Abstract
Peptide methionine sulfoxide reductase (PMSR) is a ubiquitous enzyme that repairs oxidatively damaged proteins. In Arabidopsis (Arabidopsis thaliana), a null mutation in PMSR2 (pmsr2-1), encoding a cytosolic isoform of the enzyme, exhibited reduced growth in short-day conditions. In wild-type plants, a diurnally regulated peak of total PMSR activity occurred at the end of the 16-h dark period that was absent in pmsr2-1 plants. This PMSR activity peak in the wild-type plant coincided with increased oxidative stress late in the dark period in the mutant. In pmsr2-1, the inability to repair proteins resulted in higher levels of their turnover, which in turn placed an increased burden on cellular metabolism. This caused increased respiration rates, leading to the observed higher levels of oxidative stress. In wild-type plants, the repair of damaged proteins by PMSR2 at the end of the night in a short-day diurnal cycle alleviates this potential burden on metabolism. Although PMSR2 is not absolutely required for viability of plants, the observation of increased damage to proteins in these long nights suggests the timing of expression of PMSR2 is an important adaptation for conservation of their resources.
INTRODUCTION
Protein oxidation is a frequent occurrence in cells and results in fragmentation or carbamylation of the peptide backbone (Vogt, 1995). The sulfur-containing amino acids especially are susceptible to oxidation, of which the oxidation of Met residues in proteins to Met sulfoxide (MetSO) is a well-understood posttranslational modification.
Met oxidation is caused in vivo by a variety of reactive molecules that are byproducts of aerobic metabolism (Brot and Weissbach, 1991). Met oxidation causes disruption in protein structure and function and has been implicated in a number of diseases, the onset of aging/senescence, and oxidative stress in bacteria, plants, insects, and mammals (Vogt, 1995; Moskovitz et al., 1996; Berlett and Stadtman, 1997; Gabbita et al., 1999).
The removal of oxidized proteins by proteases and their replacement by de novo synthesis as a response to oxidation of peptide residues in animals and plants has been extensively characterized (Davies, 1993; Grune and Davies, 1997). However, in addition to the turnover of damaged proteins, the enzyme peptide methionine sulfoxide reductase (PMSR; EC 1.8.4.6) can repair such proteins by catalyzing the reduction of MetSO back to Met. In recent years, the enzymatic repair of oxidized proteins has been recognized to be a key process in a wide range of aerobic organisms including bacteria, plants, and humans (Moskovitz et al., 1996, 1997, 2001). As a consequence, it has been suggested that PMSR should be part of the minimal gene set of organisms (El Hassouni et al., 1999). Evidence for the protective role of PMSR has arisen from null mutations in a diversity of organisms, such as Escherichia coli, yeast (Saccharomyces cerevisiae), and mice, and may be nonspecific in nature because the enzyme shows no substrate specificity (Moskovitz et al., 1995, 1997, 2000; Shacter, 2000). Despite the apparent universality of oxidative damage to proteins, the formation of protein MetSOs has also been shown to have specific effects. For example, their formation is associated with signal transduction processes under conditions of oxidative stress or aging, modulating both K+ channel function and activation of a plasma membrane Ca2+-ATPase (Ciorba et al., 1997; Gao et al., 1998; Tang et al., 2001). The redox regulation of protein function via Met sulfoxidation is thought to have a more long-lasting response compared with Cys oxidation. Reduction of Cys disulfide bridges occurs rapidly as soon as the cellular redox balance is regained, whereas the reversal of MetSO is enzyme dependent (Hoshi and Heinemann, 2001).
Interestingly, plants are the only organisms containing multiple PMSR isoforms. Four PMSR genes, three encoding presumed cytosolic isoforms (PMSR1 to PMSR3, At5g61640.1, At5g07460.1, and At5g07470.1, respectively; see Supplemental Figure 1 online) and one encoding a plastidial isoform (PMSR4, At4g25130.1, formerly called pPMSR; see Supplemental Figure 1 online) have been identified and localized in Arabidopsis (Arabidopsis thaliana; see Supplemental Table 1 online; Sadanandom et al., 2000). The PMSR2 gene analyzed in this study is the least expressed of three cytosolic PMSR homologs (see Supplemental Table 1 online). However, in this article, we report how a transposon insertion into the gene resulted in reduced growth and slower development of the plants under short-day conditions (8-h photoperiod). This was associated with the absence of a strong peak of expression toward the end of each dark period in the diurnal cycle. These observations provided support for the hypothesis that the role of the PMSR2 isoform in Arabidopsis is to repair oxidized proteins at a specific point in a short-day photoperiod and thus act as a means of minimizing protein turnover under conditions of limited energy provision. Furthermore, these data point to a considerable potential for oxidative damage to proteins under short photoperiods, an environmental condition which, to our knowledge, has not hitherto been regarded as particularly stressful.
RESULTS
Identification of a Null Mutation in the PMSR2 Gene
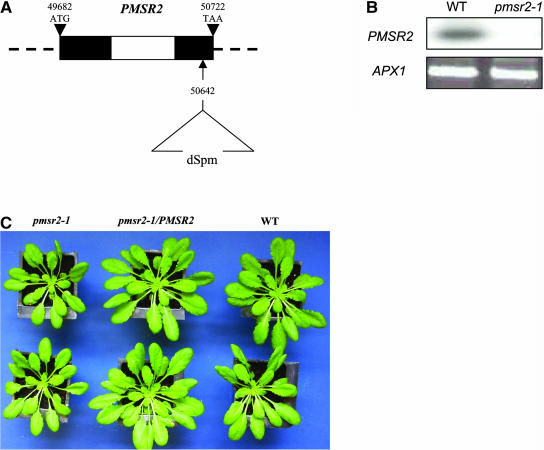
The pmsr2-1 mutation was identified through PCR-based screening of the Sainsbury Laboratory Arabidopsis Em/Spm transposon (SLAT) population (Tissier et al., 1999). A transposon insertion was located in the second exon of the gene, 80 bp upstream of the translational stop codon (Figure 1A). Homozygous pmsr2-1 were identified and shown to have no detectable PMSR2 transcript using a sensitive PCR procedure. Therefore, the transposon insertion had created a null mutation in PMSR2 (Figure 1B). Total PMSR activity was reduced by 31% in the pmsr2-1 mutant (wild-type, 0.26 ± 0.09 pmol m−2 s−1 reduced MetSO, versus pmsr2-1, 0.18 ± 0.05 pmol m−2 s−1 reduced MetSO; n = 4). The mutant was generally smaller when grown under short-day conditions (8-h photoperiod) but showed otherwise no morphological differences to wild-type plants (Table 1; Figure 1C). This size difference between pmsr2-1 and wild-type plants could not be observed when plants were grown under long-day conditions (16-h photoperiod; data not shown).
Figure 1.
The Localization and Characterization of the PMSR2 Knockout.
