Abstract
Mitogen-activated protein kinase (MAPK) cascades have crucial roles in the regulation of plant development and in plant responses to stress. Plant recognition of pathogen-associated molecular patterns or pathogen-derived effector proteins has been shown to trigger activation of several MAPKs. This then controls defence responses, including synthesis and/or signalling of defence hormones and activation of defence related genes. The MAPK cascade genes are highly complex and interconnected, and thus the precise signalling mechanisms in specific plant–pathogen interactions are still not known. Here we investigated the MAPK signalling network involved in immune responses of potato (Solanum tuberosum L.) to Potato virus Y, an important potato pathogen worldwide. Sequence analysis was performed to identify the complete MAPK kinase (MKK) family in potato, and to identify those regulated in the hypersensitive resistance response to Potato virus Y infection. Arabidopsis has 10 MKK family members, of which we identified five in potato and tomato (Solanum lycopersicum L.), and eight in Nicotiana benthamiana. Among these, StMKK6 is the most strongly regulated gene in response to Potato virus Y. The salicylic acid treatment revealed that StMKK6 is regulated by the hormone that is in agreement with the salicylic acid-regulated domains found in the StMKK6 promoter. The involvement of StMKK6 in potato defence response was confirmed by localisation studies, where StMKK6 accumulated strongly only in Potato-virus-Y-infected plants, and predominantly in the cell nucleus. Using a yeast two-hybrid method, we identified three StMKK6 targets downstream in the MAPK cascade: StMAPK4_2, StMAPK6 and StMAPK13. These data together provide further insight into the StMKK6 signalling module and its involvement in plant defence.
Introduction
Mitogen-activated protein kinase (MAPK) cascades are conserved signalling modules in eukaryotes that transduce extracellular stimuli downstream from the receptors, thus mediating the intracellular responses. The plant MAPK cascades have pivotal roles in the regulation of plant development and in responses to a variety of stress stimuli, including pathogen infection, wounding, temperature, drought, salinity, osmolarity, UV irradiation, ozone and reactive oxygen species [1].
In a general model of the MAPK signalling cascade, activation of plasma membrane receptors activates the MAPK kinase kinases (MKKKs). These are serine or threonine kinases that phosphorylate a conserved S/T-X3−5-S/T motif of the downstream MAPK kinases (MKKs), which, in turn, phosphorylate MAPKs on threonine and tyrosine residues in a conserved T-X-Y motif of their activation loop [2]. Following this MKK alteration of the phosphorylation-dependent properties of their target proteins, the activated MAPKs translate the information further, which eventually leads to changes in, e.g., gene expression, cellular redox state, or cell integrity [3].
The genome of the model plant Arabidopsis thaliana encodes 60 MKKKs, 10 MKKs and 20 MAPKs [4]. This indicates that the MAPK cascade might not simply consist of a single MKKK, MKK and MAPK connected together, but has the potential to be organised into many thousands of distinct MKKK–MKK–MAPK combinations, with some level of redundancy. To minimise unwanted cross-talk, the spatial and temporal activities of the different components must be strictly regulated [5].
Despite the potential multiplicity of MAPK cascades, only a small number of MAPK modules have been experimentally defined [6]. As the MKK family consists of a relatively small number of genes, their activity in different MAPK modules is widely dispersed [5]. In Arabidopsis, MKKs can be divided into four different groups (A–D) based on their sequence similarities [4], [7]. Group A includes Arabidopsis thaliana AtMKK1, AtMKK2 and AtMKK6. AtMKK1 and AtMKK2 act upstream of AtMAPK4 in response to cold, salinity and pathogens [8], [9]. AtMKK6 is involved in cytokinesis control and cell-cycle regulation [10]. The group B MKKs includes AtMKK3, which functions upstream of AtMAPK6 in the regulation of jasmonic acid (JA) signal transduction [11] and is involved in pathogen defence [12]; overexpression of AtMKK3 leads to enhanced tolerance to salt and increased sensitivity to abscisic acid [13]. The group C MKKs include AtMKK4 and AtMKK5, which act upstream of AtMAPK3 and AtMAPK6 in the regulation of plant development and defence responses [1], [14]–[16]. The group D MKKs include AtMKK7, AtMKK8, AtMKK9 and AtMKK10. AtMKK9 is involved in ethylene signalling [17] and in leaf senescence [18], while AtMKK7 is involved in plant basal and systemic resistance [19].
