Abstract
PABPs [poly(A)-binding proteins] bind to the poly(A) tail of eukaryotic mRNAs and are conserved in species ranging from yeast to human. The prototypical cytoplasmic member, PABP1, is a multifunctional RNA-binding protein with roles in global and mRNA-specific translation and stability, consistent with a function as a central regulator of mRNA fate in the cytoplasm. More limited insight into the molecular functions of other family members is available. However, the consequences of disrupting PABP function in whole organisms is less clear, particularly in vertebrates, and even more so in mammals. In the present review, we discuss current and emerging knowledge with respect to the functions of PABP family members in whole animal studies which, although incomplete, already underlines their biological importance and highlights the need for further intensive research in this area.
Keywords: development, gametogenesis, mRNA stability and mRNA localization, mRNA translation, neuron, poly(A)-binding protein (PABP)
Introduction
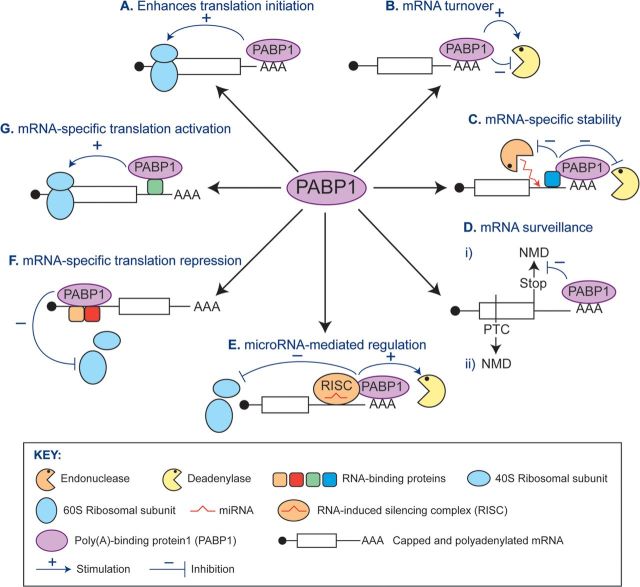
All aspects of life require tightly regulated gene expression and recent studies have highlighted both the complexity and importance of post-transcriptional control mechanisms. RNA-binding proteins play a key role in exerting and co-ordinating such regulation. One class of RNA-binding proteins that regulate numerous aspects of eukaryotic mRNA fate comprises the PABPs [poly(A)-binding proteins]. Both PABPNs (nuclear PABPs) and PABPCs (cytoplasmic PABPs) bind the poly(A) tail, but consist of very distinct domains (Figure 1A) and have different steady-state intracellular distributions and functions [1–3]. The present review discusses metazoan PABPCs (referred to hereafter as PABPs), which vary in number between organisms (Figure 1B). PABP1, the prototypical member, has four N-terminal RRMs (RNA-recognition motifs), a linker region and a highly conserved PABC (PABP C-terminal) domain (Figure 1A). Where studied, PABPs show a predominantly diffuse cytoplasmic distribution [4,5], but can be enriched at sites of localized translation, e.g. neuronal dendrites [6] or leading edges of migrating fibroblasts [7]. At least some family members shuttle to and from the nucleus [4,8] and can, during cell stress, accumulate in the nucleus [5,8] or in cytoplasmic foci, e.g. stress granules [5,9]. Studies of the molecular functions of PABPs have mainly focused on PABP1 (also called PABPC1) and have mostly used reporter mRNAs either in mammalian cell-free extracts/cell lines or in Xenopus laevis oocytes. This has revealed multiple functions that are reviewed in [1–3,10,11] (Figure 2). Briefly, the best characterized function of poly(A)-tail-bound PABP1 is enhancing translation initiation by interacting with translation initiation factors bound at the 5′-end of the mRNA (Figure 2A): this is proposed to stabilize their interaction with mRNA, thereby enhancing the recruitment of ribosomal subunits. Although considered a ‘global’ effect, the extent to which translation of individual mRNAs is stimulated can be influenced by regulation of their poly(A)-tail lengths. When bound (directly or indirectly) to sites other than the poly(A) tail, PABP1 can also activate or repress translation in an ‘mRNA-specific’ manner (Figures 2F and 2G) depending on the location of its alternative binding sites and the proteins with which it interacts [10].
Figure 1. Relatedness and domain organization of PABP family members.
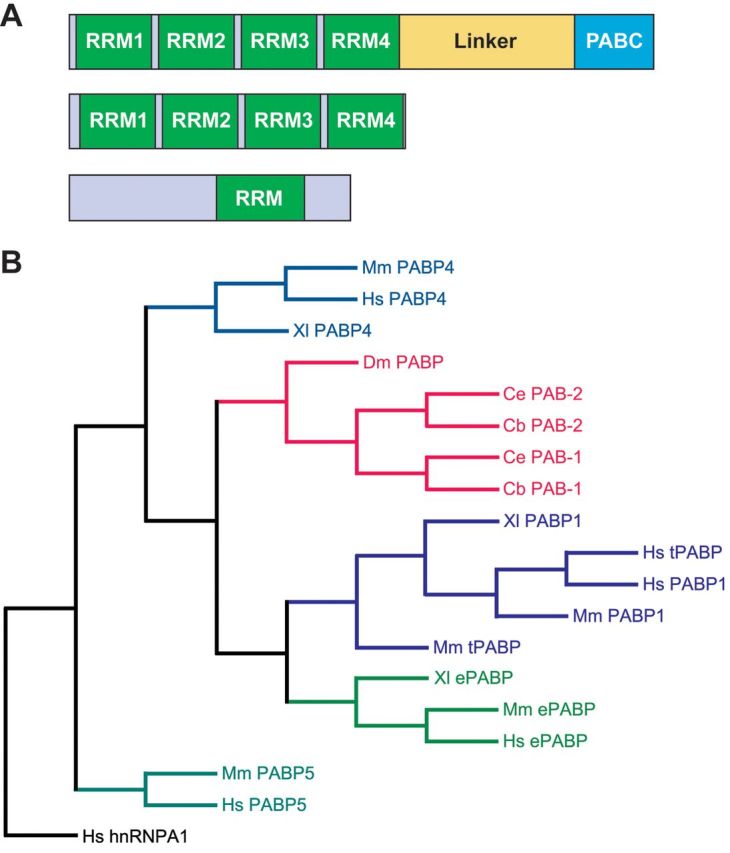
(A) Domain organization of PABPs. Top: vertebrate PABPC1, PABPC4, ePABP, tPABP (mammalian-specific), D. melanogaster dPABP and C. elegans/C. briggsae PAB-1 and PAB-2 (all predominately cytoplasmic). Middle: mammalian-specific PABPC5 (cytoplasmic). Bottom: PABPN1 (nuclear) and ePABP2 (cytoplasmic). Linker, proline/glutamine-rich variable linker region. (B) Phylogenetic tree of PABPC family proteins, rooted to human hnRNPA1 (heterogeneous nuclear ribonucleoprotein A1), an RRM-containing RNA-binding protein. Cb, C. briggsae; Ce, C. elegans; Dm, D. melanogaster; Hs, Homo sapiens; Mm, Mus musculus; Xl, X. laevis.