(A) Genomic organization of the PMSR2 gene in Arabidopsis on chromosome 5 (BAC clone K11J9) and location of the dSpm insertion in the pmsr2-1 mutant. The black and white sections are exons and introns, respectively.
(B) 3′ RACE analysis of the Arabidopsis PMSR2 gene in the wild type and the pmsr2-1 mutants. PCR products were analyzed by probing with a PMSR2 full-length cDNA. Primers for the ASCORBATE PEROXIDASE1 (APX1) gene were used as a control for cDNA synthesis.
(C) Phenotype of the pmsr2-1 mutant compared with the wild-type and pmsr2-1/PMSR2 plants. Plants were grown under short-day conditions for 4 weeks.
Table 1.
Growth Parameters of 4-Week-Old Wild-Type, pmsr2-1/PMSR2, and pmsr2-1 Plantsa
| Fresh Weight (g) | Dry Weight (g) | Leaf Number | |
|---|---|---|---|
| WT | 0.3107 ± 0.079 | 0.0281 ± 0.0092 | 15.8 ± 1.52 |
| pmsr2-1/PMSR2 | 0.2987 ± 0.056 | 0.0294 ± 0.0087 | 16.5 ± 1.55 |
| pmsr2-1 | 0.2286 ± 0.097 | 0.0207 ± 0.00808 | 13.1 ± 1.36 |
n = 20 ± sd.
The pmsr2-1 mutant was no more or less sensitive than wild-type plants to a 10-fold excess of light stress, paraquat treatment, drought, or infection by Cauliflower mosaic virus or compatible (disease-causing) Pseudomonas syringae (data not shown).
Induction of Total PMSR Activity during a 16-h Dark Period
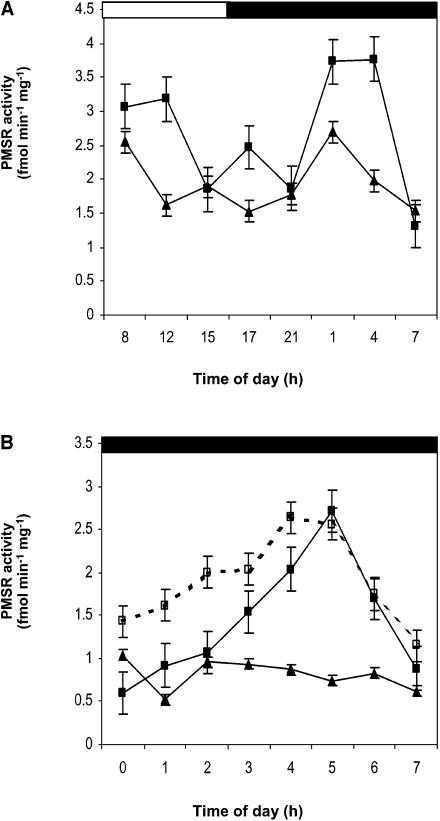
To determine if the null mutation in PMSR2 had the same effect on PMSR activity throughout a diurnal cycle, total PMSR activity was measured in short day–grown plants during a 24-h period. In wild-type plants, there were two peaks of total activity, one during the light period, which was smaller in the pmsr2-1 plants (Figure 2A). The second was a much larger increase in total PMSR activity after 11 h of darkness in wild-type plants that declined before the onset of the light period (Figures 2A and 2B). By contrast, the pmsr2-1 mutant completely lacked this second large peak of PMSR activity (Figures 2A and 2B). Complementation of pmsr2-1 was achieved with a 3.1-kb genomic clone containing the PMSR2 open reading frame and 1-kb sequences upstream of the 5′ and downstream of the 3′ untranslated regions, respectively. Complemented mutant plants (pmsr2-1/PMSR2) had PMSR activity during the dark period (Figure 2B) and restored growth rates (Table 1; Figure 1C). The steady state levels of PMSR2 mRNA over a diurnal cycle matched the PMSR activity in the wild type and was absent in the pmsr2-1 (see Supplemental Figure 1 online). The remaining members of the gene family showed no differences in the steady state level of their transcripts over a diurnal period (see Supplemental Figure 1 online).
Figure 2.
Total PMSR Activity Is Reduced in the pmsr2-1 Mutant.
(A) Total PMSR activity during a 24-h period in the wild type (closed squares) and pmsr2-1 mutant (closed triangles).
(B) PMSR activity during the second half of a 16-h dark period taken at the end of the 16-h dark period in the wild type (closed squares), pmsr2-1 mutant (closed triangles), and pmsr2-1/PMSR2 (open squares).
The Lack of PMSR Activity and Increased Oxidative Stress during the Dark Period
Because PMSR null mutations caused increased oxidative stress in other organisms (see Discussion), the degree of oxidative stress was investigated using three parameters. Changes in hydrogen peroxide (H2O2), lipid peroxidation, and the redox state of glutathione, a key cellular redox buffer (Noctor et al., 2002), were analyzed.
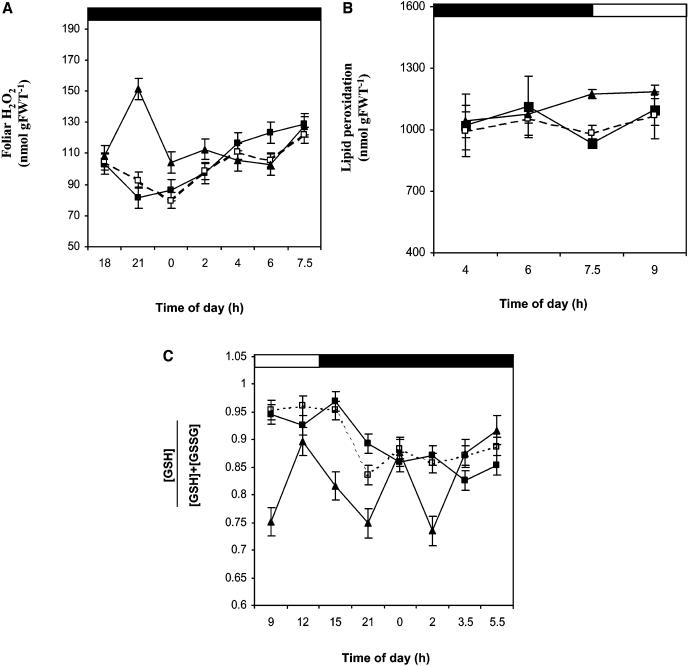
Foliar H2O2 levels were increased 1.5-fold in the pmsr2-1 plants compared with wild-type and pmsr2-1/PMSR2 plants during the early dark period (Figure 3A), whereas lipid peroxidation was increased at the end of the dark period and into the dark-to-light transition (Figure 3B). The early accumulation of a key reactive oxygen species (ROS) and the later accumulation of lipid peroxidation products strongly indicated that the pmsr2-1 plants suffered increased oxidative stress throughout the dark period compared with wild-type and pmsr2-1/PMSR2 controls. Perturbations in ROS metabolism were also reflected in highly variable fluctuations in the redox status of the glutathione pool (Figure 3C) compared with wild-type and pmsr2-1/PMSR2 plants, in which this parameter declined steadily throughout the period of the experiment. In contrast with the glutathione redox state, that of the ascorbate pool showed no significant difference between the wild type and pmsr2-1 mutant throughout the diurnal cycle (data not shown).
