., Glazebrook, J., Chang, S.H., Han, B., Zhu, T., Zou, G., and Katagiri, F. (2003). Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena, G., Asai, T., Chiu, W.-L., and Sheen, J. (2001). Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4, 392–400. [DOI] [PubMed] [Google Scholar]
- Tor, M., Gordon, P., Cuzick, A., Eulgem, T., Sinapidou, E., Mert-Turk, F., Can, C., Dangl, J.L., and Holub, E.B. (2002). Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell 14, 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen, E.A., Freddie, C.T., Kahn, K., Parker, J.E., and Jones, J.D.G. (2002). Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29, 439–451. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27, 581–590. [DOI] [PubMed] [Google Scholar]
- Yang, K.Y., Liu, Y.D., and Zhang, S.Q. (2001). Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. USA 98, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S.Q., Du, H., and Klessig, D.F. (1998). Activation of the tobacco SIP kinase by both a cell wall-derived carbohydrate elicitor and purified proteinaceous elicitins from Phytophthora spp. Plant Cell 10, 435–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S.Q., and Klessig, D.F. (1997). Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9, 809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S.Q., and Klessig, D.F. (1998. a). Resistance gene N-mediated de novo synthesis and activation of a tobacco mitogen-activated protein kinase by tobacco mosaic virus infection. Proc. Natl. Acad. Sci. USA 95, 7433–7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S.Q., and Klessig, D.F. (1998. b). The tobacco wounding-activated mitogen-activated protein kinase is encoded by SIPK. Proc. Natl. Acad. Sci. USA 95, 7225–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S.Q., and Klessig, D.F. (2001). MAPK cascades in plant defense signaling. Trends Plant Sci. 6, 520–527. [DOI] [PubMed] [Google Scholar]