Figure 2. Molecular functions associated with PABP1.
(A) PABP1 enhances global translation by binding to the poly(A) tail and interacting with factors at the mRNA 5′-end to recruit ribosomal subunits. (B) PABP1 stabilizes mRNAs by blocking access of deadenylases to the poly(A) tail, but also recruits deadenylases to the mRNA. This may be linked to translational termination. (C) An example of an mRNA-specific role in stability where PABP1 interaction with 3′-UTR-binding proteins blocks deadenylation and endonucleotic cleavage within the 3′-UTR. (D, panel i) PABP1 plays a role in correct termination at stop codons ensuring that the nonsense-mediated decay pathway is not activated. (D, panel ii) PABP1 does not participate in termination at premature termination codons (PTCs), termination is aberrant and mRNA decay ensues. (E) PABP1 enhances miRNA-mediated translational repression and deadenylation via its interaction with the miRNA-containing RISC complex. (F) An example of PABP1 acting in mRNA-specific translational repression when bound as part of a 5′-UTR repressive complex that blocks ribosome assembly. (G) PABP1 can act as an mRNA-specific activator when recruited to the 3′-UTR by other RNA-binding proteins or regulatory elements. Table 1 summarizes functions shown for different family members in the species discussed in this Figure.
PABP1 also has multiple less well-characterized roles in mRNA turnover [11,12]. Of these, its best known role is protecting the poly(A) tail from deadenylation [poly(A) removal], the first and rate-limiting step in mRNA turnover (Figure 2B). Paradoxically, PABP1 also recruits deadenylases to mRNAs (Figure 2B) and has been suggested to co-ordinate translational termination with deadenylation, thereby regulating mRNA lifespan. Similar to translation, PABP1 has mRNA-specific roles in regulating mRNA stability (Figure 2C), either as part of regulatory complexes bound to sites within mRNAs or by interacting with stabilizing or destabilizing complexes when bound to the poly(A) tail. PABP1 is also involved in miRNA-mediated translational repression and/or deadenylation (Figure 2E) and in discriminating mRNAs which should undergo nonsense-mediated decay due to the presence of premature stop codons [3,11] (Figure 2D). Unlike their Saccharomyces cerevisae counterpart, mammalian PABPs do not participate in mRNA export [5].
The molecular functions of the other PABP family members are less well-characterized, but those of two vertebrate-specific PABPs (Figure 1), ePABP (embryonic PABP, also known as ePAB and PABPC1L) and PABP4 (also known as iPABP and PABPC4) have been examined in X. laevis egg extracts and oocytes (Table 1). Germ cells and early embryos undergo periods of transcriptional quiescence and are therefore heavily reliant on changes in mRNA translation which are often associated with dynamic poly(A)-tail length regulation in the cytoplasm: shortening (deadenylation) and extension (polyadenylation) lead to translational repression and activation respectively [13]. Cytoplasmic polyadenylation facilitates PABP recruitment and occurs in multiple cell types across metazoans (e.g. oocytes, male germ cells, early embryos and neurons [13]). All three X. laevis PABPs can stimulate poly(A)-dependent and mRNA-specific translation [14–16], both PABP1 and ePABP protect mRNAs from ‘default’ deadenylation [17,18] and ePABP can enhance cytoplasmic polyadenylation [19] and retard deadenylation driven by AREs (AU-rich elements) [18]. Although the ability of X. laevis PABP4 to regulate deadenylation has not been studied, mammalian PABP4 shows preference for AU-rich sequences in addition to poly(A) [20] and enhances translation and stability of ARE-containing mRNAs in extracts and/or cell lines [21,22]. Little is known about the functions of the two mammalian-specific PABPs, tPABP (testis-specific PABP, also known as PABPC2 and PABPC3) and PABP5 (PABPC5), which lacks the linker region and the PABC domain (Figure 1A). Finally, the cytoplasmic ePABP2 (or PABPN1-like) protein is outside the scope of the present review because it resembles PABPN [23,24] (Figure 1A).
Table 1. Molecular functions of PABPs.
Functions are shown in italics where direct evidence is absent and in bold where substantive or multiple lines of evidence are available. Indirect or contradictory experimental evidence is shown in italics. Protein–protein interactions and RNA binding have not been included for brevity. PABPs are only listed where evidence is available.
| (a) Invertebrates | ||
|---|---|---|
| PABP | Molecular function | Evidence |
| D. melanogaster | Enhances translation (global and/or mRNA-specific) | • Polysome association [72] |
| dPABP | • Depletion or depletion/add-back experiments with reporter mRNAs [39,73,74] | |
| miRNA-mediated regulation | • Depletion/add-back experiments with reporter mRNAs [74] | |
| • Reporter assays using mutants that impede dPABP protein interactions [75,76] | ||
| • Overexpression experiments using tethering assays [75] | ||
| mRNA surveillance | • Reporter and tethering assays [77] | |
| • Knockdown effects on reporter and endogenous mRNA [77] | ||
| mRNA-specific stability | • Effect of pAbp hypomorphs on mRNA absundance [34] | |
| C. elegans PAB-1 | Enhances translation initiation | • Polysome association [78] |
| (b) Non-mammalian vertebrates | ||
| PABP | Molecular function | Evidence |
| X. laevis | ||
| PABP1 | Enhances translation (global and/or mRNA-specific) | • Tethering assays [14]• Knockdown studies with metabolic labelling [15] |
| Inhibits deadenylation | • Overexpression studies [17] | |
| ePABP | Enhances translation (global and/or mRNA-specific) | • Polysome association [16]• Tethering assays [16,19]• Knockdown studies with metabolic labelling [14–16] |
| Cytoplasmic polyadenylation | • Sequestration/add-back with endogenous mRNAs [19] | |
| Inhibits deadenylation | • Depletion effects on reporter mRNAs [18,79] | |
| PABP4 | Enhances translation (global and/or mRNA-specific) | • Polysome association [15]• Tethering assays [15]• Loss of polysomes following knockdown [15] |
| (c) Mammals | ||
| PABP | Molecular function | Evidence |
| PABP1 | Enhances translation (global and/or mRNA-specific) | Extensively reviewed [1–3,10] |
| mRNA turnover/mRNA-specific stability | Recently reviewed [11] | |
| mRNA surveillance | Recently reviewed [3,11,80] | |
| miRNA-mediated regulation | Recently reviewed [3,75] | |
| mRNA-specific translation repression | Recently reviewed [10,81] | |
| ePABP | Cytoplasmic polyadenylation | • Effect of knockout on endogenous mRNAs [64] |
| tPABP | Enhances translation (global and/or mRNA-specific) | • Slightly augments reporter mRNA translation in vitro, but contradicted by lack of polysome association [69] |
| PABP4 | Enhances translation (global and/or | • Slight augmentation of reporter mRNA translation [21] |
| mRNA-specific) | • Polysome association [82] | |
| mRNA-specific stability | • Effect of knockdown on endogenous mRNAs [22] | |
In the present review, we describe our current state of knowledge regarding PABP function in whole organism studies in animals, but do not address studies of plants, yeast, viral infection or parasites [3,12,25–28].