Figure 3.
The pmsr2-1 Mutant Shows Signs of Increased Oxidative Stress during the Dark.
(A) Foliar H2O2 measured during a 24-h period in the wild type (closed squares), pmsr2-1/PMSR2 (open squares), and pmsr2-1 mutant (closed triangles).
(B) Lipid peroxidation during the dark-to-light transition in the wild type (closed squares), pmsr2/PMSR2 (open squares), and pmsr2-1 mutant (closed triangles).
(C) Redox state ([GSH]/ [GSH] + [GSSG]) of the glutathione pool during a 24-h period in the wild type (closed squares), pmsr2-1/PMSR2 (open squares), and pmsr2-1 mutant (closed triangles).
Elevated Levels in ROS and Oxidative Damage in Relation to Increased Respiration and Alterations in Carbon Metabolism
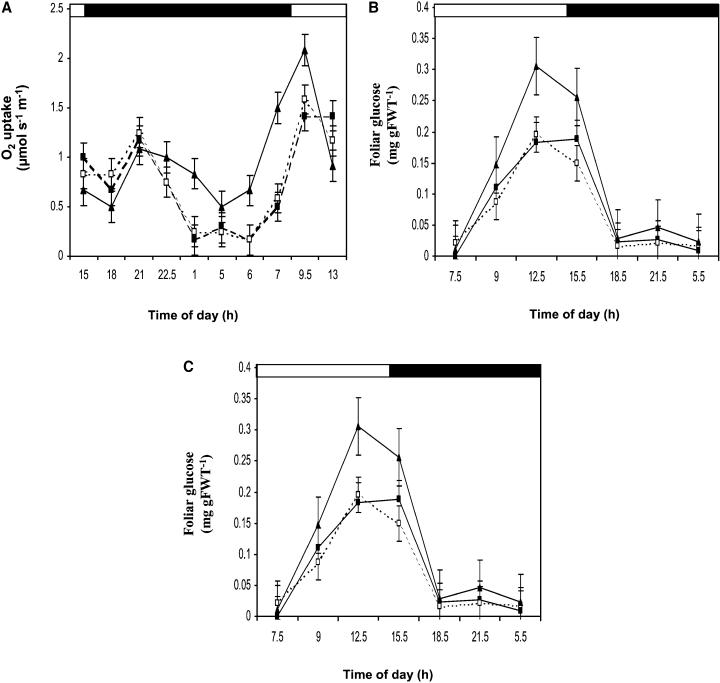
One major source of ROS production in the dark is mitochondrial respiration (Affourtit et al., 2001). Therefore, oxygen exchange of leaves was analyzed during the dark. A 1.5-fold difference in foliar respiration rates was observed in the pmsr2-1 mutant compared with wild-type and pmsr2-1/PMSR2 plants during the dark period (Figure 4A). By contrast, throughout the light period there was a 20% reduction in respiration rates in the mutant compared with the wild-type and pmsr2-1/PMSR2 plants (Figure 4A). Analysis of soluble sugars and starch showed a twofold increase in glucose levels during the light period in the pmsr2-1 plants, which declined during the dark period to wild-type and pmsr2-1/PMSR2 levels (Figures 4B and 4C). Changes in metabolism were fully restored to wild-type levels in pmsr2-1/PMSR2 plants, demonstrating that these observations were not as a result of any unrelated mutation in the pmsr2-1 background. In addition, fructose and sucrose levels showed no significant difference between wild-type, pmsr2-1/PMSR2, and pmsr2-1 plants (data not shown).
Figure 4.
The pmsr2-1 Mutant Has Altered Sugar Metabolism.
(A) Analysis of respiration (O2 uptake) during a 24-h period in the wild type (closed squares), pmsr2-1/PMSR2 (open squares), and pmsr2-1 mutant (closed triangles).
(B) Foliar glucose levels during a 24-h period in the wild type (closed squares), pmsr2-1/PMSR2 (open squares), and pmsr2-1 mutant (closed triangles).
(C) Foliar starch levels during a 24-h period in the wild type (closed squares), pmsr2-1/PMSR2 (open squares), and pmsr2-1 mutant (closed triangles).
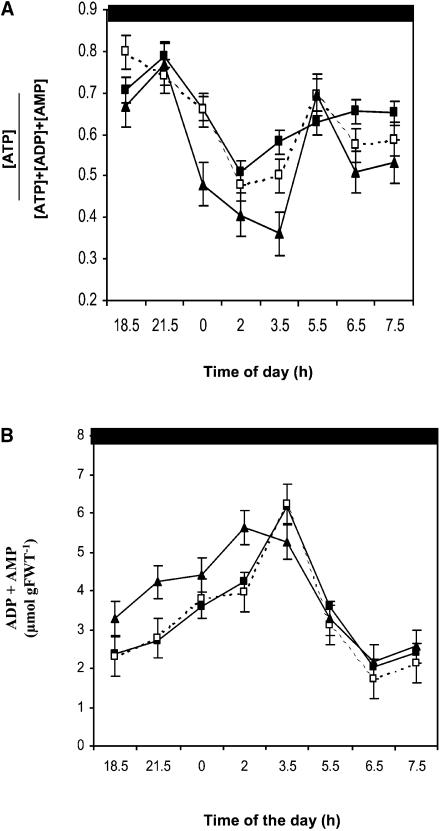
Together, these data indicated that the pmsr2-1 mutant had an approximately twofold increased rate of carbon metabolism in the dark associated with an altered diurnal pattern of photosynthesis and an increased respiration rate in the dark (see Discussion). Therefore, adenine nucleotide levels were examined in the mutant compared with the wild-type and pmsr2-1/PMSR2 plants over the diurnal cycle (Figure 5A).
Figure 5.
The pmsr2-1 Mutant Has a Reduced Energy Charge Due to Increased ADP Levels.
(A) Energy charge ([ATP]/[ATP]+[ADP]+[AMP]) during a 24-h period in the wild type (closed squares), pmsr2-1/PMSR2 (open squares), and pmsr2-1 mutant (closed triangles)
(B) ADP plus AMP levels during a 24-h period in the wild type (closed squares), pmsr2-1/PMSR2 (open squares), and pmsr2-1 mutant (closed triangles).
ATP levels were constant throughout the circadian cycle with no difference between the wild type, pmsr2-1/PMSR2, and pmsr2-1 mutant (data not shown). However, ADP and AMP levels were generally increased in the pmsr2-1 plants at the beginning of the dark period (Figure 5B). This was the cause of a decrease in energy charge ([ATP]/[ATP]+[ADP]+[AMP]; Giersch et al., 1980) in the pmsr2-1 plants at the beginning of the dark period, but they attained the same levels as did the wild-type and pmsr2-1/PMSR2 plants at the end of the dark period (Figure 5A).