During pathogen attack, MAPK signalling is an indispensable component of the host defence response, in a way that it is involved in the crosstalk between secondary messengers and hormones [20]. The key hormones in plant biotic interactions include the salicylates, jasmonates and ethylene [21], whereby their specific roles depend on the particular host–pathogen interaction. Many studies have indicated that salicylic acid (SA) is a key regulatory compound in disease resistance against fungi, bacteria and viruses (reviewed in [22]). The importance of SA in viral multiplication and symptom development has also been confirmed in potato (Solanum tuberosum L.) - Potato virus Y (PVY) interaction [23], [24]. Depending on the virus, SA can induce inhibition of viral replication and cell-to-cell or long-distance viral movement (reviewed in [25]) and in agreement with this, SA is also a key component in the directing of events during and following hypersensitive resistance (HR) [21], [24]. Hypersensitive resistance is an efficient defence strategy in plants, as it restricts pathogen growth and can be activated during host, as well as non-host, interactions. It involves programmed cell death and manifests in necrotic lesions at the site of pathogen attack (reviewed in [26]). Potato virus Y is a member of the Potyviridae family and it is an important potato pathogen worldwide. In potato, HR is conferred by the Ny genes (reviewed in [27]). The potato cultivar (cv.) Rywal carries the Ny-1 gene and it develops HR that is manifested as necrotic lesions on leaves 3 days following their inoculation with various PVY strains [28].
To date, a large number of members of the MAPK cascades from different species have been investigated, although to the best of our knowledge, there has been no systematic investigation of the MKK family and its function in defence signalling in potato. Moreover, no MAPK immune response network module has been defined for potato – PVY interactions.
We thus first performed sequence analysis of the complete MKK gene family in potato, and of its close relatives, where their genomes have been sequenced. Based on the present transcriptome data, StMKK6 was identified as the most responsive member after viral attack. We further investigated the role of StMKK6 in the response to PVY infection at the gene expression level, and studied its intracellular localisation and identified its downstream targets in the MAPK signalling cascade.
Materials and Methods
Bioinformatics analysis
Arabidopsis thaliana MKK gene family (The Arabidopsis Information Resource; TAIR: http://www.arabidopsis.org/) was blasted (tBLASTx algorithm) [29] against the potato [30], tomato (Solanum lycopersicum L.) and Nicotiana benthamiana (The SOL Genomics Network: http://solgenomics.net/) genomes to identify the MKK gene family in all three Solanaceae species. The names of the potato MKK genes were assigned based on the names of their apparent A. thaliana orthologues.
To identify and gather all of the available sequence information on StMKK6 (GenBank accession number KF837127), StMAPK4_1 (GenBank accession number KJ027594), StMAPK4_2 (GenBank accession number KJ027595), StMAPK6 (GenBank accession number KJ027596) and StMAPK13 (GenBank accession number KJ027597), several database searches were performed. Sequences of our isolated genes were blasted (tBLASTx; [29]) against NCBI (National Centre for Biotechnology Information: http://www.ncbi.nlm.nih.gov/); UniProt (Universal Protein Resource: http://www.uniprot.org/); TAIR; PlantGDB (http://www.plantgdb.org/); POCI (Potato Oligo Chip Initiative: http://pgrc-35.ipk-gatersleben.de/pls/htmldb_pgrc/f?p=194∶1); The Gene Index Project (http://compbio.dfci.harvard.edu/tgi/); and TIGR Plant Transcripts Assemblies (http://plantta.jcvi.org/search.shtml).
To identify StMKK6 orthologues we searched the UniProt database using the BLASTp algorithm [29]. Multiple-sequence alignments were performed using the MAFFT programme (version 7) [31]. Phylogenetic trees were constructed based on protein sequences using the MEGA 5 software [32], with the neighbour-joining method [33]. Bootstrap values were derived from 1,000 replicates, to quantify the relative support for branches of the inferred phylogenetic tree. To distinguish between MKK6 orthologues and other MKKs, other AtMKKs were included in the first MAFFT analysis.