Invertebrates
Drosophila melanogaster
D. melanogaster encodes one PABP1 homologue, dPABP (or PABP55B) whose molecular functions are incompletely characterized (Table 1), but which appears essential for viability since compound heterozygous deletions have an embryonic lethal phenotype at an unspecified stage [29]. Other studies, mostly using different P-element (transposon) insertions at the pAbp locus [29–34], have identified a range of phenotypes (described below). Although classified as hypomorphs (i.e. reduced gene activity or function), information concerning their actual effect on PABP function/expression (all but one lie outside the pAbp ORF) is lacking unless stated otherwise. Therefore the extent of dPABP insufficiency associated with these phenotypes remains unclear. Similarly, although some studies have used multiple mutant alleles, transgene rescue experiments to eliminate off-target effects are lacking, unless indicated otherwise.
Nonetheless, several studies suggest dPABP is important within the germline and following fertilization. D. melanogaster developing oocytes are supported by nurse cells that provide nutrients, RNAs and proteins. Localized translation of mRNAs [e.g. bcd (bicoid), osk (oskar), nos (nanos) and grk (gurken)] following transport from the nurse cells to particular sites within the oocyte, results in protein gradients that establish embryonic body axes and the germline. Correct spatiotemporal translation of these mRNAs requires multi-protein complexes to co-ordinate their localization, translational repression and activation [35].
Homozygous or compound heterozygous pAbp mutations, one of which was rescued by a dPABP transgene [34], results in arrest of oogenesis at stage 3 [31], or stages 5–6 (of 14 stages) [34], demonstrating an early requirement for dPABP during this process. Further analysis revealed that oocyte growth and positioning and egg chamber packaging were affected as mutant egg chambers contained abnormally small mis-positioned or multiple oocytes. Nurse and follicle cell development was also affected.
Some insight into dPABP function in these early oocytes comes from its association and co-localization with a protein complex that is involved in the microtubule-mediated transport of osk, bcd and grk mRNAs from nurse cells to the oocyte [35]. This association may be indirect as it is RNA-dependent, but studies in oocytes containing different compound heterozygous pAbp mutations show that dPABP is essential for posterior accumulation of osk [but not grk and bic (bicaudal)-D] mRNA [34] and for the localization of Staufen protein, which is interdependent with that of osk mRNA [36]. The specificity of dPABP for osk mRNA may be due to the presence of adenine-rich tracts within the osk mRNA 3′-UTR which bind dPABP in vitro [34].
Although a direct role for dPABP in the mRNA localization process cannot be ruled out, the failure to accumulate localized osk mRNA may be attributable to a role of dPABP in maintaining osk mRNA stability, since osk mRNA abundance is reduced in oocytes with different compound pAbp-heterozygous mutant alleles [34]. This stabilizing function may also explain why dPABP deficiency causes osk haploinsufficiency and why reducing pAbp to one copy suppresses patterning defects caused by anterior mis-localization of osk mRNA in bic-D mutants [34].
Subsequent to this early role, dPABP appears to be required for the spatiotemporal control of other mRNAs that establish embryonic protein gradients, e.g. via localized Grk synthesis. Eggs from several homozygous and compound heterozygous pAbp hypomorphs display patterning defects suggestive of reduced dorso-anterior Grk protein levels [31], and heterozygous pAbp hypomorphs exacerbate patterning defects in grk mutants [31]. Interestingly, dPABP can be co-isolated with Cup and Encore (Enc), proteins that function in grk mRNA localization and either its translational repression or activation respectively [31,37]. pAbp and cup mutants lay eggs with opposite patterning defects (‘ventralized’ compared with ‘dorsalized’ respectively), whereas a heterozygous pAbp hypomorph enhances the ventralized and collapsed egg phenotypes of enc mutants [31]. This suggests that dPABP may antagonize Cup-mediated repression and, similar to Enc, is required to promote grk mRNA translation.
dPABP may also contribute to translational repression of cad (caudal) mRNA [33] by a Bcd-containing complex which establishes the posterior–anterior gradient of Caudal [38]. Bin-3 (bicoid-interacting protein 3) is also implicated in this repression and the levels of embryonic lethality of a bin3 bcd double mutant are enhanced by a pAbp heterozygous hypomorph [33], but it is not clear whether dPABP contribution to cad mRNA repression is direct.
A later role in determining body morphology is suggested by studies of wing size. Heterozygous pAbp hypomorph flies or those overexpressing an inhibitor of translation that sequesters dPABP, called dPaip2 (D. melanogaster PABP-interacting protein 2), in wing-imaginal discs have reduced wing size [39]. Importantly, co-overexpression of dPABP with dPaip2 rescues wing cell number and size [39].
In addition to affecting oogenesis and body pattern, dPABP is also required for spermatogenesis [30,32] as several compound heterozygous pAbp mutant flies are male sterile and display aberrant meiosis (only undergoing one meiotic division), spermatid elongation and/or cytokinesis [30,32]. Although the molecular events leading to these defects are unknown, the pAbp meiotic phenotype partially overlaps with that of Larp (La-related protein), a factor thought to regulate mRNA stability in Caenorhabditis elegans and translation of certain mRNAs in mammalian cells [40–42], which interacts biochemically and genetically with dPABP [30].