Changes in Dark Metabolism and Alteration in Photosynthetic Electron Transport
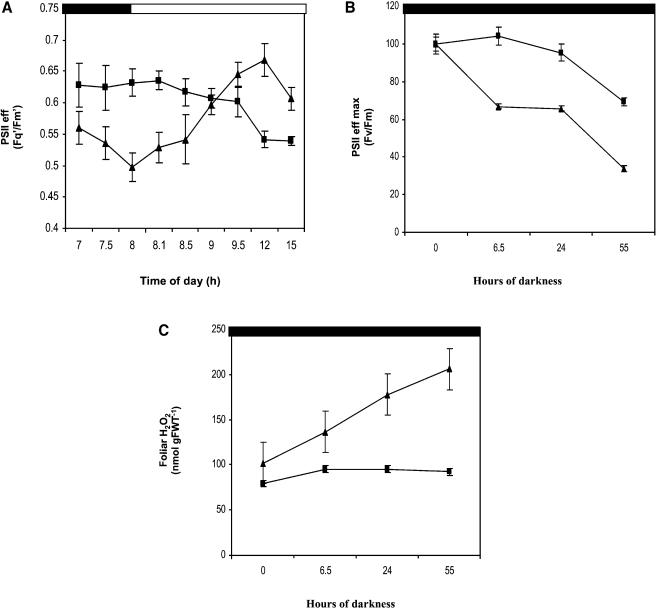
To determine whether the changes in respiration and sugar levels had an effect on photosynthetic electron transport, chlorophyll fluorescence measurements were performed during the light period. There was no difference in the maximum photosystem II (PSII) efficiency between the wild-type and pmsr2-1 mutant plants (data not shown), taken as an indication that damage to PSII was not occurring in the mutant. At the onset of the light period, the operating efficiency of PSII (Fq′/Fm′) was lower in the mutant than in wild-type plants, which was reversed within 1 h of illumination. Thereafter, Fq′/Fm′ was significantly increased during the remainder of the light period in the mutant compared with the wild-type plants (Figure 6A).
Figure 6.
Photosynthetic Capacity Is Altered in the pmsr2-1 Mutant.
(A) Operating efficiency of PSII (Fq′/Fm′) during the dark-to-light transition in the wild type (closed squares) and pmsr2-1 mutant (closed triangles).
(B) The effect of prolonged dark periods on photosynthesis was analyzed measuring maximum efficiency of PSII (Fv/Fm) during a 55-h dark period in the wild type (closed squares) and pmsr2-1 mutant (closed triangles).
(C) The effect of prolonged dark periods on H2O2 levels during a 55-h dark period in the wild type (closed squares) and pmsr2-1 mutant (closed triangles).
Extended Dark Periods and Oxidative Stress in pmsr2-1 Plants
To confirm that oxidative stress increased during a dark period, pmsr2-1 plants were exposed to an artificially long dark period of 55 h. Under these conditions, the maximum efficiency of PSII (Fv/Fm) decreased during extended dark period from 0.82 to 0.5 in both the wild type and mutant. However, the operating efficiency of leaves of pmsr2-1 plants declined by 35% more than the wild-type plants during the course of the experiment (Figure 6B). Over the same period, H2O2 levels increased up to 2.5-fold in the pmsr2-1 mutant compared with the wild type (Figure 6C). Plants were transferred back to a short-day cycle after the extended dark period, and the pmsr2-1 plants started flowering within 2 d.
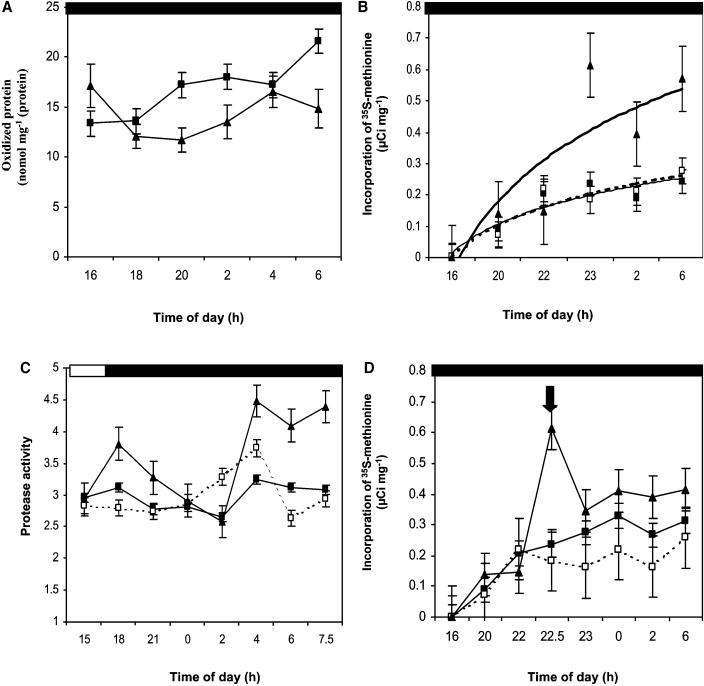
Oxidative Stress during the Dark Period Caused Increased Protein Oxidation and Protein Turnover
Analyzing the content of protein oxidation in the wild-type and mutant plants tested the hypothesis that PMSR is essential for the repair of oxidized proteins. Generally, oxidized proteins, if not repaired, are replaced with functional proteins causing protein turnover. Therefore, a proteasome inhibitor (MG132) was fed through the petiole of detached Arabidopsis leaves to prevent the turnover of oxidized proteins. During the treatment with MG132, protein oxidation increased significantly in the pmsr2-1 plants but not in the wild type during the dark period (Figure 7A). It should be realized that this is a measure of general protein oxidation and not Met sulfoxidation. As a consequence of these preliminary experiments with the proteasome inhibitor, we predicted that increased protein breakdown and synthesis should occur in the pmsr2-1 mutant during the dark period. Therefore, proteinase activity and the incorporation and elimination of 35S-Met into proteins during the dark period were analyzed (Figure 7). A higher incorporation of l-[35S]-Met into proteins suggested increased protein synthesis (Figure 7B). At the same time, proteinase activity increased significantly in the pmsr2-1 mutant during the dark period (Figure 7C), resulting in an increased elimination of 35S-Met during the chase experiment (Figure 7D). Together, these results suggested that there was increased protein turnover in the pmsr2-1 mutant compared with the wild type in the dark period.
Figure 7.
Evidence for Increased Protein Turnover in the pmsr2-1 Mutant.
(A) Protein oxidation (carbamylation) after MG132 treatment during the dark in the wild type (closed squares) and pmsr2-1 mutant (closed triangles).
(B) Incorporation of 35S-Met into total protein during the dark in the wild type (closed squares), pmsr2-1/PMSR2 (open squares), and pmsr2-1 mutant (closed triangles).