Information on the targets of AtMKK6 was collected from the Arabidopsis Interactome Network Map [34]. The corresponding targets in potato were assigned by A. thaliana-potato orthologue and potato paralogue connections [35].
The location of the StMKK6 gene (PGSC0003DMG403005720) within the potato genome was identified using the Ensembl Plants portal (http://plants.ensembl.org/index.html). The promoter sequences were obtained from the Solanum_tuberosum-PGSC_DM_v34_superscaffolds database. Promoter sequences were analysed using the PlantCARE web service [36].
Localisation predictions of the A. thaliana and potato MKK6 proteins were performed using PredictProtein [37] and the data on the whole A. thaliana MKK family using the SUBA3 web services [38].
Metadata analysis
The expression data of the StMKK genes in developmental tissues and under stress conditions were collected through the use of the Bio-Analytic Resource for Plant Biology (http://bar.utoronto.ca/welcome.htm) and the Potato eFP browser [39]. The developmental tissues were vegetative and reproductive organs from greenhouse-grown plants. For each MKK, the RNA expression signals (FPKM values) based on the RNA sequencing of double monoploid S. tuberosum Group Phureja clone DM1-3 (DM) [40] and the genome sequence and analysis of the tuber crop potato [30] were collected. AtMKK6 transcription data from various datasets were analysed using Genevestigator [41] and only two-fold changes were regarded as significant.
We collected data from previously reported microarray expression analyses of the potato HR cv. Rywal and the SA deficient NahG-Rywal infected with PVYN-Wi ([24], GEO: GSE46180), for all of the potato MKKs and MAPKs. Microarray identifiers were connected to the PGSC gene identifiers [35] and only the probes with less than four single nucleotide polymorphisms compared to the PGSC transcripts were used.
Co-expression analysis of microarray expression datasets was carried out with Biolayout Express3D 3.0 software [42], with ratios of PVY/mock normalised signals. The network was constructed using the Pearson correlation coefficient threshold of 0.98, and the graph was clustered using the Markov clustering algorithm with inflation 2.3; the other parameters were set at the default values.
Gene expression analysis
Total RNA from control and SA- treated samples was extracted using MagMAX-96 Total RNA Isolation Kit (Life Technologies) according to manufacturer's instructions. RNA concentration was quantified by UV absorption at 260 nm using a NanoDrop ND1000 spectrophotometer (Nanodrop technologies). Integrity and purity of the RNA samples were determined by agarose gel electrophoresis and OD 260/280 nm absorption ratios. To confirm microarray results the same RNA samples previously analysed in microarray experiments [24] were used. Reverse transcription was performed on 1 µg of total RNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The expression of StMKK6 was assayed by real-time PCR (qPCR). The primers and probe (MKK6_F, MKK6_R and MKK6_S) targeting StMKK6 were designed by Primer Express 2.0 (Applied Biosystems) (Table S1 in File S1). The qPCR was performed using TaqMan chemistry, with Cq values determined as described previously [24]. All reactions were run on a 7500 Fast Real-Time PCR System (Applied Biosystems) in 5 µl volume with 300 nM concentration of primers and 150 nM of probe and performed in duplicate. Linearity (R2) and efficiency (E = 10[−1/slope]) [43] of each reaction were compared to the accepted values. The standard curve method was used for relative gene expression quantification. Target gene accumulation was normalised to two endogenous control genes, cytochrome oxidase (COX) [44] and elongation factor 1 (EF-1) [45]. To determine differentially expressed genes, the Student t-test was performed.
Cloning
The full-length sequences of StMKK6, StMAPK4_1, StMAPK4_2, StMAPK6 and StMAPK13 were amplified from potato cv. Rywal cDNA with the primers listed in Table S1 in File S1. The fragments were inserted into the pJET 1.2 blunt cloning vector (Thermo Scientific) and sequenced (GATC Biotech). For the localisation studies, StMKK6 was cloned into the pENTR/D TOPO vector (Invitrogen), using the LR reaction, following the manufacturer protocol, and recombined into the binary destination vectors pH7YWG2 (YFP) and PH7CWG2 (CFP) [46], to produce proteins with C-terminal YFP/CFP fusion.