Many mRNA-binding proteins that function in the germline also appear to be important in neurons. Neurons pass signals to other cells via synapses and localized translation at these sites is thought to regulate synaptic plasticity [43]. Aggregates containing dPABP and eIF4E (eukaryotic initiation factor 4E) coincide with polyribosome clusters within subsynaptic compartments of larval NMJs (neuromuscular junctions), posited sites of localized translation [29]. Mutants that increase synaptic activity, or larvae overexpressing pAbp mRNA show increased occurrence of subsynaptic dPABP/eIF4E aggregates, altered levels of some synaptic proteins, significantly larger NMJs and more efficient neurotransmission [29], consistent with modified subsynaptic translation. However, similar observations were made with reduced pAbp mRNA levels (heterozygous pAbp hypomorph) and neither increased nor decreased pAbp mRNA levels were reflected in dPABP protein levels [29], making the observed alterations in dPABP/eIF4E aggregates and synaptic activity difficult to explain.
Studies of dFMR1, the D. melanogaster homologue of FMRP (fragile-X mental retardation protein), also link dPABP to synaptic plasticity. FMRP is an RNA-binding protein that regulates mRNA localization, translation and stability [44] and is important for cognition. dFMR1 is required for long-term memory [45] and a genetic screen for genes involved in dFMR1-mediated translational repression identified pAbp [46]. dPABP co-localizes with dFMR1-positive neuritic RNPs (ribonucleoproteins) and its overexpression inhibits dendritic branching, suggestive of a function in translational repression [46], but this activity awaits confirmation.
Interestingly, the phenotypes of several D. melanogaster models of human neurodegenerative diseases are also affected by dPABP. For instance, neurodegeneration in SCA3 (spinal cerebellar ataxia 3) models is exacerbated by heterozygous pAbp deletion and reduced by dPABP overexpression [47]. Similarly, PABP1 accumulates in cytoplasmic inclusions in motor neurons from ALS (amyotrophic lateral sclerosis) patients [48] and siRNA-mediated dPABP knockdown in fly models of ALS suggests it is required for inclusion formation [48].
C. elegans and Caenorhabditis briggsae
C. elegans and C. briggsae encode two PABPCs, PAB-1 and PAB-2 ([49]; WormBase) whose molecular functions largely await characterization (Table 1). In C. elegans, individual pab-1 (detailed in [50]) or pab-2 knockdown leads to limited somatic defects such as abnormally protruding vulva (high and low penetrance respectively; also seen in a pab-1 nonsense mutation), low penetrance ruptured vulva, and flaccid body morphology (pab-1 only) [49,51].
In both species, pab-2 is X-chromosomal. Consistent with germline X-chromosome inactivation, its knockdown does not affect C. elegans fertility [49]. However, despite conservation of chromosome silencing [52], knockdown in C. briggsae drastically affects embryo number and mortality [49], which is suggestive of reproductive and/or developmental defects.
In contrast, pab-1 is essential for fertility in both species [49,51,53–56]. C. elegans is a hermaphrodite, which first makes sperm and then switches to oogenesis, and knockdown at different stages of post-embryonic development revealed PAB-1 is required throughout gametogenesis. Even late-stage PAB-1 depletion is deleterious, leading to defective oogenesis, high embryonic death and infertility of surviving progeny [54]. This suggests that PAB-1 is likely to regulate mRNAs that function at different stages of germline development [54]. PAB-1 is present in, and reportedly required for the formation of, P-granules [54], germ-cell specific cytoplasmic foci, which are considered centres of post-transcriptional regulation as they are rich in RNA-binding proteins and mRNA [57]. Although P-granules are implicated in both germline proliferation and gametogenesis, it is unclear whether the association of PAB-1 with these foci accounts for its essential germline function.
Germ cell ablation can lead to longevity and gigantism [58,59] and pab-1 nonsense alleles increase lifespan 1.3–1.5-fold [49,53] and body volume 1.4-fold [49], which may be a consequence of the described germline defects. Curiously, pab-2 deletion alters relative lifespan when worms are fed on different bacterial species [60].
Vertebrate phenotypes associated with loss of PABP function
X. laevis
X. laevis PABPs exhibit distinct distributions: in adult tissues, PABP1 is widely expressed albeit at variable levels [16,61], and PABP4 mRNA is widely distributed [15], whereas ePABP is restricted to the gonads [16,61]. In oocytes and early embryos, ePABP is the predominant PABP [15,16,18]. Changes in ePABP phosphorylation during oocyte maturation, when fully grown oocytes become fertilization competent, suggest regulation of its activity coincident with changes in the poly(A)-tail length and translation of many mRNAs [19]. Oocyte maturation is impeded by ectopic xPAIP2 (X. laevis PABP-interacting protein 2)-mediated PABP sequestration which blocks the cytoplasmic polyadenylation of mRNAs whose translational activation is required for maturation. Rescue by overexpression of ePABP, but not a form in which several phospho-residues have been mutated, demonstrates the importance of ePABP and these phosphorylations for oocyte maturation [19].
Each PABP is essential for X. laevis embryonic development and viability [15]. Morpholino-mediated knockdown of PABP1 leads to a range of morphological phenotypes in tadpoles (e.g. abnormal development of the eye, cement gland, tail and fin, and body curvature), problems with movement and embryonic death by stage 30/31 (out of 66 developmental stages) [15]. ePABP knockdown results in similar morphological and movement defects, but, surprisingly, death occurs later, by stage 35, perhaps due to the higher levels of ePABP in early embryos delaying effective knockdown [15]. In contrast, PABP4 knockdown results mainly in anterior morphological defects (e.g. cephalic and ventral oedema, malformation of the head, poor eye development, and digestive tract deformities) and abnormal swimming motions. PABP4 phenotypes become apparent later than those of PABP1 and ePABP- and PABP4-deficient embryos do not die until stage 50 [15]. Importantly, the respective phenotypes were recapitulated with multiple morpholinos and could be effectively rescued [15].
For each of these knockdowns, the developmental defects were accompanied by significant decreases in global protein synthesis, suggesting a potential molecular basis for the phenotypes [15]. However, despite this commonality, cross-rescue experiments showed that neither ePABP nor PABP4 could fully rescue PABP1 knockdown, indicating functional differences must also exist between individual PABP family members, which domain-swap experiments showed to be conferred by multiple domains [15]. The ability of all three PABPs to stimulate global translation [15] (Table 1) suggests that their non-redundant functions may relate to individual roles in regulating mRNA-specific translation or mRNA decay.
Mouse
Insight into the phenotypic consequences of loss of PABP function in mammals is only available for Epab. Murine Epab mRNA is only present in male and female germ cells and one- and two-cell stage embryos [62,63]. In contrast with ePABP-deficient X. laevis, Epab−/− mice display no growth or developmental abnormalities, a difference that may reflect reduced reliance on post-transcriptional regulation in mouse embryos since zygotic transcription begins at the two-cell stage in mice rather than at mid-blastula transition in X. laevis.