(C) Protease activity measurements during a 24-h period in the wild type (closed squares), pmsr2-1/PMSR2 (open squares), and pmsr2-1 mutant (closed triangles).
(D) Elimination of 35S-Met from total protein during the dark period in the wild type (closed squares), pmsr2-1/PMSR2 (open squares), and pmsr2-1 mutant (closed triangles). The arrow indicates the transfer of 35S-labeled leaves into nonlabeled solution.
DISCUSSION
Plants appear to be unique among aerobic organisms in possessing a small multigene family encoding PMSR isoforms (Sadanandom et al., 1996, 2000). Therefore, it was surprising to find that the loss of cytosolic PMSR2 could not be compensated for by the presence of two very similar isoforms present in the same subcellular compartment (see supplemental data online). Total PMSR activity was induced in wild-type Arabidopsis but not the pmsr2-1 null mutant at the end of a 16-h dark period (Figure 2). It was concluded that the PMSR2 isoform was responsible for the activity peak during the dark period because replacing the gene by transformation resulted in restoration of the peak of PMSR activity absent in the null mutant (Figure 2B). A failure to induce dark-period PMSR2 activity was associated with a reduced growth habit under short-day conditions (Table 1; Figure 1C). Complementation of the pmsr2-1 mutant with the wild-type gene resulted in a restoration of the growth and all of the other key parameters discussed below.
The starting point for considering the role of PMSR2 is that the pmsr2-1 mutant showed increased oxidative stress in the dark phase of short-day conditions. The increased production of H2O2, lipid peroxidation, and the erratic variations in the glutathione pool support this suggestion (Figure 3). Furthermore, the production of H2O2 could be raised to 1.5-fold higher than wild-type levels if pmsr2-1 plants were kept in extended dark periods (up to 55 h; Figure 6C). A further indication of increased stress during long dark periods in the pmsr2-1 plants was the production of shoots and flowers within 2 d after transfer back to their short photoperiod conditions (see Results). In support of these observations, knockout mutants in the single PMSR genes of E. coli, yeast, and mice show an increased sensitivity to oxidative stress, lower survival rates, and high levels of protein-bound MetSO (Moskovitz et al., 1995, 1997, 2001). However, the pmsr2-1 mutant was not more sensitive to externally applied oxidative stress (see Results), and it is important to realize that the loss of a repair function for proteins does not directly explain the observed increases in ROS, lipid peroxidation, or the behavior of the glutathione pool. We considered it more likely that the increase in oxidative stress parameters in the pmsr2-1 mutant may have been an indirect consequence of a failure to reduce MetSO in cytoplasmic proteins.
In the dark, the production of ROS would occur as a byproduct of heterotrophic metabolism, the most likely source being mitochondrial electron transport to O2 (Affourtit et al., 2001). The increased rates of respiration in pmsr2-1 and consumption of glucose and starch (Figure 4) make this the likely source of the observed increase in oxidative stress parameters (Figure 3). It is possible that respiration could be a source of oxidative stress in the pmsr2-1 mutant and in wild-type plants under certain conditions. Support for this argument comes from two sources. First, the feeding of increased levels of glucose to heterotrophic Nicotiana tabacum (tobacco) suspension culture cells led to increased respiration rates and a measurable increase in oxidative stress (Bowler et al., 1989). From the data presented here on pmsr2-1, calculations of the rate of glucose disappearance (2.3-fold higher in the mutant) agree with the rates of respiration (1.9-fold higher in the mutant). The amount of H2O2 produced under such circumstances would account for 30% of O2 consumption (calculated from the data in Figures 3A and 4A, assuming that 510 mg is 0.2 m2 of leaf). Therefore, together these data support the notion that the increased oxidative stress in the mutant can be explained by the increased ROS arising as a consequence of stimulated starch catabolism.
A second line of evidence comes from studies on dark-induced senescence in plants. Senescence is an oxidative process that shows an increase in the generation of ROS and causes deterioration of cellular metabolism (Pastori and Del Rio, 1997; Del Rio et al., 2003). Leaf senescence can also be induced by prolonged dark periods, and it has been shown that at physiological and molecular levels both natural and dark-induced senescence have many aspects in common (Pastori and Del Rio, 1994a, 1994b, 1997). Similarly, senescence of root nodules of legumes can be induced by several days of dark treatment, leading to the accumulation of oxidized proteins and alterations in carbon metabolism and respiration (Matamoros et al., 1999). Furthermore, it has also been shown that the expression of some dark-inducible genes is related to sugar starvation occurring in leaf cells during dark periods (Fujiki et al., 2001). All these phenomena (increased protein oxidation and alterations in carbon metabolism and respiration) occur in the pmsr2-1 mutant during the dark period (Figure 4).
During the dark, metabolic processes in a plant cell are comparable to processes occurring in nonphotosynthesizing eukaryotic cells, and parallels can be drawn to well-characterized systems in bacteria, yeast, and mammals. Recent evidence has shown that the only human PMSR isoform is located in the mitochondrion, the major site of ROS production (Hansel et al., 2002). Mitochondrial respiration is a vital factor in ROS production and increased oxidative damage during aging in mammals (Lee and Wei, 1997). Especially in humans, the connection between the generation of ROS, Met sulfoxidation in proteins, and the onset of aging and diseases has been clearly demonstrated. The occurrence of these deleterious processes usually corresponds to lower PMSR activities (Moskovitz et al., 1996; Chao et al., 1997; Gao et al., 1998; Gabbita et al., 1999).
Oxidative damage to proteins appears to be of a general nature, in which the specificity of substrates cannot be easily determined, and PMSR activity measurements using an unspecific substrate suggest that complex protein folding is not required (see Methods; Abrams et al., 1981; Shacter, 2000). This means that in wild-type plants in extended dark periods, the oxidized Met residues in a range of cytoplasmic proteins are reduced by PMSR2, thus conserving their functions. The consequence of a failure to repair oxidized proteins is that the pmsr2-1 mutant would have to replace such proteins with new ones. It is clearly capable of doing so because it completes a normal life cycle in short-day conditions, albeit with slightly reduced growth rate (Table 1; Figure 1C). The increased levels of protease activity (Figure 7C) and increased incorporation or depletion of 35S-Met into protein (Figures 7B and 7D) support the argument that there were increased rates of protein turnover in the pmsr2-1 mutant in the dark period. Increased protein turnover would require increased rates of metabolism in the dark to support the higher demand in energy. The elevated rates of respiration (Figure 4A), increased consumption of both glucose and starch (Figures 4B and 4C), and the depressed foliar energy charge over a 24-h cycle (Figure 5A) support this view. Thus, the increased oxidative stress in pmsr2-1 brought about by increased rates of respiration would be an indirect consequence of the need to increase protein turnover in the absence of a dark-induced PMSR2-catalyzed protein repair mechanism.
