Abstract
The Gram-negative saprophytic bacterium Burkholderia pseudomallei is the causative agent of melioidosis, a severe infectious disease of both humans and animals. Severity of the disease is thought to be dependent on both the health status of the host, including diabetes mellitus and kidney disease, and bacterial-derived factors. To identify the bacterial factors important during an acute infection, gene expression profiles in the spleen, lung, and liver of BALB/c (Th2 prototype) and C57BL/6 mice (Th1 prototype) were determined using DNA microarrays. This analysis identified BPSS1521 (bprD), a predicted transcriptional regulator located in the type III secretion system (T3SS-3) operon, to be up regulated, specifically in C57BL/6 mice. BALB/c mice infected with a bprD mutant showed a shorter time to death and increased inflammation, as determined by histopathological analysis and enumeration of bacteria in the spleen. Elevated numbers of multinucleated giant cells (MNGCs), which is the hallmark of melioidosis, were detected in both the wild-type and the bprD mutants; a similar elevation occurs in melioidosis patients. One striking observation was the increased expression of BPSS1520 (bprC), located downstream of bprD, in the bprD mutant. BprC is a regulator of T6SS-1 that is required for the virulence of B. pseudomallei in murine infection models. Deletion of bprD led to the overexpression of bprC and a decreased time to death. bprD expression was elevated in C57BL/6 —as compared to BALB/c—mice, suggesting a role for BprD in the natural resistance of C57BL/6 mice to B. pseudomallei. Ultimately, this analysis using mice with different immune backgrounds may enhance our understanding of the outcomes of infection in a variety of models.
Introduction
Burkholderia pseudomallei is a motile, aerobic, non-spore-forming, Gram-negative soil saprophytic bacterium [1]. It is the causative agent of melioidosis, an infectious disease endemic to Southeast Asia and northern Australia but present worldwide [2]. B. pseudomallei has been classified as a potential agent for bioterrorism (Tier 1 agent) by the U.S. Centers for Disease Control and Prevention, and has been suggested to be an emerging infectious disease [3]. In northeast Thailand, melioidosis accounts for 20% of community-acquired septicemias, with a 40% mortality rate even with appropriate treatment [4]. The clinical manifestations of melioidosis are diverse, and can include acute fulminant septicemia, localized lesions, and chronic disease. The disease is also associated with a high rate of relapse and affects all host organ systems [5]. The lungs are the most commonly affected organ, displaying primary lung abscesses or pneumonia followed by septicemia (blood-borne pneumonia) [1]. Although several virulence factors have been identified, the mechanism of pathogenesis is not fully understood. The ability of B. pseudomallei to invade, survive, and replicate intracellularly allows it to persist in the body during latent, chronic infection [6], [7], [8]. A number of virulence factors have been identified for B. pseudomallei infection of mammalian cells; these include type III (T3SSs) and type VI (T6SSs) secretion systems, quorum-sensing molecules, capsular polysaccharide, lipopolysaccharide, flagella, type IV pili, siderophores, and secreted proteins such as hemolysin, lipases and proteases [9], [10], [11].
As there is a broad spectrum of clinical outcomes and the severity of infection is associated with the diversity of the B. pseudomallei genome, the various virulence factors would expected to be differentially expressed under specific growth conditions. To identify and characterize bacterial genes whose products are involved in pathogenesis, two mouse strains with defined immunological backgrounds were used. The murine melioidosis models of acute (BALB/c: Th2 prototype) and chronic (C57BL/6: Th1 prototype) infections were used, as they have been shown previously to mimic the corresponding disease stages in humans [12]. BALB/c mice exhibit increased levels of proinflammatory cytokines, such as TNF-α, IL-1β, and IFN-γ, in addition to early infiltration of neutrophils, which contribute to the development of acute disease. Symptoms of acute disease include tissue destruction, multiple organ failure, and septic shock. In contrast, C57BL/6 mice, Th1 prototype, can effectively control B. pseudomallei infection, as demonstrated by moderate increases in cytokine levels and greater macrophage infiltration, allowing time for an adaptive immune response to occur [13], [14], [15], [16]. At present, the relative importance of the cell-mediated and humoral arms of the innate and adaptive immune responses is unclear [17], [18], [12].
In this study, B. pseudomallei 1909a, a highly virulent isolate from a severely septic patient, was used to establish an acute infection model in BALB/c and C57BL/6 mice. Gene expression profiles of bacteria replicating in the spleen, lung, and liver of mice at 4 days post-infection were compared with those of bacteria cultured in vitro using the whole genome Burkholderia mallei/pseudomallei DNA microarray from the Pathogen Functional Genomics Resource Center. Genes with unknown function and increased expression levels in at least two organs (lungs, spleen, or liver) of C57BL/6 mice were initially targeted for further analysis, as they may be associated with specific bacterial defense responses during acute infection.
Results
Expression of bprD is increased in the lungs and livers of C57BL/6 compared to BALB/c mice
To identify the bacterial factors important for acute B. pseudomallei infection, gene expression profiles in the lungs, livers, and spleens of BALB/c and C57BL/6 mice were determined. For this analysis, RNA was isolated at 4 days post-infection from the spleens, lungs, and livers of infected mice as well as from B. pseudomallei, 1909a cultured in vitro to the exponential phase.
Gene expression profiles determined using DNA microarrays obtained from the Pathogen Functional Genomics Resource Center (pfgrc.jcvi.org) showed that 5.0% of the B. pseudomallei genome (3.3% and 1.7% increased and decreased expression, respectively) in BALB/c mice, and 5.5% of the B. pseudomallei genome (3.3% and 2.3% increased and decreased expression, respectively) in C57BL/6 mice, were differentially expressed during acute infection (intraperitoneal injection), as compared to bacteria cultured in vitro. Genes with altered expression patterns were distributed equally between chromosome I—which encodes genes with core functions, such as metabolism and growth; and chromosome II—which encodes genes with accessory functions required for survival, and virulence factors [19].
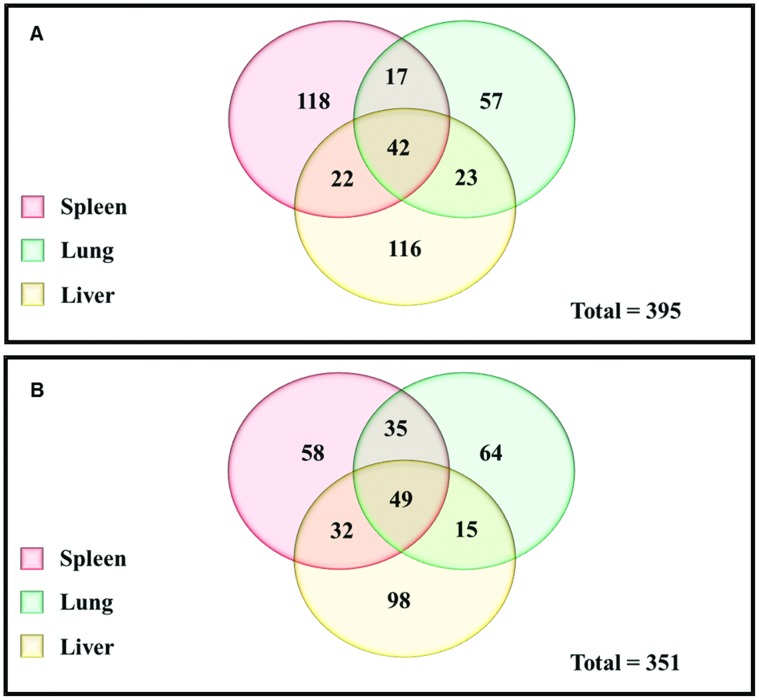
To reduce the complexity of this dataset, genes whose expression was increased in all three of the target organs (lungs, liver, and spleen) were selected initially. This analysis resulted in: 1) 42 genes in BALB/c mice (Figure 1A); 2) 49 genes in C57BL/6 mice (Figure 1B); and 3) 34 genes whose expression was increased in both mouse strains. The majority of the genes in these three datasets were metabolic or housekeeping genes.
Figure 1. Venn diagrams showing the number of genes differentially expressed in each organ.
Genes expressed at higher levels in the spleen (pink); lung (green); and liver (yellow) of BALB/C (A) and C57BL/6 (B) mice compared to in vitro.