Epab−/− females, but not males, are sterile [64,65]. In mammals, hormonal signals trigger the development and maturation of oocytes within follicles that progress through a series of developmental stages and contain somatic cells that respond to and support the growing oocyte. Epab−/− mice have normal oestrus and normal follicle numbers at all stages, with the exception of secondary follicles which are overly abundant, but their oocytes fail to mature either in vivo or in vitro. Similar to what was described previously for X. laevis [19], this maturation defect appears to be due, at least in part, to abrogated cytoplasmic polyadenylation, resulting in reduced expression of proteins required for oocyte maturation [64]. However, ePABP may also be required earlier in oogenesis, since injection of Epab mRNA into fully grown Epab−/− oocytes failed to rescue maturation in vitro [64].
Following ovulation, follicles become corpora lutea which, in superovulated Epab−/− mice, show increased retention of oocytes, indicative of defective ovulation [64]. This phenotype appears to result from somatic cell defects in the follicle which are likely indirect as these cells do not express Epab [64].
Cell-based models of red blood cell maturation
In a recent study, a potential role for mammalian PABP4 in red blood cell maturation was identified [22]. Mature red blood cells lack a nucleus making their terminal differentiation highly dependent on post-transcriptional control; this can be modelled by dimethylsulfoxide treatment of MEL (lymphoma-derived murine erythroleukaemia) cells. Intriguingly, shRNA-mediated depletion of PABP4 in MEL cells increased or decreased the abundance of limited mRNA subsets and hindered terminal MEL cell differentiation [22]. Although the underlying mechanism requires clarification, it was suggested that AREs in some of these mRNAs may aid PABP4 binding to impede their rapid decay following deadenylation.
Perspectives
Although recent work has provided fascinating insight into the complexity of the biological processes in which PABP family members are involved, their molecular multi-functionality suggests that we have only scratched the surface. Indeed, even in invertebrates where PABPs are already implicated in diverse phenotypes, they may be involved in additional processes, e.g. circadian rhythm and transposon silencing [66–68]. Our knowledge is more limited in vertebrates, which encode a greater diversity of PABPs, with only their roles in early development and oogenesis having been explored. In mammals, the critical role of ePABP in oocytes is conserved, but information on other PABPs is not available although extrapolating from non-mammalian studies suggests that mammalian PABP1 may be essential. It remains to be determined whether mammalian PABP4 plays an analogous role to its X. laevis counterpart in development and whether its effects on mammalian erythroid differentiation are recapitulated in vivo. There is no insight into the roles of tPABP and PABP5 from cell lines or other models, but tPABP is only expressed in a subset of male germ cells, indicating its function is restricted to the male germline [69], where it may be redundant with PABP1. Consistent with this idea, tPABP interacts with translation factors and can stimulate reporter mRNA translation in vitro [69] (Table 1). However, tPABP appears not to be polysome-associated and discrepancy exists as to whether its distribution within cytoplasmic foci called chromatoid bodies is distinct from that of PABP1 [69,70]. Little is known about the expression of PABP5, whose domain structure (Figure 1A) suggests distinct roles from other PABPs, although intriguingly, a truncated PABP5 isoform is present in mitochondria, suggesting a potential function in these organelles [71]. In conclusion, further investigation into the roles of PABPs in whole organisms, complementing molecular studies that underscore their central role in cytoplasmic mRNA metabolism, should uncover the full extent of their importance in both normal and diseased states.
Acknowledgments
We apologize to those whose work we have not cited owing to space constraints. We thank Matt Brook, Simon Bullock, Lenka Hrabalkova, Judith Kimble, Jessica Scanlon and Gavin Wilkie for critical discussions.
Abbreviations
- ARE
AU-rich element
- Bcd
Bicoid
- Bic-D
Bicaudal-D
- Bin
Bicoid-interacting protein
- Cad
Caudal
- dFMR1
D. melanogaster FMRP1
- dPABP
D. melanogaster PABP
- dPaip2
D. melanogaster PABP-interacting protein 2
- eIF
eukaryotic initiation factor
- Enc
Encore
- ePABP
embryonic PABP
- FMRP
fragile-X mental retardation protein
- Grk
Gurken
- MEL
lymphoma-derived murine erythroleukaemia
- NMJ
neuromuscular junction
- Osk
Oskar
- PABC
PABP C-terminal
- PABP
poly(A)-binding protein
- PABPC
cytoplasmic PABP
- PABPN
nuclear PABP
- RRM
RNA-recognition motif
- tPABP
testis-specific PABP
Footnotes
RNA UK 2014: An Independent Meeting held at Low Wood Hotel, Windermere, U.K., 24–26 January 2014. Organized and Edited by Niki Gray, Gracjan Michlewski and Steve West (University of Edinburgh, U.K.).
Funding
Work within the laboratory is currently funded by the BBSRC (Biotechnology and Biological Sciences Research Council), the MRC (Medical Research Council) and Tommy’s.