The StMKK6 promoter was amplified using the genomic library from potato cv. Santé as the template, which was constructed using GenomeWalker Universal kit (BD Biosciences Clontech), as described previously [47]. In the genome walking PCR amplifications, the Advantage 2 Polymerase Mix (Clontech) was used with the PCR conditions suggested by the manufacturer. The adaptor primer AP1 and a nested primer AP2, that were provided by the manufacturer were paired with the nested reverse gene-specific primers MKK6_AP1 and MKK6_AP2 for amplification of the region upstream from the StMKK6 gene. The fragment was inserted into the pJET 1.2 blunt cloning vector (Thermo Scientific) and sequenced. The 200-bp region of the StMKK6 promoter (identical in all three of the investigated genotypes) was afterwards cloned in front of the StMKK6 gene in the pH7YWG2 vector, using QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies), with the MAPKprom-YFP_F and MKK6prom-YFP_R primers. The StMKK6 promoter region from potato cv. Rywal was amplified from the genomic DNA using the primers listed in Table S1 in File S1.
For the yeast two-hybrid analysis, the coding regions of the selected genes were cloned into pGBKT7 (StMKK6) or pGADT7 (StMAPK4_1, - 4_2, - 6, -13) using QuikChange II Site-Directed Mutagenesis kit (Agilent Technologies). The primers used are listed in Table S1 in File S1.
Hormonal treatment
Two different potato genotypes were used in this study including 1 non-transgenic cv. Rywal and 1 transgenic line NahG, transgenic plants of cv. Rywal (NahG-Rywal) [24]. Potato plants were grown in stem node tissue culture. Two weeks after node segmentation they were transferred to soil and grown as previously described by Baebler et al. 2009 [45].
SA treatment was performed spraying plants with SA-analog 300 µM INA (98% 2,6-Dichloroisonicotinic acid, Aldrich) in distilled water. Control plants were sprayed with distilled water alone. Leaves were harvested after 24 h of treatment and immediately frozen in liquid nitrogen. Two biological replicates per treatment were analysed.
Localisation studies
N. benthamiana plants were grown in a growth chamber at 22/20°C under a 16/8 h light/dark cycle. PVYNTN (isolate NIB-NTN, AJ585342) inoculation of N. benthamiana was performed as described by Baebler et al. 2009 [45]. Plasmids pH7YWG2 (YFP), pH7CWG2 (CFP), pH7YWG2::StMKK6-YFP, pH7CWG2::StMKK6-CFP under the 35S promoter, and pH7YWG2::StMKK6 under the control of the native promoter, were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. The A. tumefaciens cells were cultured, harvested by centrifugation, and re-suspended in 0.2 mM acetosyringone solution, and infiltrated into 4–5-week-old N. benthamiana leaves, 8 days after PVY/mock inoculation.
After 48 h to 72 h, YFP/CFP were visualized with a Leica TCS SP5 laser-scanning microscope mounted on a Leica DMI 6000 CS inverted microscope (Leica Microsystems, Germany), with an HC PL FLUOTAR 10.0×0.30 DRY objective. For excitation, the 458 nm (enhanced cyan fluorescent protein) and 514 nm (enhanced yellow fluorescent protein) lines of an Argon laser were used. The enhanced cyan fluorescent protein emission was measured from 475 nm to 495 nm, and the enhanced yellow fluorescent protein from 525 nm to 550 nm. Autofluorescence emission was measured from 690 nm to 750 nm. Differential interference contrast (DIC) images were captured using the transmission light detector of the confocal microscope. All of the images were acquired with the 10× objective or with an additional 3.64× zoom. The acquired images were processed using the Leica LAS AF Lite software (Leica Microsystems). pH7YWG2 and pH7CWG2 without the inserted StMKK6 were used as the controls.
Yeast two-hybrid assay
The coding regions of selected genes were cloned into pGBKT7 (StMKK6) or pGADT7 (StMAPK4_1, -4_2, -6, -13). To introduce insertions into plasmids the QuikChange II Site-Directed Mutagenesis Kit (AgilentTechnologies) was used. The primers used are listed in Table S1 in File S1.