As this study aimed to identify novel B. pseudomallei genes important for virulence, the data analysis focused initially on genes of unknown function and those in known virulence operons, particularly the T3SSs and T6SSs operons [20]. This targeted analysis identified a total of 36 genes (Table 1), with 7 genes in T6SSs and 6 in T3SSs, the most interesting of which was BPSS1521 (bprD). bprD expression was 7.2-, 21.0-, and 18.6-fold, and 8.8-, 3.5-, and 3.5-fold, higher in the spleen, lung, and liver of infected C57BL/6 and BALB/c mice, respectively, as compared to bacteria cultured in vitro. Moreover, the fold changes in bprD expression in the lungs and livers of C57BL/6 mice were considerably higher than those in BALB/c mice (Table 1). Additional genes of interest were identified in the T6SS-1 operon, the most interesting of which was BPSS1512 (tssM). This gene showed higher expression levels in the lungs of infected C57BL/6 mice and the livers of infected BALB/c mice, as compared to bacteria cultured in vitro. tssM has been reported to suppress host immune responses during infection, thus increasing bacterial survival and decreasing the time to death of tssM-mutant-infected mice [21]. The expression patterns of both genes were validated by qRT-PCR. The in vivo/in vitro gene expression ratios of B. pseudomallei in the lungs of BALB/c and C57BL/6 mice were consistent with the microarray data (Figure S1).
Table 1. List of the B. pseudomallei genes that have >2SD fold change in expression (in vivo/in vitro).
| Locus Tag | Gene Symbol (NCBI) | Gene description (NCBI) | Fold Change | |||||
| Spleen | Lung | Liver | ||||||
| BALB/c | C57BL/6 | BALB/c | C57BL/6 | BALB/c | C57BL/6 | |||
| BPSL0226 | fliJ | Flagella fliJ protein | *6.33 | *4.65 | ND | ND | 1.91 | 3.39 |
| BPSL0274 | flgF | Flagella basal body rod protein FlgF | *4.52 | 1.81 | 1.19 | 1.36 | 1.01 | 1.45 |
| BPSL0029 | fliO | Flagella protein | 2.21 | 3.31 | 3.61 | 1.91 | *13.55 | 2.7 |
| BPSL0272 | flgD | Flagella basal body rod modification protein | 2.9 | 1.4 | 0.77 | *9.58 | 0.79 | 1.81 |
| BPSL0277 | flgI | Flagella basal body P-ring protein | 1.0 | *8.26 | 2.23 | *5.72 | 2.65 | 1.09 |
| BPSL0808 | Peptidase | 1.2 | 1.82 | 1.5 | *7.12 | 1.57 | 0.83 | |
| BPSL1505 | rpoS | RNA polymerase sigma factor RpoS | 1.42 | 1.56 | *4.03 | 3.63 | 2.07 | 1.07 |
| BPSL1819 | Fimbriae-assembly-like protein | 2.64 | 2.09 | 1.83 | *4.43 | 2.55 | 2.6 | |
| BPSL1820 | Fimbriae assembly-like protein | 3.15 | 1.76 | 0.97 | 1.85 | *4 | 3.27 | |
| BPSL1902 | Hypothetical protein | 2.52 | 1.42 | *3.71 | 2.36 | 2.05 | 1.28 | |
| BPSL2403 | plcN | Non-hemolytic phospholipase C | 0.58 | *7.9 | 0.9 | 2.21 | 3.4 | 0.43 |
| BPSL2686 | rmlB | dTDP-glucose 4,6-dehydratase | *3.9 | 1.04 | 2.28 | 2.54 | 1.57 | 1.18 |
| BPSL3099 | Outer membrane protein | 0.84 | 0.88 | 0.98 | 0.98 | *4.9 | 0.77 | |
| BPSL3172 | Hypothetical protein | 1.1 | 1.5 | 2.6 | *4.3 | 0.78 | 2.74 | |
| BPSL3294 | flhA | Flagella biosynthesis protein FlhA | 0.68 | 0.66 | 0.36 | 1.28 | *4.66 | *3.89 |
| BPSL3304 | tsr | Methyl-accepting chemotaxis protein I | *3.91 | 2.49 | 0.78 | *13.9 | 1.93 | 3.49 |
| BPSS0029 | Transport-related membrane protein | 2.86 | 1.38 | 0.78 | *6.12 | 0.67 | *3.82 | |
| BPSS0417 | Hypothetical protein | 1.23 | 2.74 | 0.98 | *6.15 | 1.24 | *3.94 | |
| BPSS0520 | Hypothetical protein | 1.51 | 0.84 | 0.65 | 1.72 | 1.31 | *6.03 | |
| BPSS0524 | Hypothetical protein | 1 | *4.29 | 2.19 | 2.23 | 2.31 | 1.14 | |
| BPSS0860 | Flagella hook-associated protein | 1.41 | 2.14 | 1.29 | *3.92 | 2.44 | 1.86 | |
| BPSS0989 | Hypothetical protein | 2.94 | *3.75 | 1.71 | 2.46 | 3.09 | 1.65 | |
| BPSS1264 | Hypothetical protein | 2.62 | 2.35 | *9.47 | 1.7 | *94.6 | 6.24 | |
| BPSS1405 | sctS | Type III secretion-associated protein | *3.92 | 2.16 | 1.47 | 2.08 | 1.93 | 3.49 |
| BPSS1507 | ttssI | Hypothetical protein | 2.6 | *3.79 | 0.75 | 2.24 | 1.13 | *4.0 |
| BPSS1512 | tssM | Hypothetical protein | 1.9 | 2.07 | 1.24 | *7.36 | *7.4 | 2.6 |
| BPSS1521 | bprD | Hypothetical protein | *8.8 | *7.17 | 3.49 | *21.0 | 3.46 | *18.6 |
| BPSS1524 | bopA | Intercellular spread protein | 0.45 | 0.52 | *10.4 | 0.72 | 2.23 | 0.82 |
| BPSS1531 | bipC | Cell invasion protein | *3.89 | 3.35 | *3.92 | 3.63 | 3.15 | 1.75 |
| BPSS1533 | bicA | Surface presentation of antigens protein | 3.08 | *4.6 | 2.51 | 2.66 | 2.47 | 2.527 |
| BPSS1538 | bsaV | Surface presentation of antigens protein | 1.1 | 1.52 | *3.82 | 1.05 | *4.45 | 1.26 |
| BPSS1543 | bsaQ | Type III secretion system protein | 1.25 | *24.05 | 2.17 | *8.19 | 2.63 | 0.83 |
| BPSS1833 | udg2 | UDP-glucose 6-dehydrogenase 2 | *4.17 | 1.15 | 1.84 | 1.9 | 1.44 | 1.07 |
| BPSS1875 | Chemotaxis-related protein | *3.84 | *4.56 | 2.04 | *7.54 | 1.4 | 3.49 | |
| BPSS2095 | Hypothetical protein | 2.45 | 1.59 | 0.42 | *4.83 | 1.14 | 2.34 | |
| BPSS2103 | Hypothetical protein | *6.74 | 1.65 | 3.5 | *4.2 | *4.0 | 2.28 | |
* Expression level change >2SD in the organs of mice and ND = no data.
BPSS1521 (bprD) mutation has no effect on growth rate in vitro but alters the expression of downstream genes
bprD (BPSS1521) is located in the T3SS-3 operon between BPSS1522 (bprB) and BPSS1520 (bprC). bprB and bprC are predicted to be response regulators; bprC has been reported to regulate T6SS-1 [9], [22]; however, the function of bprD is unknown [19], [23]. To investigate the function of bprD, a mutant strain was constructed using a double homologous recombination of a tetracycline cassette in B. pseudomallei strain K96243 [19], as the sequence of the 1909a strain is unknown. The K96243 strain is virulent in both BALB/c and C57BL/6 mice via the IP route of infection and is used commonly for cloning and investigation in numerous animal models [24], [25], [26], [27], [28]. The LD50 of the K96243 strain in BALB/c mice is 103, compared to 20 for the 1909a strain. No significant differences in growth of the two B. pseudomallei strains were observed after in vitro culture in a rich medium (Luria–Bertani, LB) (Figure S2).