References
- 1.Kuhn U., Wahle E. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Gorgoni B., Gray N.K. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: a developmental perspective. Brief Funct. Genomic Proteomic. 2004;3:125–141. doi: 10.1093/bfgp/3.2.125. [DOI] [PubMed] [Google Scholar]
- 3.Goss D.J., Kleiman F.E. Poly(A) binding proteins: are they all created equal? Wiley Interdiscip. Rev. RNA. 2013;4:167–179. doi: 10.1002/wrna.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afonina E., Stauber R., Pavlakis G.N. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 1998;273:13015–13021. doi: 10.1074/jbc.273.21.13015. [DOI] [PubMed] [Google Scholar]
- 5.Burgess H.M., Richardson W.A., Anderson R.C., Salaun C., Graham S.V., Gray N.K. Nuclear relocalisation of cytoplasmic poly(A)-binding protein (PABP) 1 and 4 in response to UV irradiation reveals mRNA-dependent export of metazoan PABPs. J. Cell Sci. 2011;124:3344–3355. doi: 10.1242/jcs.087692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muddashetty R., Khanam T., Kondrashov A., Bundman M., Iacoangeli A., Kremerskothen J., Duning K., Barnekow A., Huttenhofer A., Tiedge H., Brosius J. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J. Mol. Biol. 2002;321:433–445. doi: 10.1016/S0022-2836(02)00655-1. [DOI] [PubMed] [Google Scholar]
- 7.Woods A.J., Roberts M.S., Choudhary J., Barry S.T., Mazaki Y., Sabe H., Morley S.J., Critchley D.R., Norman J.C. Paxillin associates with poly(A)-binding protein 1 at the dense endoplasmic reticulum and the leading edge of migrating cells. J. Biol. Chem. 2002;277:6428–6437. doi: 10.1074/jbc.M109446200. [DOI] [PubMed] [Google Scholar]
- 8.Kumar G.R., Shum L., Glaunsinger B.A. Importin α-mediated nuclear import of cytoplasmic poly(A) binding protein occurs as a direct consequence of cytoplasmic mRNA depletion. Mol. Cell Biol. 2011;31:3113–3125. doi: 10.1128/MCB.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kedersha N.L., Gupta M., Li W., Miller I., Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgess H.M., Gray N.K. mRNA-specific regulation of translation by poly(A)-binding proteins. Biochem. Soc. Trans. 2010;38:1517–1522. doi: 10.1042/BST0381517. [DOI] [PubMed] [Google Scholar]
- 11.Brook M., Gray N.K. The role of mammalian poly(A)-binding proteins in co-ordinating mRNA turnover. Biochem. Soc. Trans. 2012;40:856–864. doi: 10.1042/BST20120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mangus D.A., Evans M.C., Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charlesworth A., Meijer H.A., de Moor C.H. Specificity factors in cytoplasmic polyadenylation. Wiley Interdiscip. Rev. RNA. 2013;4:437–461. doi: 10.1002/wrna.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray N.K., Coller J.M., Dickson K.S., Wickens M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorgoni B., Richardson W.A., Burgess H.M., Anderson R.C., Wilkie G.S., Gautier P., Martins J.P., Brook M., Sheets M.D., Gray N.K. Poly(A)-binding proteins are functionally distinct and have essential roles during vertebrate development. Proc. Natl. Acad. Sci. U.S.A. 2011;108:7844–7849. doi: 10.1073/pnas.1017664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkie G.S., Gautier P., Lawson D., Gray N.K. Embryonic poly(A)-binding protein stimulates translation in germ cells. Mol. Cell. Biol. 2005;25:2060–2071. doi: 10.1128/MCB.25.5.2060-2071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wormington M., Searfoss A.M., Hurney C.A. Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J. 1996;15:900–909. [PMC free article] [PubMed] [Google Scholar]
- 18.Voeltz G.K., Ongkasuwan J., Standart N., Steitz J.A. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 2001;15:774–788. doi: 10.1101/gad.872201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friend K., Brook M., Bezirci F.B., Sheets M.D., Gray N.K., Seli E. Embryonic poly(A)-binding protein (ePAB) phosphorylation is required for Xenopus oocyte maturation. Biochem. J. 2012;445:93–100. doi: 10.1042/BJ20120304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sladic R.T., Lagnado C.A., Bagley C.J., Goodall G.J. Human PABP binds AU-rich RNA via RNA-binding domains 3 and 4. Eur. J. Biochem. 2004;271:450–457. doi: 10.1046/j.1432-1033.2003.03945.x. [DOI] [PubMed] [Google Scholar]
- 21.Okochi K., Suzuki T., Inoue J., Matsuda S., Yamamoto T. Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly(A)-binding protein: implication of Tob in translational control. Genes Cells. 2005;10:151–163. doi: 10.1111/j.1365-2443.2005.00826.x. [DOI] [PubMed] [Google Scholar]
- 22.Kini H.K., Kong J., Liebhaber S.A. Cytoplasmic poly(A) binding protein C4 serves a critical role in erythroid differentiation. Mol. Cell. Biol. 2014;34:1300–1309. doi: 10.1128/MCB.01683-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Good P.J., Abler L., Herring D., Sheets M.D. Xenopus embryonic poly(A) binding protein 2 (ePABP2) defines a new family of cytoplasmic poly(A) binding proteins expressed during the early stages of vertebrate development. Genesis. 2004;38:166–175. doi: 10.1002/gene.20015. [DOI] [PubMed] [Google Scholar]
- 24.Cosson B., Braun F., Paillard L., Blackshear P., Beverley Osborne H. Identification of a novel Xenopus laevis poly (A) binding protein. Biol. Cell. 2004;96:519–527. doi: 10.1016/j.biolcel.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 25.Amrani N., Dong S., He F., Ganesan R., Ghosh S., Kervestin S., Li C., Mangus D.A., Spatrick P., Jacobson A. Aberrant termination triggers nonsense-mediated mRNA decay. Biochem. Soc. Trans. 2006;34:39–42. doi: 10.1042/BST0340039. [DOI] [PubMed] [Google Scholar]
- 26.Guerra N., Vega-Sendino M., Perez-Morgado M.I., Ramos E., Soto M., Gonzalez V.M., Martin M.E. Identification and functional characterization of a poly(A)-binding protein from Leishmania infantum (LiPABP) FEBS Lett. 2011;585:193–198. doi: 10.1016/j.febslet.2010.11.042. [DOI] [PubMed] [Google Scholar]
- 27.Smith R.W.P., Gray N.K. Poly(A)-binding protein (PABP): a common viral target. Biochem. J. 2010;426:1–12. doi: 10.1042/BJ20091571. [DOI] [PubMed] [Google Scholar]
- 28.Tuteja R., Pradhan A. Isolation and functional characterization of eIF4F components and poly(A)-binding protein from Plasmodium falciparum. Parasitol. Int. 2009;58:481–485. doi: 10.1016/j.parint.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Sigrist S.J., Thiel P.R., Reiff D.F., Lachance P.E., Lasko P., Schuster C.M. Postsynaptic translation affects the efficacy and morphology of neuromuscular junctions. Nature. 2000;405:1062–1065. doi: 10.1038/35016598. [DOI] [PubMed] [Google Scholar]