In summary, these observations on the pmsr2-1 mutant strongly suggest that in wild-type plants the repair of oxidized proteins in extended night periods by the timely induction of PMSR2 activity conserves the energy and carbon resources of the plant. Such resources would otherwise be needed to support an increased rate of protein turnover. In the mutant, the increased respiration rates would promote a higher degree of oxidative stress, a situation that does not arise in wild-type plants under short days as a consequence of the function of PMSR2. In contrast with the metabolic requirements for the replacement of damaged proteins, PMSRs require only a source of reductant mediated by their interaction with thioredoxins (Lowther et al., 2000; Moskovitz et al., 2000; Boschi-Müller et al., 2001). Thus, despite the pmsr2-1 mutant being viable in the laboratory, we propose that PMSR2 is an essential fitness component for plants in the external environment. The requirement for PMSR2 could be part of a seasonal adaptation to extend viability of the plant in increasing length of dark periods, thus helping to control the onset of senescence. This might explain why Arabidopsis and other plants harbor a multigenic family encoding PMSR isoforms (see supplemental data online) because different isoforms could be required for a range of distinct physiological states.
METHODS
Plant Material
Arabidopsis (ecotype Columbia [Col-0]) were grown to mature rosette stage (5 weeks after germination) under controlled environmental conditions (PPFD 150 μmol m−2 s−1) with either a 16-h photoperiod (long day) or 8-h photoperiod (short day) at 25°C and a relative humidity of 50%. The pmsr2-1 mutant was grown under the same conditions, and selection for phosphinothricin resistance was performed by spraying twice per week with BASTA (2 mg L−1).
Plant Growth Measurements
The leaf material of 20 plants was harvested, and residual soil and water was removed carefully. The leaves were weighed (fresh weight), and the material was frozen in liquid nitrogen and freeze-dried overnight. The weight was determined, and plants were further dried at 120°C until the weight remained constant (dry weight).
PCR and DNA Gel Blot Analysis of the Transposon Insertion
DNA was extracted using the DNeasy plant mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Twenty nanograms was used in a restriction ligation, followed by inverse PCR using transposon-specific primers to identify the insertion of the transposon (dSpm1, 5′-CTTATTTCAGTAAGAGTGTGGGGTTTTGG-3′; and dSpm5, 5′-CGGGATCCGACACTCTTTAATTAACTGACACTC-3′). For DNA gel blot analysis, 20 μg of DNA was digested with BstYI and HindIII and run on a vertical 0.8% agarose gel. The DNA was transferred overnight onto nitrocellulose and probed with a bar (Hellens et al., 2000) probe to analyze transposon copy number.
RNA Extraction and 3′ RACE PCR
Total RNA was extracted using the RNeasy plant mini kit (Qiagen) according to the manufacturer's instructions. A total of 5 μg of RNA was used for cDNA synthesis using a Poly(A) specific primer (rapid amplification of cDNA 3′ ends [RACE] procedure; Frohman et al., 1988). PCR was performed using a PMSR2-specific primer (cpm2a, 5′-TGGATCCTTCTCCCAATCGCAC-3′). PCR products were separated on an agarose gel and transferred onto a Hybond N+ membrane (Amersham Biosciences, Amersham, UK) and probed with a PMSR2 cDNA probe.
Complementation of the PMSR2 Null Mutation by Transformation of Arabidopsis
A 3.1-kb genomic fragment containing the PMSR2 open reading frame and 1000 bp at the 3′ and 5′ untranslated region was synthesized by PCR (gen1, 5′-TCATCATATCGTTTGCC-3′; and gen3, 5′-AATCTCCTCGTCCAATAGTTG-3′) and cloned into pGreen 0029 (Hellens et al., 2000). The pmsr2-1 plants were transformed using the floral dip method (Clough and Bent, 1998). The transformants were selected on MS media containing kanamycin (50 μg/mL). Transformants were analyzed by 3′ RACE PCR and total PMSR activity measurements.
Leaf Chlorophyll Fluorescence Measurements
Chlorophyll a fluorescence was measured using a fluorescence monitoring system (FMS1; Hansatech Instruments, Kings Lynn, UK) connected to Windows software (Microsoft, Reading, UK). The equipment was used according to the manufacturer's instructions. The parameters Fv/Fm and Fq′/Fm′ were calculated as described in Baker et al. (2001). Fv/Fm and Fq′/Fm′ provide an estimate of the maximum efficiency and operating efficiency of PSII photochemistry, respectively.
H2O2, Lipid Peroxidation, and Enzyme Activities
A total of 0.1 g plant material was extracted in 1 mL of ice-cold buffer containing 50 mM Hepes, pH 7.5, 50 mM MgCl2, and 1 mM EDTA using a mortar and pestle. The extract was centrifuged at 12,000g, and the supernatant was used directly for enzyme assays and analysis of H2O2 and lipid peroxide content. H2O2 was assayed as described in Jimenez et al. (2002). Lipid peroxidation was measured by a colorimetric assay for the decomposition products of oxidized polyunsaturated fatty acids, malondialdehyde, and 4-hydroxyalkenals using the Bioxytech LPO-586 Assay (Oxis International, Portland, OR) following the manufacturer's instructions.
Total PMSR Activity
Total PMSR activity was measured in plant crude extracts using an artificial substrate N-acetyl l-[methyl-14C] MetSO. The synthesis of the substrate and the procedures for the enzyme assay were according to a method by Brot and Weissbach (1982). A 300-μL reaction containing 270 pmol N-acetyl 14C L-MetSO, 25 mM Tris-HCl, pH 8.0, 15 mM DTT, and 10 mM MgCl2 was started with 50 μL of plant extract. The reaction was incubated at 37°C and stopped after 0, 20, and 60 min by the addition of 0.5M HCl. The N-acetyl 14C Met product was extracted in 3 mL of ethyl acetate, and the ethyl acetate phase was assayed for radioactivity in a liquid scintillation counter (Wallac 1219 RACKBETA liquid scintillation counter; Turku, Finland). PMSR activity was expressed as pmol min−1 mg−1 protein. Protein concentration was estimated by the method of Bradford (Bradford, 1976).
Metabolite Analysis
A total of 0.2 g of plant material was extracted in 1.5 mL of 0.7 M HClO4 (soluble sugars) or 10% (v/v) HClO4 (nucleotides) and left for 30 min on ice. The extract was centrifuged at 12,000g, and the supernatant was neutralized with 2M KOH/0.4M KCl to pH 6 to 7. The resulting potassium HClO4 precipitate was pelleted by centrifugation as above, and the supernatant was analyzed for soluble sugars, starch, and nucleotides as described by Stitt et al. (1989).
Respiration
Respiration was analyzed using an LD2 leaf disc oxygen electrode (Hansatech Instruments) based on a design by Delieu and Walker (1981). The consumption of oxygen was measured over a 20-min time period. A 1-mV change equals the consumption of 10 μmol O2. Respiration rates are expressed as μmol O2 m−2 s−1.