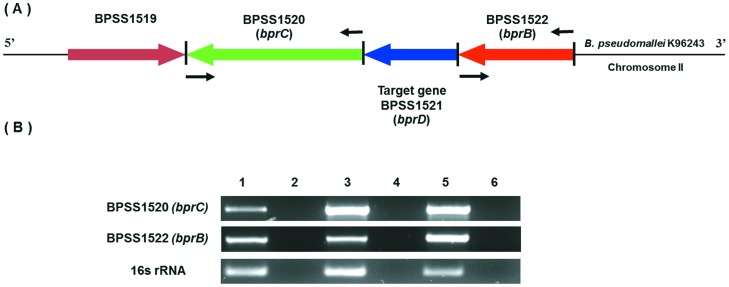
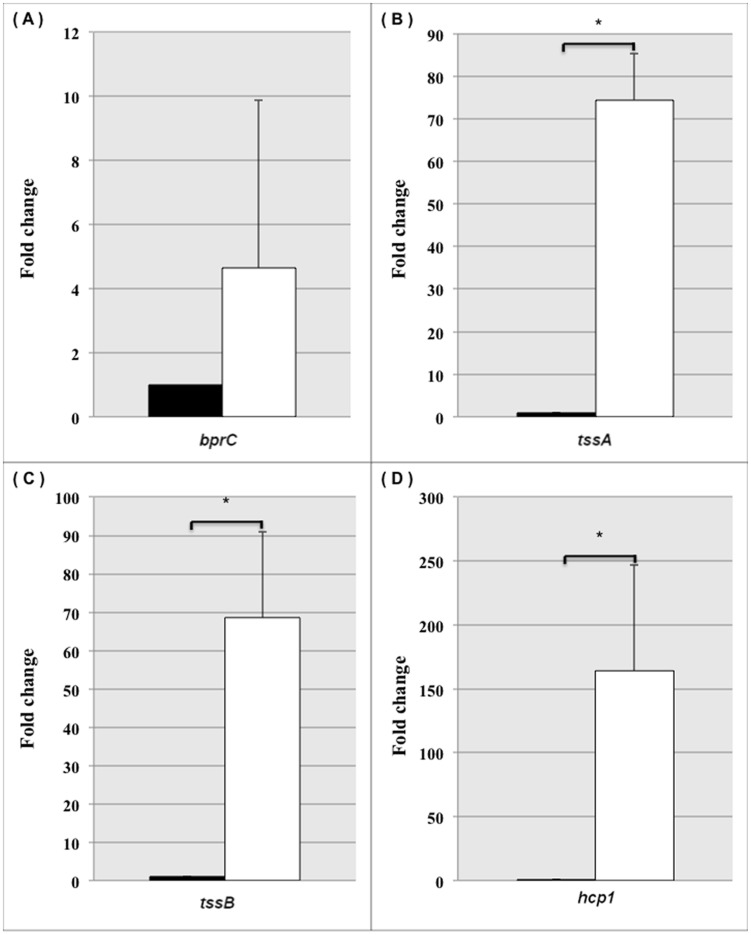
As BPSS1521 is the second gene in the T3SS-3 operon, whether deletion of this gene altered the expression of the flanking genes, BPSS1520 and BPSS1522, was determined (Figure 2A). Expression levels of the individual genes were assessed using RT-PCR (Figure 2B) and qRT-PCR (data not shown). The results showed that bprD mutation did not affect the expression level of the upstream gene (BPSS1522 - bprB), but altered the expression of the downstream gene (BPSS1520 - bprC) (Figure 2B). BPSS1520 (bprC), an AraC regulator that is required for the expression of T6SS-1, was upregulated in the bprD mutant. To confirm upregulation of BPSS1520 in the bprD mutant strain, the T6SS genes, BPSS1496 (tssA), BPSS1497 (tssB), and BPSS1498 (hcp1), under the control of bprC in vitro [20] were analyzed using qRT-PCR. All T6SS genes were found to be expressed at higher levels in the bprD mutant (Figure 3).
Figure 2. Schematic diagram of the B. pseudomallei K96243 bprD gene and agarose gel of RT-PCR products.
Schematic diagram of the genomic organization of the B. pseudomallei K96243 region containing bprD (A), and RT-PCR analysis of expression of genes in this region (B). The arrows show the position and direction of genes. The positions of RT-PCR primers are indicated by black arrows. Lane 1, RT-PCR product from B. pseudomallei K96243 wild-type cDNA; lane 2, negative control for wild-type or DNase-treated wild-type RNA (to evaluate contamination of wild-type RNA with gDNA); lane 3, B. pseudomallei K96243 bprD mutant cDNA; lane 4, negative control for mutant or DNase-treated bprD mutant RNA (to evaluate contamination of mutant RNA with gDNA); lane 5, B. pseudomallei K96243 wild-type genomic DNA control; and lane 5, No DNA control.
Figure 3. Fold changes in gene expression in the B. pseudomallei K96243 wild-type and bprD-mutant strains.
The fold changes in expression in the B. pseudomallei K96243 wild-type (▪) and bprD-mutant (□) strains of the BPSS1520 (bprC) (A), BPSS1496 (tssA) (B), BPSS1497 (tssB) (C), and BPSS1498 (hcp1) (D) genes at mid-logarithmic phase in LB medium were measured by qRT-PCR. The bars indicate means ± standard error of two experiments; * significant difference.
Finally, to confirm that the insertion of the tetracycline marker in bprD did not result in a polar effect on bprC, a markerless allele replacement method was used to re-construct the B. pseudomallei K96243 bprD mutant [29]. Expression analysis in this mutant strain confirmed the increased expression of bprC, tssA, tssB and hcp1in the K96243 bprD mutant (data not shown).
BPSS1521 (bprD) mutant results in a shorter time to death than the wild-type in BALB/c mice
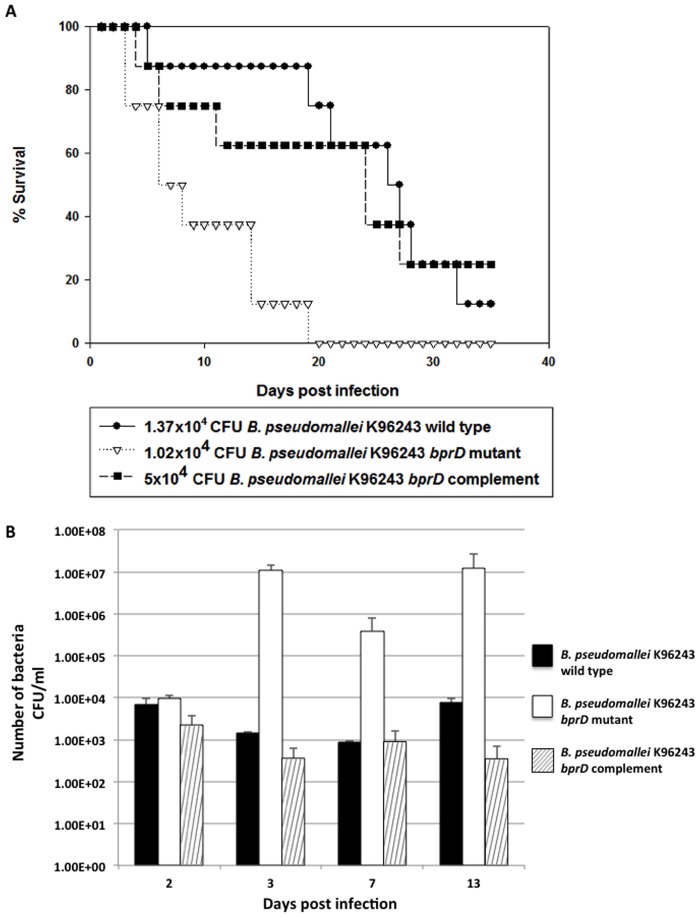
Both T3SS-3 and T6SS-1 are important for intracellular growth and survival. The T6SS-1 regulators (bsaN, bprC, and virAG) are essential for B. pseudomallei virulence in mice [20]. The virulence of the bprD mutant in mice was therefore evaluated. Because C57BL/6 mice are relatively resistant to B. pseudomallei infection and mimic chronic outcomes, while BALB/c mice are relatively sensitive and mimic acute infection, BALB/c mice were used for comparison of virulence. BALB/c mice, intraperitoneally infected with ∼104 CFU of the B. pseudomallei K96243 bprD mutant showed a significantly shorter mean survival time (10 days), as compared to mice infected with wild-type K96243 (>25 days) (Figure 4A) (p<0.0015). Complementation with a functional copy of bprD (pBBR1MCSbprD) restored survival outcomes to wild-type levels (mean survival >21 days) (Figure 4A). The expression plasmid, pBBR1MCSbprD, in the B. pseudomallei bprD mutant was found to be stable throughout the course of the experiment (>21 days).
Figure 4. Survival curves and numbers of bacteria in the spleen of BALB/c mice after infection with B. pseudomallei.