- 30.Blagden S.P., Gatt M.K., Archambault V., Lada K., Ichihara K., Lilley K.S., Inoue Y.H., Glover D.M. Drosophila Larp associates with poly(A)-binding protein and is required for male fertility and syncytial embryo development. Dev. Biol. 2009;334:186–197. doi: 10.1016/j.ydbio.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 31.Clouse K.N., Ferguson S.B., Schupbach T. Squid, Cup, and PABP55B function together to regulate gurken translation in Drosophila. Dev. Biol. 2008;313:713–724. doi: 10.1016/j.ydbio.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pertceva J.A., Dorogova N.V., Bolobolova E.U., Nerusheva O.O., Fedorova S.A., Omelyanchuk L.V. The role of Drosophila hyperplastic discs gene in spermatogenesis. Cell Biol. Int. 2010;34:991–996. doi: 10.1042/CBI20100105. [DOI] [PubMed] [Google Scholar]
- 33.Singh N., Morlock H., Hanes S.D. The Bin3 RNA methyltransferase is required for repression of caudal translation in the Drosophila embryo. Dev. Biol. 2011;352:104–115. doi: 10.1016/j.ydbio.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 34.Vazquez-Pianzola P., Urlaub H., Suter B. Pabp binds to the osk 3′UTR and specifically contributes to osk mRNA stability and oocyte accumulation. Dev. Biol. 2011;357:404–418. doi: 10.1016/j.ydbio.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 35.Kugler J.M., Lasko P. Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during Drosophila oogenesis. Fly (Austin) 2009;3:15–28. doi: 10.4161/fly.3.1.7751. [DOI] [PubMed] [Google Scholar]
- 36.Jenny A., Hachet O., Zavorszky P., Cyrklaff A., Weston M.D., Johnston D.S., Erdelyi M., Ephrussi A. A translation-independent role of oskar RNA in early Drosophila oogenesis. Development. 2006;133:2827–2833. doi: 10.1242/dev.02456. [DOI] [PubMed] [Google Scholar]
- 37.Hawkins N.C., Van Buskirk C., Grossniklaus U., Schupbach T. Post-transcriptional regulation of gurken by encore is required for axis determination in Drosophila. Development. 1997;124:4801–4810. doi: 10.1242/dev.124.23.4801. [DOI] [PubMed] [Google Scholar]
- 38.Lasko P. Posttranscriptional regulation in Drosophila oocytes and early embryos. Wiley Interdiscip. Rev. RNA. 2011;2:408–416. doi: 10.1002/wrna.70. [DOI] [PubMed] [Google Scholar]
- 39.Roy G., Miron M., Khaleghpour K., Lasko P., Sonenberg N. The Drosophila poly(A) binding protein-interacting protein, dPaip2, is a novel effector of cell growth. Mol. Cell. Biol. 2004;24:1143–1154. doi: 10.1128/MCB.24.3.1143-1154.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nykamp K., Lee M.H., Kimble J. C. elegans La-related protein, LARP-1, localizes to germline P bodies and attenuates Ras-MAPK signaling during oogenesis. RNA. 2008;14:1378–1389. doi: 10.1261/rna.1066008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zanin E., Pacquelet A., Scheckel C., Ciosk R., Gotta M. LARP-1 promotes oogenesis by repressing fem-3 in the C. elegans germline. J. Cell Sci. 2010;123:2717–2724. doi: 10.1242/jcs.066761. [DOI] [PubMed] [Google Scholar]
- 42.Tcherkezian J., Cargnello M., Romeo Y., Huttlin E.L., Lavoie G., Gygi S.P., Roux P.P. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev. 2014;28:357–371. doi: 10.1101/gad.231407.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holt C.E., Schuman E.M. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron. 2013;80:648–657. doi: 10.1016/j.neuron.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Rubeis S., Fernandez E., Buzzi A., Di Marino D., Bagni C. Molecular and cellular aspects of mental retardation in the Fragile X syndrome: from gene mutation/s to spine dysmorphogenesis. Adv. Exp. Med. Biol. 2012;970:517–551. doi: 10.1007/978-3-7091-0932-8. [DOI] [PubMed] [Google Scholar]
- 45.Bolduc F.V., Bell K., Cox H., Broadie K.S., Tully T. Excess protein synthesis in Drosophila fragile X mutants impairs long-term memory. Nat. Neurosci. 2008;11:1143–1145. doi: 10.1038/nn.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cziko A.M., McCann C.T., Howlett I.C., Barbee S.A., Duncan R.P., Luedemann R., Zarnescu D., Zinsmaier K.E., Parker R.R., Ramaswami M. Genetic modifiers of dFMR1 encode RNA granule components in Drosophila. Genetics. 2009;182:1051–1060. doi: 10.1534/genetics.109.103234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lessing D., Bonini N.M. Polyglutamine genes interact to modulate the severity and progression of neurodegeneration in Drosophila. PLoS Biol. 2008;6:e29. doi: 10.1371/journal.pbio.0060029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim H.J., Raphael A.R., LaDow E.S., McGurk L., Weber R.A., Trojanowski J.Q., Lee V.M., Finkbeiner S., Gitler A.D., Bonini N.M. Therapeutic modulation of eIF2α phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat. Genet. 2014;46:152–160. doi: 10.1038/ng.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maciejowski J., Ahn J.H., Cipriani P.G., Killian D.J., Chaudhary A.L., Lee J.I., Voutev R., Johnsen R.C., Baillie D.L., Gunsalus K.C., et al. Autosomal genes of autosomal/X-linked duplicated gene pairs and germ-line proliferation in Caenorhabditis elegans. Genetics. 2005;169:1997–2011. doi: 10.1534/genetics.104.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Green R.A., Kao H.L., Audhya A., Arur S., Mayers J.R., Fridolfsson H.N., Schulman M., Schloissnig S., Niessen S., Laband K., et al. A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell. 2011;145:470–482. doi: 10.1016/j.cell.2011.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ciosk R., DePalma M., Priess J.R. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development. 2004;131:4831–4841. doi: 10.1242/dev.01352. [DOI] [PubMed] [Google Scholar]
- 52.Kelly W.G., Schaner C.E., Dernburg A.F., Lee M.H., Kim S.K., Villeneuve A.M., Reinke V. X-chromosome silencing in the germline of C. elegans. Development. 2002;129:479–492. doi: 10.1242/dev.129.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ko S., Park J.H., Lee A.R., Kim E., Jiyoung K., Kawasaki I., Shim Y.H. Two mutations in pab-1 encoding poly(A)-binding protein show similar defects in germline stem cell proliferation but different longevity in C. elegans. Mol. Cells. 2010;30:167–172. doi: 10.1007/s10059-010-0103-2. [DOI] [PubMed] [Google Scholar]