Protein Oxidation and Proteinase Activity
Protein oxidation (carbonyl content) was measured according to a method described by Prasad (1996). Proteinase activity was measured using 0.2% (w/v) azocasein as substrate according to a method described by Galleschi et al. (2002). One unit of activity was defined as the amount of enzyme required to produce an absorbance change of 1 in a 1-cm cuvette. Proteinase activity was expressed as unit mg−1 protein.
In Vitro Labeling of Plant Proteins with 35S-Met
Detached leaves were incubated in water containing 51.8 kBq/μL label (L-35S-Met; Amersham Biosciences; specific activity 37 TBq/mmol) for 8 h at the beginning of the dark period. The labeled leaves were subsequently placed into water during the remaining 8 h of the dark period, and samples were taken every 2 h. Alternatively, leaves were labeled during the entire dark period, and samples were taken twice hourly. Proteins of the labeled leaves were extracted in 0.1 M sodium phosphate buffer, pH 7.0, containing a protease inhibitor cocktail (Sigma-Aldrich, Poole, UK). Proteins were precipitated by centrifugation at 3000g at 4°C after incubation in four volumes of acetone overnight at −20°C. The pellet was washed twice with ice-cold acetone and was then air-dried and resuspended in 0.1 M sodium carbonate and 0.1 M DTT. Radioactivity was determined by liquid scintillation counting. Total protein concentration was estimated as described above.
Glutathione Analysis
Glutathione was extracted in 0.1 M HCl by grinding 0.1 g of frozen leaf material in 1 mL of acid and incubating the extract on ice for 30 min. After centrifugation at 20,000g for 10 min, the supernatant was used to measure the content of glutathione (as GSH) by HPLC using the monobromobimane derivatization method (Newton et al., 1981) as described by Creissen et al. (1999). GSSG was analyzed by treatment with DTT and determined as total glutathione as described by Creissen et al. (1999).
Supplementary Material
Acknowledgments
U.B. is grateful for the support by a John Innes Foundation studentship. We thank Jonathan Jones (Sainsbury Laboratory, Norwich, UK) for the provision of the SLAT library. This work was supported by a Biotechnology and Biological Sciences Research Council Core Strategic grant to the John Innes Centre.
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Philip M. Mullineaux (phil.mullineaux@bbsrc.ac.uk).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.015818.
References
- Abrams, W.R., Weinbaum, G., Weissbach, L., Weissbach, H., and Brot, N. (1981). Enzymatic reduction of oxidized a1-proteinase inhibitor restores biological activity. Proc. Natl. Acad. Sci. USA 78, 7483–7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affourtit, C., Krab, K., and Moore, A.L. (2001). Control of plant mitochondrial respiration. Biochim. Biophys. Acta 1504, 58–69. [DOI] [PubMed] [Google Scholar]
- Baker, N.R., Oxborough, K., Lawson, T., and Morisson, J.I. (2001). High resolution imaging of photosynthetic activities of tissues, cells and chloroplasts in leaves. J. Exp. Bot. 52, 615–621. [PubMed] [Google Scholar]
- Berlett, B.S., and Stadtman, E.R. (1997). Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 372, 20313–20316. [DOI] [PubMed] [Google Scholar]
- Boschi-Müller, S., Azza, S., and Branlant, G. (2001). E. coli methionine sulfoxide reductase with a truncated N terminus or C terminus, or both, retains the ability to reduce methionine sulfoxide. Protein Sci. 10, 2272–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler, C., Alliotte, T., De Loose, M., Van Montagu, M., and Inze, D. (1989). The induction of manganese superoxide dismutase in response to stress in Nicotiana plumbaginifolia. EMBO J. 8, 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brot, N., and Weissbach, H. (1982). Reduction of N-acetyl methionine sulfoxide: A simple assay for peptide methionine sulfoxide reductase. Anal. Biochem. 122, 291–294. [DOI] [PubMed] [Google Scholar]
- Brot, N., and Weissbach, H. (1991). Biochemistry of methionine sulfoxide residues in proteins. Biofactors 3, 91–96. [PubMed] [Google Scholar]
- Chao, C.-C., Ma, Y.-S., and Stadtman, E.R. (1997). Modification of protein surface hydrophobicity and methionine oxidation by oxidative systems. Proc. Natl. Acad. Sci. USA 94, 2969–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciorba, M.A., Heinemann, S.H., Weissbach, H., Brot, N., and Hoshi, T. (1997). Modulation of potassium channel function by methionine oxidation and reduction. Proc. Natl. Acad. Sci. USA 94, 9932–9937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Creissen, G., Firmin, J., Fryer, M., Kular, B., Leyland, N., Reynolds, H., Pastori, G., Wellburn, F., Baker, N., Wellburn, A., and Mullineaux, P. (1999). Elevated glutathione biosynthetic capacity in the chloroplasts of transgenic tobacco plants paradoxically causes increased oxidative stress. Plant Cell 11, 1277–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, K.J.A. (1993). Protein modification by oxidation and the role of proteolytic enzymes. Biochem. Soc. Trans. 21, 346–352. [DOI] [PubMed] [Google Scholar]
- Delieu, T., and Walker, D.A. (1981). Polarographic measurement of photosynthetic O2 evolution by leaf discs. New Phytol. 89, 156–175. [Google Scholar]
- Del Rio, L.A., Sandalio, L.M., Altomare, D.A., and Zilinskas, B.A. (2003). Mitochondrial and peroxisomal manganese superoxide dismutase: Differential expression during leaf senescence. J. Exp. Bot. 54, 923–933. [DOI] [PubMed] [Google Scholar]
- El Hassouni, M.E., Chambost, J.P., Expert, D., van Gijsegem, F., and Barras, F. (1999). The minimal gene set member msrA, encoding peptide methionine sulfoxide reductase, is a virulence determinant of plant pathogen Erwinia chrysanthemi. Proc. Natl. Acad. Sci. USA 96, 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman, M.A., Dush, M.K., and Martin, G.R. (1988). Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki, Y., Yoshikawa, Y., Sato, T., Inada, N., Ito, M., Nishida, I., and Watanabe, A. (2001). Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol. Plant. 111, 345–352. [DOI] [PubMed] [Google Scholar]
- Gabbita, S.P., Aksenov, M.Y., Lovell, M.A., and Markesbery, W.R. (1999). Decrease in methionine sulfoxide reductase in Alzheimer's disease brain. J. Neurochem. 73, 1660–1666. [DOI] [PubMed] [Google Scholar]
- Galleschi, L., Capocchi, A., Ghiringehelli, S., and Saviozzi, F. (2002). Antioxidants, free radicals, storage proteins, and proteolytic activities in wheat (Triticum durum) seeds during accelerated aging. J. Agric. Food Chem. 50, 5450–5457. [DOI] [PubMed] [Google Scholar]