(A) The virulence of the B. pseudomallei K96243 wild-type and bprD mutant strains was compared in BALB/c mice, which are highly susceptible to infection. X-axis, days after infection; Y-axis, % survival. BALB/c mice (groups of eight) were intraperitoneally infected with ∼104 CFU of the B. pseudomallei K96243 wild-type (•), bprD mutant (∇) or bprD complemented (▪) strain, and their survival was monitored daily. The survival of mice infected with the wild-type strain was significantly different from those infected with the bprD mutant (P = 0.0015). (B) The numbers of B. pseudomallei K96243 wild-type, bprD-mutant, and bprD-complemented strains in the spleen of BALB/c mice. X-axis, days after infection; Y-axis, number of bacteria (CFU). A total of 1×104 CFU of the wild-type (▪) and bprD-mutant (□) strains, and 0.5×104 CFU of the bprD-complemented (  ) strain, were intraperitoneally injected into BALB/c mice. The number of bacteria in the spleen on days 2, 3, 7 and 13 was determined. The experiment was performed twice; average values of data are shown.
) strain, were intraperitoneally injected into BALB/c mice. The number of bacteria in the spleen on days 2, 3, 7 and 13 was determined. The experiment was performed twice; average values of data are shown.
BPSS1521 (bprD) mutant infection elicits an increased inflammatory response
As a splenic abscess is the most common complication of B. pseudomallei infection [1], bacterial growth and histological alterations resulting from bprD mutant replication in intraperitoneally infected BALB/c mice were evaluated. The bacterial burden in the spleen was similar at 2 days post infection with either the wild-type or the bprD mutant (6.9×103 and 9.6×103 CFU, respectively) (Figure 4B). Thereafter, the bprD mutant replicated rapidly, reaching 1.1×107 CFU on day 3, and remained stable (∼1.2×107 CFU) until day 13. In contrast, the burden of the wild-type strain decreased. Functional bprD complementation partially restored the behavior of the mutant strain (Figure 4B).
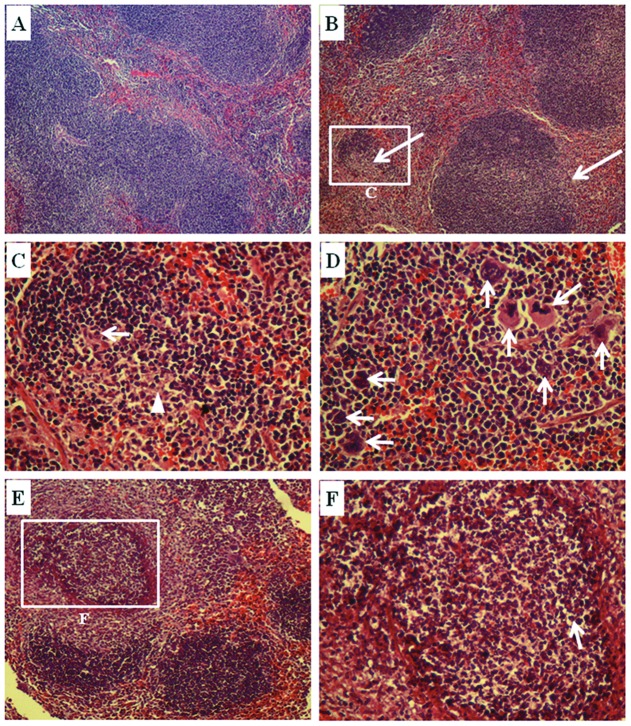
As the number of bacteria in the spleen at 3 days post infection differed significantly between the bprD mutant and the wild-type, a histological analysis of the spleens of infected mice was performed. The results indicated multifocal areas of inflammatory cell infiltration and neutrophil abscess formation (Figure 5B–C). Pathological scores were as follows: 1–4: 0 = within normal limits; 1 = <25%; 2 = 25–50%; 3 = 50–75%; and 4 = >75% inflammatory cell infiltration and neutrophil abscess formation. The pathological score of spleens collected from mice infected with B. pseudomallei K96243 wild-type was 1.5, whereas that of mice infected with the bprD mutant was 3.0. Normal histology of spleens from non-infected BALB/c mice is shown in Figure 5A. Spleens from mice infected with either the wild-type (Figure 5D) or the bprD mutant displayed multinucleated giant cells. Spleens from mice infected with the bprD mutant showed multifocal areas of granulomatous reactions with necrotic centers (Figure 5E–F). These lesions contained a mixture of macrophages and neutrophils surrounding the central areas, which comprised necrotic cells and nuclear debris. Larger abscesses were found in mice infected with the bprD mutant (Figure 5E) as compared to the wild type (Figure 5B).
Figure 5. Photomicrographs of hematoxylin-and-eosin-stained spleens from BALB/c mice infected with B. pseudomallei.
The spleens were collected on day 3 from non-infected BALB/c mice (A; ×100) and mice infected with B. pseudomallei (B–F). The spleen of mice infected with the B. pseudomallei K96243 wild-type strain showed multifocal areas of inflammatory cell infiltration and neutrophil abscess formation (arrows in B; ×100). The neutrophils (arrow) and necrotic cells (arrow head) are shown at high magnification (C; ×400). Numerous multinucleated giant cells are observed (arrows in D; ×400). Mice infected with the B. pseudomallei bprD mutant showed multifocal to coalescent pyogranulomatous splenitis (E; ×100). Necrotic areas with neutrophils (arrow) are shown at high magnification (F; ×400).
The liver pathology of mice infected with the bprD mutant differed only slightly from those infected with the wild type; however, micro-necrosis appeared to be more predominant in the livers of mice infected with the bprD mutant. No inflammation was observed in the alveolar spaces of lungs from both groups of mice at day 3 after infection (data not shown).
Discussion
The pathogenesis of B. pseudomallei and how it interacts with and responds to the host immune system remain poorly understood. Both the host background and the diversity of bacteria contribute to the wide range of the disease outcomes. More than 50% of B. pseudomallei-infected patients are immunocompromised, with diabetes being the most prevalent underlying disease [30], suggesting that host immune status is important for the onset of disease. In this current study, B. pseudomallei gene expression profiles during acute infection in the spleen, lung, and liver of BALB/c (Th2-biased response, relatively susceptible to B. pseudomallei infection) and C57BL/6 (Th1-biased response, relatively resistant to B. pseudomallei infection) were assessed using whole-genome microarrays. The microarray data were in concordance with gene expression profiles from a hamster model of acute melioidosis [31] and a rat lung model of acute and chronic infection [32]. The majority of genes with altered expression levels (+/− 2SD compared with bacteria cultured in vitro) in both murine backgrounds in all three target organs (lungs, livers, and spleens) were related to metabolic function. The expression levels of a subset of T3SS-3 and T6SS-1 virulence factors were elevated in C57BL/6, as compared to BALB/c, mice. This suggests the influence of a strong Th1 response or a compensatory mechanism for bacterial survival.
T3SS-3 and T6SS-1 are essential for bacterial intracellular survival and contribute to the virulence of B. pseudomallei [20], [33]. Comparison of B. pseudomallei gene expression profiles in mice showed increased expression levels of BPSS1521 (bprD) in all target organs in C57BL/6 mice (7–21-fold changes), and in the lungs and livers of BALB/c mice (3–9-fold changes), as quantified by qRT-PCR.
bprD is located in the T3SS-3 operon between BPSS1522 (bprB) and BPSS1520 (bprC). BPSS1522 (bprB), BPSS1520 (bprC) and BPSS1521 (bprD) are located in the same operon, and BPSS1530 (bprA) is located downstream of these genes. Deletion of either bprA (ΔbprA::FRT) or all of bprB, bprC and bprD (ΔbprBDC::FRT) did not change T3SS-3 expression or secretion [22]. Chen and colleagues impressively uncovered the control cascade between T3SS-3 and T6SS-1 in murine RAW264.7 macrophages, and reported attenuated virulence of the bprC, virAG and tssAB mutants in BALB/c mice [20]. In this study, bprC expression was elevated in the bprD mutant, together with a shorter time to death, in BALB/c mice. Therefore, it was hypothesized that bprD acts as a negative regulator of bprC. Additionally, involvement of T6SS-1, which is required for intracellular growth, correlates with our findings of an increased bacterial burden in the spleens of mice infected with the bprD mutant at 4 days post-infection.