- 54.Ko S., Kawasaki I., Shim Y.H. PAB-1, a Caenorhabditis elegans poly(A)-binding protein, regulates mRNA metabolism in germline by interacting with CGH-1 and CAR-1. PLoS ONE. 2013;8:e84798. doi: 10.1371/journal.pone.0084798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lall S., Piano F., Davis R.E. Caenorhabditis elegans decapping proteins: localization and functional analysis of Dcp1, Dcp2, and DcpS during embryogenesis. Mol. Biol. Cell. 2005;16:5880–5890. doi: 10.1091/mbc.E05-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simmer F., Moorman C., van der Linden A.M., Kuijk E., van den Berghe P.V., Kamath R.S., Fraser A.G., Ahringer J., Plasterk R.H. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Updike D., Strome S. P granule assembly and function in Caenorhabditis elegans germ cells. J. Androl. 2010;31:53–60. doi: 10.2164/jandrol.109.008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lapierre L.R., Hansen M. Lessons from C. elegans: signaling pathways for longevity. Trends Endocrinol. Metab. 2012;23:637–644. doi: 10.1016/j.tem.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patel M.N., Knight C.G., Karageorgi C., Leroi A.M. Evolution of germ-line signals that regulate growth and aging in nematodes. Proc. Natl. Acad. Sci. U.S.A. 2002;99:769–774. doi: 10.1073/pnas.012511099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coolon J.D., Jones K.L., Todd T.C., Carr B.C., Herman M.A. Caenorhabditis elegans genomic response to soil bacteria predicts environment-specific genetic effects on life history traits. PLoS Genet. 2009;5:e1000503. doi: 10.1371/journal.pgen.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cosson B., Couturier A., Le Guellec R., Moreau J., Chabelskaya S., Zhouravleva G., Philippe M. Characterization of the poly(A) binding proteins expressed during oogenesis and early development of Xenopus laevis. Biol. Cell. 2002;94:217–231. doi: 10.1016/S0248-4900(02)01195-4. [DOI] [PubMed] [Google Scholar]
- 62.Ozturk S., Guzeloglu-Kayisli O., Demir N., Sozen B., Ilbay O., Lalioti M.D., Seli E. Epab and Pabpc1 are differentially expressed during male germ cell development. Reprod. Sci. 2012;19:911–922. doi: 10.1177/1933719112446086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seli E., Lalioti M.D., Flaherty S.M., Sakkas D., Terzi N., Steitz J.A. An embryonic poly(A)-binding protein (ePAB) is expressed in mouse oocytes and early preimplantation embryos. Proc. Natl. Acad. Sci. U.S.A. 2005;102:367–372. doi: 10.1073/pnas.0408378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guzeloglu-Kayisli O., Lalioti M.D., Aydiner F., Sasson I., Ilbay O., Sakkas D., Lowther K.M., Mehlmann L.M., Seli E. Embryonic poly(A)-binding protein (EPAB) is required for oocyte maturation and female fertility in mice. Biochem. J. 2012;446:47–58. doi: 10.1042/BJ20120467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ozturk S., Guzeloglu-Kayisli O., Lowther K.M., Lalioti M.D., Sakkas D., Seli E. Epab is dispensable for spermatogenesis and male fertility in mice. Mol. Reprod. Dev. 2014;81:390. doi: 10.1002/mrd.22319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lim C., Allada R. ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science. 2013;340:875–879. doi: 10.1126/science.1234785. [DOI] [PubMed] [Google Scholar]
- 67.Vastenhouw N.L., Fischer S.E., Robert V.J., Thijssen K.L., Fraser A.G., Kamath R.S., Ahringer J., Plasterk R.H. A genome-wide screen identifies 27 genes involved in transposon silencing in C. elegans. Curr. Biol. 2003;13:1311–1316. doi: 10.1016/S0960-9822(03)00539-6. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y., Ling J., Yuan C., Dubruille R., Emery P. A role for Drosophila ATX2 in activation of PER translation and circadian behavior. Science. 2013;340:879–882. doi: 10.1126/science.1234746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kimura M., Ishida K., Kashiwabara S., Baba T. Characterization of two cytoplasmic poly(A)-binding proteins, PABPC1 and PABPC2, in mouse spermatogenic cells. Biol. Reprod. 2009;80:545–554. doi: 10.1095/biolreprod.108.072553. [DOI] [PubMed] [Google Scholar]
- 70.Meikar O., Vagin V.V., Chalmel F., Sostar K., Lardenois A., Hammell M., Jin Y., Da Ros M., Wasik K.A., Toppari J., et al. An atlas of chromatoid body components. RNA. 2014;20:483–495. doi: 10.1261/rna.043729.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kazak L., Reyes A., Duncan A.L., Rorbach J., Wood S.R., Brea-Calvo G., Gammage P.A., Robinson A.J., Minczuk M., Holt I.J. Alternative translation initiation augments the human mitochondrial proteome. Nucleic Acids Res. 2013;41:2354–2369. doi: 10.1093/nar/gks1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Satterfield T.F., Pallanck L.J. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum. Mol. Genet. 2006;15:2523–2532. doi: 10.1093/hmg/ddl173. [DOI] [PubMed] [Google Scholar]
- 73.Duncan K.E., Strein C., Hentze M.W. The Sxl-Unr corepressor complex uses a PABP-mediated mechanism to inhibit ribosomal recruitment to msl-2 mRNA. Mol. Cell. 2009;36:571–582. doi: 10.1016/j.molcel.2009.09.042. [DOI] [PubMed] [Google Scholar]
- 74.Moretti F., Kaiser C., Zdanowicz-Specht A., Hentze M.W. PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat. Struct. Mol. Biol. 2012;19:603–608. doi: 10.1038/nsmb.2309. [DOI] [PubMed] [Google Scholar]
- 75.Braun J.E., Huntzinger E., Izaurralde E. The role of GW182 proteins in miRNA-mediated gene silencing. Adv. Exp. Med. Biol. 2013;768:147–163. doi: 10.1007/978-1-4614-5107-5. [DOI] [PubMed] [Google Scholar]
- 76.Huntzinger E., Kuzuoglu-Ozturk D., Braun J.E., Eulalio A., Wohlbold L., Izaurralde E. The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res. 2013;41:978–994. doi: 10.1093/nar/gks1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Behm-Ansmant I., Gatfield D., Rehwinkel J., Hilgers V., Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 2007;26:1591–1601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scheckel C., Gaidatzis D., Wright J.E., Ciosk R. Genome-wide analysis of GLD-1-mediated mRNA regulation suggests a role in mRNA storage. PLoS Genet. 2012;8:e1002742. doi: 10.1371/journal.pgen.1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim J.H., Richter J.D. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes Dev. 2007;21:2571–2579. doi: 10.1101/gad.1593007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schweingruber C., Rufener S.C., Zund D., Yamashita A., Muhlemann O. Nonsense-mediated mRNA decay–mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim. Biophys. Acta. 2013;1829:612–623. doi: 10.1016/j.bbagrm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 81.Bag J., Bhattacharjee R.B. Multiple levels of post-transcriptional control of expression of the poly (A)-binding protein. RNA Biol. 2010;7:5–12. doi: 10.4161/rna.7.1.10256. [DOI] [PubMed] [Google Scholar]
- 82.Burgess H.M., Gray N.K. An integrated model for the nucleo-cytoplasmic transport of cytoplasmic poly(A)-binding proteins. Commun. Integr. Biol. 2012;5:243–247. doi: 10.4161/cib.19347. [DOI] [PMC free article] [PubMed] [Google Scholar]