- Gao, J., Yin, D., Yao, Y., Williams, T.D., and Squier, T.C. (1998). Progressive decline in the ability of calmodulin isolated from aged brain to activate the plasma membrane Ca2+-ATPase. Biochemistry 37, 9536–9548. [DOI] [PubMed] [Google Scholar]
- Giersch, C., Heber, U., Kobayashi, Y., Inoue, Y., Shibata, K., and Heldt, H.W. (1980). Energy charge, phosphorylation and proton motive force in chloroplasts. Biochim. Biophys. Acta 590, 59–73. [DOI] [PubMed] [Google Scholar]
- Grune, D., and Davies, K.J. (1997). Breakdown of oxidized proteins as a part of secondary antioxidant defenses in mammalian cells. Biofactors 6, 165–172. [DOI] [PubMed] [Google Scholar]
- Hansel, A., Kuschel, L., Hehl, S., Lemke, C., Agricola, H.J., Hoshi, T., and Heinemann, S.H. (2002). Mitochondrial targeting of human peptide methionine sulfoxide reductase (MSRA), an enzyme involved in the repair of oxidized proteins. FASEB J. 16, 911–913. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Hoshi, T., and Heinemann, S.H. (2001). Regulation of cell function by methionine oxidation and reduction. J. Physiol. 531, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez, A., Creissen, G., Kular, B., Firmin, J., Robinson, S., Verhoeyen, M., and Mullineaux, P. (2002). Changes in oxidative processes and components of the antioxidant system during tomato fruit ripening. Planta 214, 751–758. [DOI] [PubMed] [Google Scholar]
- Lee, H.C., and Wei, Y.H. (1997). Role of mitochondria in human aging. J. Biomed. Sci. 4, 319–326. [DOI] [PubMed] [Google Scholar]
- Lowther, W.T., Brot, N., Weissbach, H., and Matthews, B.W. (2000). Structure and mechanism of peptide methionine sulfoxide reductase, an “anti-oxidation” enzyme. Biochemistry 39, 13307–13312. [DOI] [PubMed] [Google Scholar]
- Matamoros, M.A., Baird, L.M., Escuredo, P.R., Dalton, D.A., Minchin, F.R., Iturbe-Ormaetxe, I., Rubio, M.C., Moran, J.F., Gordon, A.J., and Becana, M. (1999). Stress-induced legume root nodule senescence. Physiological, biochemical, and structural alterations. Plant Physiol. 121, 97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz, J., Bar-Noy, S., Williams, W.M., Requena, J., Berlet, B.S., and Stadtman, E.R. (2001). Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc. Natl. Acad. Sci. USA 98, 12920–12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz, J., Berlett, B.S., Poston, J.M., and Stadtman, E.R. (1997). The yeast peptide-methionine sulfoxide reductase functions as antioxidant in vivo. Proc. Natl. Acad. Sci. USA 94, 9585–9589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz, J., Jenkins, N.A., Gilbert, D.J., Copeland, N.G., Jursky, R., Weissbach, H., and Brot, N. (1996). Chromosomal localization of the mammalian peptide-methionine sulfoxide reductase gene and its differential expression in various tissues. Proc. Natl. Acad. Sci. USA 93, 3205–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz, J., Poston, J.M., Berlet, B.S., Nosworthy, N.J., Szcepanowski, R., and Stadtman, E.R. (2000). Identification and characterization of a putative active site for peptide methionine sulfoxide reductase (MsrA) and its substrate stereospecificity. J. Biol. Chem. 276, 14267–14272. [DOI] [PubMed] [Google Scholar]
- Moskovitz, J., Rahman, M.A., Strassman, J., Yancey, S.O., Kushner, S.R., Brot, N., and Weissbach, H. (1995). Escherichia coli peptide methionine sulfoxide reductase gene: Regulation of expression and role in protecting against oxidative damage. J. Bacteriol. 177, 502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, G.L., Dorian, R., and Fahey, C. (1981). Analysis of biological thiols: Derivatization with monobromobimane and separation by reverse-phase high-performance liquid chromatography. Anal. Biochem. 114, 383–387. [DOI] [PubMed] [Google Scholar]
- Noctor, G., Gornez, L., Vanacker, H., and Foyer, C.H. (2002). Interactions between biosynthesis, compartmentation and transport in the control of glutathione homeostasis and signaling. J. Exp. Bot. 53, 1283–1304. [DOI] [PubMed] [Google Scholar]
- Pastori, G.M., and Del Rio, L.A. (1994. a). Activated oxygen species and superoxide dismutase activity in peroxisomes from senescent pea leaves. Proc. R. Soc. Edinb. 102B, 505–509. [Google Scholar]
- Pastori, G.M., and Del Rio, L.A. (1994. b). An activated-oxygen-mediated role for peroxisomes in the mechanism of senescence of Pisum sativum L. leaves. Planta 193, 385–391. [Google Scholar]
- Pastori, G.M., and Del Rio, L.A. (1997). Natural senescence in pea leaves. An activated oxygen-mediated function for peroxisomes. Plant Physiol. 113, 411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad, T.K. (1996). Mechanism of chilling-induced oxidative stress injury and tolerance in developing maize seedlings: Changes in antioxidant system, oxidation of proteins and lipids, and protease activities. Plant J. 10, 1017–1026. [Google Scholar]
- Sadanandom, A., Piffanelli, P., Knott, R., Robinson, C., Sharpe, A., Lydiate, D., Murphy, D.J., and Fairbairn, D.J. (1996). Identification of a peptide methionine sulphoxide reductase gene in an oleosin promoter from Brassica napus. Plant J. 10, 235–242. [DOI] [PubMed] [Google Scholar]
- Sadanandom, A., Poghosyan, Z., Fairbairn, D.J., and Murphy, D.J. (2000). Differential regulation of plastidial and cytosolic isoforms of peptide methionine sulfoxide reductase in Arabidopsis. Plant Physiol. 123, 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shacter, E. (2000). Quantification and significance of protein oxidation in biological samples. Drug Metab. Rev. 32, 307–326. [DOI] [PubMed] [Google Scholar]
- Stitt, M., Iilley, R.M.C., Gerhardt, R., and Heldt, H.W. (1989). Determination of metabolite levels in specific cells and subcellular compartments of plant leaves. Methods Enzymol. 174, 518–522. [Google Scholar]
- Tang, X.D., Daggett, H., Hanner, M., Garcia, M.L., McManus, O.B., Brot, N., Weissbach, H., Heinemann, S.H., and Hoshi, T. (2001). Oxidative regulation of large conductance calcium-activated potassium channels. J. Gen. Physiol. 117, 253–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier, A.F., Marillonet, S., Klimyuk, V., Patel, K., Torres, M.A., Murphy, G., and Jones, J.D.G. (1999). Multiple independent defective supressor-mutator transposon insertions in Arabidopsis. Plant Cell 11, 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt, W. (1995). Oxidation of methionyl residues in proteins: Tools, targets and reversal. Free Radic. Biol. Med. 18, 93–105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.