Pathological studies in BALB/c mice infected with wild-type B. pseudomallei showed numerous multinucleated giant cells (MNGCs), a hallmark of acute B. pseudomallei infection in cell culture [7] and in melioidosis patients [34]. It was shown previously that the T3/6SS components, virAG, bprC and tssAB, are required for MNGC formation. Our data indicate slightly increased numbers of MNGCs in the spleen of BALB/c mice infected with the bprD mutant, as compared to those infected with the wild-type. The bprD mutant showed increased areas of multifocal granulomatous reaction with necrotic centers, which contributed to a higher overall pathological score. The decreased expression of bprC due to suppression via BprD controls tssAB in T6SS-1, resulting in the establishment of a chronic infection in C57BL/6 mice. Further investigation is required to determine whether the Th1-biased immune response in C57BL/6 mice is involved in the altered expression levels of bprD. This phenomenon is similar to that reported for the sseL mutant in Salmonella typhimurium and tssM mutants in B. pseudomallei KHW [21], [35]. tssM (BPSS1512) is located in the T6SS-1 (BPSS1496-BPSS1512) operon of B. pseudomallei [36], and was reported by Tan and colleagues to play a role in suppressing the host innate immune response. Mice infected with the B. pseudomallei KHW tssM mutant showed a reduced overall time to death and an increased inflammatory response in the spleen [21].
In summary, the negative regulation of bprC by BprD sheds further light on the complexity of regulation between T3SS-3 and T6SS-1, and suggests that further investigation of the possibility of suppressing T6SS-1—which is the most important B. pseudomallei virulence factor —is warranted.
Materials and Methods
Ethics Statement
BALB/c and C57BL/6 mice were obtained from the National Laboratory Animal Center, Mahidol University, Bangkok, Thailand. The maintenance and care of the experimental animals complied with the National Animal Center guidelines (polycarbonate cages with stainless steel wire-bar lid feeders in which food is provided ad libitum, sterile wood shavings as bedding material, fed a commercial diet (No. CP 082; Perfect Companion Group Co. Ltd., Thailand). Reverse-osmosis (RO) drinking water was provided ad libitum and contained 10–12 ppm chlorine. The animals were maintained under a 12∶12-h light: dark cycle. The B. pseudomallei mouse infection protocol was approved by the Animal Ethics Committee, Khon Kaen University, Khon Kaen, Thailand (Reference No. AE06/53) based on the ethics of animal experimentation guidelines of the National Research Council of Thailand. All surgeries and terminations were performed under isoflurane anesthesia, and all efforts were made to minimize suffering. All clinical bacterial isolates were anonymized.
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Tables 2 and 3. The B. pseudomallei K96243 wild-type, bprD mutant, and bprD complemented strains, and the Escherichia coli strain used for construction of the B. pseudomallei mutant and expression strains were cultured in Luria–Bertani (LB) agar or broth with shaking at 200 rpm overnight. When necessary, antibiotics were supplemented as follows (in µg/ml): for E. coli, ampicillin (Ap), 100; chloramphenicol (Cm), 30; tetracycline (Tc), 25; and for B. pseudomallei, Cm, 30; Tc, 50. The requirement for approval of the use of all clinical isolates in a study of this nature was waived by the Human Ethics Committee of Khon Kaen University.
Table 2. Bacterial strains used in this study.
| Bacterial strain | Description | Source or Reference | ||
| B. pseudomallei | 1909a | Clinical isolate from sputum of 34-year-old male with diabetes mellitus who is a rice farmer in Ubonratchathani province, Thailand | Kindly provided by Dr. Narisara Chantratita, Mahidol Oxford Tropical Medicine Research Unit (MORU), Faculty of Tropical Medicine, Mahidol University. | |
| B. pseudomallei | K96243 | Clinical isolate from a patient admitted to Khon Kaen provincial hospital, Khon Kaen, Thailand | [21] | |
| B. pseudomallei | K96243 | bprD mutant | K96243 derivative; DbprD; Tcr | This study |
| B. pseudomallei | K96243 | bprD complemented | bprD mutant carrying pBBR1MCSbprD; Tcr Cmr | This study |
| B. pseudomallei | K96243 | bprD clean mutant | K96243 derivative; DbprD | This study |
| E. coli | DH5α | General cloning | [46] | |
| E. coli | SM10lpir | Mobilizing strain, SM10 with a l prophage carrying the gene encoding the π protein; Cms Tcs Gms Kmr Tps Pxs | [47] | |
| E. coli | S17-llpir | Mobilizing strain, S17-1 with a l prophage carrying the gene encoding the π protein; Cms Tcs Gms Kms Tpr Pxs Smr | [49] |
Cm, chloramphenicol; Gm, gentamicin; Ap, ampicillin; Tc, tetracycline; Km, kanamycin; Tp, Trimethoprim; Px, polymyxin; Sm, streptomycin; r = resistance; s = sensitive.
Table 3. Plasmids used in this study.
| Plasmids | Description | Source or Reference |
| pDM4 | Suicide vector; sacBR oriT oriR6K Cmr | [48] |
| pDM4Δ bprD | pDM4::ΔbprD B. pseudomallei K96243; Cmr | This study |
| pDM4Δ bprD :: Tcr | pDM4::ΔbprD::Tcr B. pseudomallei K96243; Cmr Tcr | This study |
| pBBR1MCS | A Cmr bhr plasmid of 4.7 kb, and contains 16 unique cloning sites within the lacZ∼ gene, Kmr, Apr, Tcr, and Gmr | [38] |
| pBBR1MCS bprD | pBBR1MCS harboring the bprD gene | This Study |
| pUTminiTn5Tc | R6K-based suicide delivery plasmid; Tcr | Kindly provided by Prof. Ben Adler |
Cm, chloramphenicol; Gm, gentamicin; Ap, ampicillin; Tc, tetracycline; Km, kanamycin; Tp, Trimethoprim; Px, polymyxin; Sm, streptomycin; r = resistance; s = sensitive.
Biosecurity aspects
Concerning biosecurity aspects, both animal and general bacterial laboratory facilities were operated following all the security and safety regulations of our university. Animal experiments were carried out in the Northeast Animal Laboratory Center located in Faculty of Medicine, Khon Kaen University under the national procedure for infectious agents. This is a BSL2 plus facility that is currently being upgraded to BSL3 practices. Microbiological experiments were carried out under supervision of the director of the Melioidosis Research Center who received certificate of completion for BSL3 Executive Train-the-Trainer Program (Emory University, NSITE Applied Biosafety Training Program).
Construction of the BPSS1521 (bprD) mutant and complemented strains
The genome sequence of B. pseudomallei 1909a is not yet available; furthermore, its high virulence prohibited comparison of its virulence with that of the mutant strain. Both 1909a and K96243 strains can infect and cause disease in BALB/c and C57BL/6 mice via the IP route. Therefore, the BPSS1521 (bprD) mutant was constructed in B. pseudomallei K96243 by double-crossover allelic exchange using the λpir-dependent pDM4 vector [48], and the constructed plasmids were introduced into B. pseudomallei K96243 by conjugation using the E. coli SM10λpir (kindly provided by Prof. Ben Adler, Monash University, Australia) as described by Milton in 1996 [37] and Songsri in 2012 [38]. Procedures for clone construction are described in Figure S4. The plasmids and primer sequences used in this study are listed in Tables 3 and 4. The clean deletion construct was confirmed by DNA sequencing.
Table 4. Primers used in this study.
| GenBank accession number | Gene symbol | Gene Description | Primer name | Primer sequence | Purpose | Amplicon size (bp) |
| YP_111519.1 | BPSS1512 | Hypothetical protein | BPSS1512-F | GGACAAACGCTGGAAGTGAT | Internal region | 129 |
| YP_111519.1 | BPSS1512 | Hypothetical protein | BPSS1512-R | GTTCAGAAAGAACGCCTTGG | Internal region | 129 |
| YP_111526.1 | BPSS1520 | AraC family transcriptional regulator | BPSS1520-F | GGACGAGCTCGATTACATGC | Internal region | 153 |
| YP_111526.1 | BPSS1520 | AraC family transcriptional regulator | BPSS1520-R | GGCAGATGAAGATGCTGCTC | Internal region | 153 |
| YP_111527.1 | BPSS1521 | Hypothetical protein | BPSS1521-F | TCGATCTTCTCGCTGACCTC | Internal region | 100 |
| YP_111527.1 | BPSS1521 | Hypothetical protein | BPSS1521-R | TCAAGGAGATCCGCTTCAAC | Internal region | 100 |
| YP_111527.1 | BPSS1521 | Hypothetical protein | bprD-PFUp | TTAATTTCTAGAGCATCGGAGCAACAAGAATC | Upstream BPSS1521 | 927 |
| YP_111527.1 | BPSS1521 | Hypothetical protein | bprD-PRUp | TTATGAAGATCTTTTCCTGTCGTGAACATTGG | Upstream BPSS1521 | 927 |
| YP_111527.1 | BPSS1521 | Hypothetical protein | bprD-PFDown | TTGTCAAGATCTGCGTAACGTGTGACGTGTTG | Downstream BPSS1521 | 929 |
| YP_111527.1 | BPSS1521 | Hypothetical protein | bprD-PRDown | ATAATACCCGGGGATGAAGATGCTGCTCGATG | Downstream BPSS1521 | 929 |
| YP_111527.1 | BPSS1521 | Hypothetical protein | bprD-PFComp | ATACTTGGTACCAGGAAAGACATCATGAAGCTC | Full-length BPSS1521 | 456 |
| YP_111527.1 | BPSS1521 | Hypothetical protein | bprD-PRComp | TTAACATCTAGATCACGGCGCCGGGCGCTG | Full-length BPSS1521 | 456 |
| AY_305818.1 | 16S ribosomal RNA | 16s rRNA-F | GGCTAATACCCGGAGTGGA | Internal region | 194 | |
| AY_305818.1 | 16S ribosomal RNA | 16s rRNA-R | CTAGCCTGCCAGTCACCAA | Internal region | 194 | |
| YP_111503.1 | BPSS1496 | Hypothetical protein | BPSS1496-F | AGCGGGTCAACATCGTCTAT | Internal region | 163 |
| YP_111503.1 | BPSS1496 | Hypothetical protein | BPSS1496-R | ACGTCGTTGAAGTCGTCCTT | Internal region | 163 |
| YP_111504.1 | BPSS1497 | Hypothetical protein | BPSS1497-F | GCTCGCTGAAGTTTCTCGTC | Internal region | 151 |
| YP_111504.1 | BPSS1497 | Hypothetical protein | BPSS1497-R | TCGGCCGTATAGACCTTCTG | Internal region | 151 |
| YP_111505.1 | BPSS1498 | Hypothetical protein | BPSS1497-F | GTCATGACGGGAAAATCCAC | Internal region | 134 |
| YP_111505.1 | BPSS1498 | Hypothetical protein | BPSS1497-R | CGACGATCTGTCCATTTCCT | Internal region | 134 |
| pDM4 | BAP3771 | TAACGGCAAAAGCACCGCCGGACATCA | pDM4 MCS flanking region | 177 | ||
| pDM4 | BAP3772 | ACATGTGGAATTGTGAGCGGATAACAA | pDM4 MCS flanking region | 177 | ||
| Tcr cassette | BAP5118 | GCGTAGTCGATAGTGGCTCC | Internal region | |||
| Tcr cassette | BAP5116 | ATCAGGGACAGCTTCAAGGA | Internal region |
MCS, multiple cloning site.
For complementation, the full length of bprD together with the ribosomal binding site (RBS) was amplified using bprD-PFComp and bprD-PRComp primers (Table 4), and then cloned into pBBR1MCS [39] at KpnI and XbaI restriction sites to generate pBBR1MCSbprD. The constructed plasmids were then transformed into E. coli S17-1λpir and conjugated with the B. pseudomallei K96243 bprD mutant to generate the B. pseudomallei K96243 bprD-complemented strain.
The stability of pBBR1MCSbprD in the B. pseudomallei bprD mutant was investigated by culturing the B. pseudomallei K96243 bprD complemented with pBBR1MCSbprD in LB broth without antibiotics at 37°C overnight and sampling to LB agar plates daily for up to 21 days to obtain colonies. Ten colonies per day were PCR amplified using BPSS1521-F/BPSS1521-R primers (Table 4) to detect bprD.
The B. pseudomallei K96243 bprD clean deletion mutant was constructed by the allelic replacement system based on a non-replicative plasmid, pEXKm5 (kindly provided by Prof. Herbert P Schweizer, Colorado State University, USA), as described by Lopez in 2009 [29]. This mutant was used in the RNA-sequencing experiment to confirm the gene expression profiles of the B. pseudomallei K96243 wild-type and bprD mutant strains; this mutant will also be used for further study.
The bprD mutant and complement strain were originally constructed and examined for virulence. Subsequently, we constructed a clean deletion mutant confirming the role of bprD in virulence. Our initial animal studies clearly showed a role for this gene in virulence, therefore retesting the clean deletion mutant for altered virulence was not carried as it would replicate existing data and inappropriately increase the number of animals used.
Animal infection and organ harvest
For the DNA microarray assay, BALB/c and C57BL/6 mice (males, 4–5 weeks old, 15–16 g, n = 15 mice each to be adequate for total RNA extraction) were intraperitoneally injected with 100-µl 1× PBS containing 103 CFU of B. pseudomallei 1909a. Animals were then humanely euthanized with isoflurane, and the spleen, lung, and liver were harvested at day 4 post-infection (when bacterial numbers in the lung and liver are similar (Figure S3), and subjected to bacterial RNA isolation.
For virulence assessment, BALB/c mice, eight per group, were intraperitoneally inoculated with 100 µl of 104 CFU/ml logarithmic-phase cultures of the B. pseudomallei K96243 wild-type, bprD mutant, or bprD-complemented strain. The experiment was performed independently in duplicate and viable bacteria were enumerated by the plate count technique. After injection, survival was monitored daily during 8:30 am–6 pm for 35 days, and mice showing severe signs of disease were euthanized for humane reasons when possible. After 35 days, the remaining animals were humanely euthanized with isoflurane.
Histopathology and bacterial counts in spleen
Three groups of BALB/c mice, eight mice per group, were intraperitoneally injected with 103 CFU of the B. pseudomallei K96243 wild-type, bprD-mutant or bprD-complemented strains. Two mice on each of days 2, 3, 7 and 13 were humanely euthanized with isoflurane, and bacteria in the spleens were enumerated. The average numbers of bacteria in the spleens from two independent experiments were reported.
For histopathology, two groups of BALB/c mice, three mice per group, were intraperitoneally injected with 103 CFU of the B. pseudomallei K96243 wild-type or bprD mutant strain; 3 days later (when bacteria were present in the organs), they were humanely euthanized with isoflurane and a portion of the spleen, lung, and liver was fixed in 10% buffered formalin and embedded in paraffin. Serial 4-µm-thick sections from each organ were cut and placed on microscope slides. Dried slides were deparaffinized and stained with hematoxylin and eosin (H&E). The sections were visualized using a Nikon ECLIPSE 80i microscope and photographed with a Nikon DS-Fi1 digital camera (Nikon, Japan). Lesions were scored on the basis of inflammatory cell infiltration and neutrophil abscess formation using the scoring system described in the results section.
RNA Manipulation
(a) Total RNA extraction from bacterial cultures
Total RNA from mid-log phase (0.4–0.5 OD600 nm) B. pseudomallei 1909a cultured in LB broth, at 37°C with shaking at 200 rpm was extracted using TRIzol Reagent (Invitrogen, USA), as recommended by the manufacturer. The RNA in DEPC-treated water was treated with RQ1 RNase-Free DNase (Promega, USA) to remove contaminating genomic DNA.
(b) Extraction of bacterial RNA from the organs of infected mice
The organs of infected BALB/c and C57BL/6 mice (spleen, lungs, and liver) (n = 15 from each mouse) were harvested, five mice at a time, under sterile conditions, transferred immediately to RNALater (Ambion, USA), and chilled on ice. Each organ was rapidly homogenized separately in TRIzol reagent by grinding and sieving through a sterile stainless-steel sieve device, and then RNA was extracted as recommended by the manufacturer. The total RNA used for DNA microarrays was extracted from two independent groups of animals.
cDNA synthesis
Total RNA extracted from each mouse organ during infection with B. pseudomallei (in vivo) or during the exponential phase of bacterial culture (in vitro) was converted to cDNA. In brief, 15 µg of RNA were mixed with 10 µg of Genome Direct Primers (bpGDPs) (1 µg/µl) to prime the transcription reactions, rather than using random primers to preferentially transcribe bacterial mRNA in the mixed RNA sample [40]. The mixture was heated at 70°C for 5 min, immediately chilled on ice and spun briefly to bottom down the solution. Then, 5 µl of 10 mM dNTPs, 5 µl of 5× first-strand buffer, 25 units of Recombinant RNasin Ribonuclease inhibitor, 1 µl of M-MLV RT (200 units/µl), and nuclease-free water were added to a 20-µl final volume. The mixture was mixed gently and incubated at 37°C for 60 min before being heat-inactivated at 70°C for 15 min. Unincorporated nucleotides were removed by gel filtration (Illustra MicroSpin G-50 Columns, GE Healthcare Bio-Sciences, NJ, USA).
DNA labeling and microarray slide hybridizations
The whole-genome DNA microarray slides of B. mallei and B. pseudomallei version 2.0, received from the Pathogen Functional Genomics Research Center, MD, USA, were used in this study [41]. The gDNA of B. pseudomallei K96243 (10–15 µg) was labeled with Cy3-dCTP fluorescent dye by the standard nick translation reaction according to the manufacturer's protocol (Promega, WI, USA) [42]. For cDNA labeling, Cy5-labeled was incorporated using the Pronto Universal Hybridization kit (Corning, NY, USA), as described in the manufacturer's manual.
For hybridization, the genomic DNA was used to normalize each transcript signal to improve the signal-to-noise ratio [43]. Four micrograms of Cy5-labeled cDNA from each sample were mixed with 2 µg of Cy3-labeled gDNA of the K96243, dried in a SpeedVac concentrator for 30 min, and re-suspended in 50-µl Corning hybridization solution (Corning, NY, USA). Post-processed slides were hybridized while the LifterSlip Microarray Coverslips (Erie Scientific, USA) were placed over the array slides, incubated at 42°C in a water bath for 16–20 h, and post-hybridization washed according to the protocol recommended by Corning before scanning. DNA microarray slides, after hybridization and washing, were scanned by a GenePix 4000B laser scanner (Molecular Devices, Sunnyvale, CA, USA) and initially analyzed using the GenePix Pro 6.1 software to determine the fluorescence intensities of the two dyes for each spot.
Microarray data analysis
The data analysis was performed as described by Bartpho et al., 2012 [42]. Hybridization signals from cDNA samples were compared to the gDNA hybridization signals to estimate the relative gene expression levels for normalization (genomic normalization) [43]. Data from replicate genes were averaged before gene identification. An average of two replicate hybridizations from two independent biological replicates was used to calculate gene expression levels. The log2 expression values in vivo and in vitro were used to identify those genes with mean log2 [in vivo]/[in vitro] ratios that deviated more than an overall mean of ±2 SD as being significantly differentially expressed in vivo. B. pseudomallei gene expression profiles in the lungs, spleens, and livers of BALB/c and C57BL/6 mice were performed as hierarchical clustering as groups of similarity in gene expression using a free software package [44], and mapped to COG functional categories [45].
The microarray data have been deposited in the NCBI Gene Expression Omnibus [46]. All data are MIAME compliant (Minimum Information About a Microarray Experiment) and gene expression profiles are accessible through GEO Series accession number GSE51369.
Gene validation (qRT-PCR)
The selected genes were validated by quantitative real-time RT-PCR (qRT-PCR) using the LightCycler FastStart DNA MasterPLUS SYBR Green I with the LightCycler Carousel-Based System (Roche Diagnostics, USA). The specific primers for all selected genes, BPSS1520, BPSS1521, BPSS1522, BPSS1512, BPSS1496, BPSS1496 and 16s rRNA (an internal control) are listed in Table 4. All data were analyzed using the 2−ΔΔCT method. The results were expressed as target/reference ratios of each sample divided by the target/reference ratio of the calibrator. When the target samples were the B. pseudomallei genes expressed in vivo, the target calibrators were the B. pseudomallei genes expressed in vitro and the reference was the 16s rRNA gene. Results were reported as means of triplicate samples with standard deviations.
Supporting Information
Fold changes in gene expression in vivo / in vitro . Fold changes in the expression of the BPSS1521 (A) and BPSS1512 (B) genes as determined by DNA microarray (upper) and validated by qRT-PCR (lower) in the lungs of BALB/c (▪) and C57BL/6 mice (□). The difference in the fold change determined by qRT-PCR between BALB/c and C57BL/6 mice was not significant (p = 0.13 and 0.16, respectively).
(TIF)
Growth curves of the B. pseudomallei K96243 wild-type (•) and bprD mutant (□) strains; no significant difference was evident ( p = 0.23).
(TIF)
Numbers of bacteria in the spleen (A), lung (B), and liver (C) of BALB/c (♦) and C57BL/6 (▪) mice on days 3 to 5.
(TIF)
Schematic diagrams of the construction of the B. pseudomallei K96243 bprD mutant using pDM4. (A) Small arrows represent primer sites used to generate the upstream and downstream fragments to clone into pDM4 together with the Tcr cassette from pUTminiTn5Tc, to generate the pDMΔbprD::Tcr plasmid. (B1) Dotted cross indicates the first and second recombination steps used to replace bprD on the B. pseudomallei K96243 chromosome with the Tcr cassette from pDMΔbprD::Tcr, resulting in generation of the bprD mutant. (B2) Dotted cross indicates the first and second recombination steps at the same site leading to abortive allelic exchange and generation of the wild type rather than the bprD mutant.
(TIF)
Acknowledgments
We thank the Pathogen Functional Genomics Research Center, MD, USA for donating the DNA microarray slides and Prof. Ben Adler, Monash University, Australia for providing pDM4, E. coli SM10λpir and pUTminiTn5Tc for this study. We also thank Prof. Herbert P Schweizer, Colorado State University, USA for providing pEXKm5 for clean deletion construction. We wish to acknowledge the assistance of the Khon Kaen University Publication Clinic, Research and Technology Transfer Affairs, Khon Kaen University.
Funding Statement
This research work was supported by the Thailand Research Fund through the Royal Golden Jubilee Ph.D. Program (Grant no. PHD/0166/2549) to SC and RW, the Commission on Higher Education (CHE) Thailand, and Faculty of Medicine, Khon Kaen University, Khon Kaen, Thailand. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. White NJ (2003) Melioidosis. Lancet 361: 1715–1722. [DOI] [PubMed] [Google Scholar]
- 2. Dance DA (2000) Melioidosis as an emerging global problem. Acta Trop 74: 115–119. [DOI] [PubMed] [Google Scholar]
- 3. Pappas G, Panagopoulou P, Christou L, Akritidis N (2006) Category B potential bioterrorism agents: bacteria, viruses, toxins, and foodborne and waterborne pathogens. Infect Dis Clin North Am 20: 395–421, x. [DOI] [PubMed] [Google Scholar]
- 4. Chaowagul W, White NJ, Dance DA, Wattanagoon Y, Naigowit P, et al. (1989) Melioidosis: a major cause of community-acquired septicemia in northeastern Thailand. J Infect Dis 159: 890–899. [DOI] [PubMed] [Google Scholar]
- 5. Currie BJ, Mayo M, Anstey NM, Donohoe P, Haase A, et al. (2001) A cluster of melioidosis cases from an endemic region is clonal and is linked to the water supply using molecular typing of Burkholderia pseudomallei isolates. Am J Trop Med Hyg 65: 177–179. [DOI] [PubMed] [Google Scholar]
- 6. Jones AL, Beveridge TJ, Woods DE (1996) Intracellular survival of Burkholderia pseudomallei . Infect Immun 64: 782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kespichayawattana W, Rattanachetkul S, Wanun T, Utaisincharoen P, Sirisinha S (2000) Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: a possible mechanism for cell-to-cell spreading. Infect Immun 68: 5377–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pruksachartvuthi S, Aswapokee N, Thankerngpol K (1990) Survival of Pseudomonas pseudomallei in human phagocytes. J Med Microbiol 31: 109–114. [DOI] [PubMed] [Google Scholar]
- 9. Stevens MP, Wood MW, Taylor LA, Monaghan P, Hawes P, et al. (2002) An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol Microbiol 46: 649–659. [DOI] [PubMed] [Google Scholar]
- 10. Galyov EE, Brett PJ, DeShazer D (2010) Molecular insights into Burkholderia pseudomallei and Burkholderia mallei pathogenesis. Annu Rev Microbiol 64: 495–517. [DOI] [PubMed] [Google Scholar]
- 11. Wiersinga WJ, van der Poll T, White NJ, Day NP, Peacock SJ (2006) Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei . Nat Rev Microbiol 4: 272–282. [DOI] [PubMed] [Google Scholar]
- 12. Lazar Adler NR, Govan B, Cullinane M, Harper M, Adler B, et al. (2009) The molecular and cellular basis of pathogenesis in melioidosis: how does Burkholderia pseudomallei cause disease? FEMS Microbiol Rev 33: 1079–1099. [DOI] [PubMed] [Google Scholar]
- 13. Barnes JL, Ulett GC, Ketheesan N, Clair T, Summers PM, et al. (2001) Induction of multiple chemokine and colony-stimulating factor genes in experimental Burkholderia pseudomallei infection. Immunol Cell Biol 79: 490–501. [DOI] [PubMed] [Google Scholar]
- 14. Hoppe I, Brenneke B, Rohde M, Kreft A, Haussler S, et al. (1999) Characterization of a murine model of melioidosis: comparison of different strains of mice. Infect Immun 67: 2891–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ulett GC, Ketheesan N, Hirst RG (2000) Cytokine gene expression in innately susceptible BALB/c mice and relatively resistant C57BL/6 mice during infection with virulent Burkholderia pseudomallei . Infect Immun 68: 2034–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ulett GC, Ketheesan N, Hirst RG (2000) Proinflammatory cytokine mRNA responses in experimental Burkholderia pseudomallei infection in mice. Acta Trop 74: 229–234. [DOI] [PubMed] [Google Scholar]
- 17. Cheng AC, Currie BJ (2005) Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev 18: 383–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gan YH (2005) Interaction between Burkholderia pseudomallei and the host immune response: sleeping with the enemy? J Infect Dis 192: 1845–1850. [DOI] [PubMed] [Google Scholar]
- 19. Holden MT, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, et al. (2004) Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei . Proc Natl Acad Sci U S A 101: 14240–14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Y, Wong J, Sun GW, Liu Y, Tan GY, et al. (2011) Regulation of type VI secretion system during Burkholderia pseudomallei infection. Infect Immun 79: 3064–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan KS, Chen Y, Lim YC, Tan GY, Liu Y, et al. (2010) Suppression of host innate immune response by Burkholderia pseudomallei through the virulence factor TssM. J Immunol 184: 5160–5171. [DOI] [PubMed] [Google Scholar]
- 22. Sun GW, Chen Y, Liu Y, Tan GY, Ong C, et al. (2010) Identification of a regulatory cascade controlling Type III Secretion System 3 gene expression in Burkholderia pseudomallei . Mol Microbiol 76: 677–689. [DOI] [PubMed] [Google Scholar]
- 23. Sun GW, Gan YH (2010) Unraveling type III secretion systems in the highly versatile Burkholderia pseudomallei . Trends Microbiol 18: 561–568. [DOI] [PubMed] [Google Scholar]
- 24. Cuccui J, Milne TS, Harmer N, George AJ, Harding SV, et al. (2012) Characterization of the Burkholderia pseudomallei K96243 capsular polysaccharide I coding region. Infect Immun 80: 1209–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sarkar-Tyson M, Thwaite JE, Harding SV, Smither SJ, Oyston PC, et al. (2007) Polysaccharides and virulence of Burkholderia pseudomallei . J Med Microbiol 56: 1005–1010. [DOI] [PubMed] [Google Scholar]
- 26. Burtnick MN, DeShazer D, Nair V, Gherardini FC, Brett PJ (2011) Burkholderia mallei cluster 1 type VI secretion mutants exhibit growth and actin polymerization defects in RAW 264.7 murine macrophages. Infect Immun 78: 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. AuCoin DP, Reed DE, Marlenee NL, Bowen RA, Thorkildson P, et al. (2012) Polysaccharide specific monoclonal antibodies provide passive protection against intranasal challenge with Burkholderia pseudomallei . PLoS One 7: e35386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Muller CM, Conejero L, Spink N, Wand ME, Bancroft GJ, et al. (2012) Role of RelA and SpoT in Burkholderia pseudomallei virulence and immunity. Infect Immun 80: 3247–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lopez CM, Rholl DA, Trunck LA, Schweizer HP (2009) Versatile dual-technology system for markerless allele replacement in Burkholderia pseudomallei . Appl Environ Microbiol 75: 6496–6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Suputtamongkol Y, Chaowagul W, Chetchotisakd P, Lertpatanasuwun N, Intaranongpai S, et al. (1999) Risk factors for melioidosis and bacteremic melioidosis. Clin Infect Dis 29: 408–413. [DOI] [PubMed] [Google Scholar]
- 31. Tuanyok A, Tom M, Dunbar J, Woods DE (2006) Genome-wide expression analysis of Burkholderia pseudomallei infection in a hamster model of acute melioidosis. Infect Immun 74: 5465–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Schaik E, Tom M, DeVinney R, Woods DE (2008) Development of novel animal infection models for the study of acute and chronic Burkholderia pseudomallei pulmonary infections. Microbes Infect 10: 1291–1299. [DOI] [PubMed] [Google Scholar]
- 33. Stevens MP, Friebel A, Taylor LA, Wood MW, Brown PJ, et al. (2003) A Burkholderia pseudomallei type III secreted protein, BopE, facilitates bacterial invasion of epithelial cells and exhibits guanine nucleotide exchange factor activity. J Bacteriol 185: 4992–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wong KT, Puthucheary SD, Vadivelu J (1995) The histopathology of human melioidosis. Histopathology 26: 51–55. [DOI] [PubMed] [Google Scholar]
- 35. Le Negrate G, Faustin B, Welsh K, Loeffler M, Krajewska M, et al. (2008) Salmonella secreted factor L deubiquitinase of Salmonella typhimurium inhibits NF-kappa B, suppresses Ikappa B alpha ubiquitination and modulates innate immune responses. J Immunol 180: 5045–5056. [DOI] [PubMed] [Google Scholar]
- 36. Schell MA, Ulrich RL, Ribot WJ, Brueggemann EE, Hines HB, et al. (2007) Type VI secretion is a major virulence determinant in Burkholderia mallei . Mol Microbiol 64: 1466–1485. [DOI] [PubMed] [Google Scholar]
- 37. Milton DL, O'Toole R, Horstedt P, Wolf-Watz H (1996) Flagellin A is essential for the virulence of Vibrio anguillarum . J Bacteriol 178: 1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Songsri J, Proungvitaya T, Wongratanacheewin S, Homchampa P (2012) Tn5-OT182 should not be used to identify genes involved in biofilm formation in Burkholderia pseudomallei . Southeast Asian J Trop Med Public Health 43: 124–128. [PubMed] [Google Scholar]
- 39. Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, et al. (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166: 175–176. [DOI] [PubMed] [Google Scholar]
- 40. Talaat AM, Hunter P, Johnston SA (2000) Genome-directed primers for selective labeling of bacterial transcripts for DNA microarray analysis. Nat Biotechnol 18: 679–682. [DOI] [PubMed] [Google Scholar]
- 41. Burkholderia mallei/pseudomallei microarray version 2 Description. Available: http://pfgrc.jcvi.org/index.php/microarray/array_description/burkholderia_mallei/version2.html. Accessed 20 Jul 2010.
- 42. Bartpho T, Wongsurawat T, Wongratanacheewin S, Talaat AM, Karoonuthaisiri N, et al. (2012) Genomic islands as a marker to differentiate between clinical and environmental Burkholderia pseudomallei . PLoS One 7: e37762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Talaat AM, Howard ST, Hale Wt, Lyons R, Garner H, et al. (2002) Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res 30: e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28: 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166: 557–580. [DOI] [PubMed] [Google Scholar]
- 48. Miller VL, Mekalanos JJ (1988) A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170: 2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Simon R, Preifer U, Uhler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio Technol 1: 784–91. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fold changes in gene expression in vivo / in vitro . Fold changes in the expression of the BPSS1521 (A) and BPSS1512 (B) genes as determined by DNA microarray (upper) and validated by qRT-PCR (lower) in the lungs of BALB/c (▪) and C57BL/6 mice (□). The difference in the fold change determined by qRT-PCR between BALB/c and C57BL/6 mice was not significant (p = 0.13 and 0.16, respectively).
(TIF)
Growth curves of the B. pseudomallei K96243 wild-type (•) and bprD mutant (□) strains; no significant difference was evident ( p = 0.23).
(TIF)
Numbers of bacteria in the spleen (A), lung (B), and liver (C) of BALB/c (♦) and C57BL/6 (▪) mice on days 3 to 5.
(TIF)
Schematic diagrams of the construction of the B. pseudomallei K96243 bprD mutant using pDM4. (A) Small arrows represent primer sites used to generate the upstream and downstream fragments to clone into pDM4 together with the Tcr cassette from pUTminiTn5Tc, to generate the pDMΔbprD::Tcr plasmid. (B1) Dotted cross indicates the first and second recombination steps used to replace bprD on the B. pseudomallei K96243 chromosome with the Tcr cassette from pDMΔbprD::Tcr, resulting in generation of the bprD mutant. (B2) Dotted cross indicates the first and second recombination steps at the same site leading to abortive allelic exchange and generation of the wild type rather than the bprD mutant.
(TIF)