Yeast two-hybrid analysis was performed using the Matchmaker GAL4-based two-hybrid assay (Clontech). The vector pGBKT7, containing bait protein fused to Gal DNA-binding domain, was used to transform the yeast strain Y2H Gold through the polyethylene glycol/LiAc-based method (Clontech), according to manufacturer instructions. After selection on SD/-Leu medium, the transformed colonies were used for co-transformation following the protocol described by Clontech, with the pGADT7 vector. It was used as a prey and contained target genes fused to Gal DNA-activation domain. Co-transformants were selected on SD/-Leu/-Trp (DDO) medium. Positive interaction transformants were selected on SD/-Leu/-Trp/-His/-Ade/x-a-Gal/Aba (QDO/X/A) medium. All of the genes were previously tested for autoactivation and toxicity, plating yeast colonies transformed with pGBKT7_StMKK6 on SD/-Trp/x-a-Gal/Aba and pGADT7_StMAPK4_1, -4_2, -6, -13 on SD/-Leu/x-a-Gal/Aba. Positive (pGBKT7_SV40 large T-antigen -pGADT7_53) and negative (pGBKT7_SV40 large T-antigen -pGADT7_Lam) interaction controls, included in the Matchmaker Gold Yeast Two-Hybrid System (Clontech), were performed in parallel.
Results
The MKK family in potato
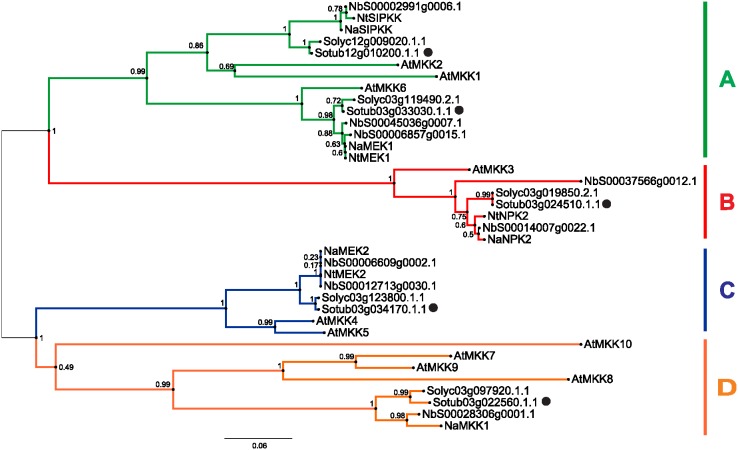
Our genome analysis of the potato, tomato and Nicotiana attenuata genomes identified five MKK genes in each, with four in the tobacco Nicotiana tabacum L. genome, and eight in the N. benthamiana genome (Figure 1). Based on the phylogenetic tree of Arabidopsis and five Solanaceae species, for only two of the A. thaliana genes (AtMKK3, AtMKK6) a specific orthologue could be assigned from potato, tomato or Nicotiana spp., while the other MKKs form larger orthologue groups. The MKK family in potato, tomato and Nicotiana spp. can also be divided into four groups: group A (A. thaliana and N. benthamiana, three genes; potato, tomato, N. tabacum and N. attenuata, two genes); B (N. benthamiana, two genes; the rest, one gene each); C (A. thaliana and N. benthamiana, two genes; potato, tomato, N. tabacum and N. attenuata, one gene each); and D (A. thaliana, four genes; Solanaceae spp., one gene each, except N. tabacum which has none). Out of the five examined Solanaceae spp., N. benthamiana is the only one of these in which duplication of the MKK genes occurred after speciation and thus N. benthamiana has possible paralogues for MKK3 (NbS00014007g0022.1, NbS00037566g0012.1), MKK4/5 (NbS00012713g0030.1, NbS00006609g0002.1) and MKK6 (NbS00006857g0015.1, NbS00045036g0007.1) (Figure 1).
Figure 1. Phylogenetic tree of MKK family in A. thaliana and five Solanaceae species.
The species are A. thaliana (At), potato (Sotub), tomato (Solyc), N. benthamiana (Nb), N. attenuata (Na) and N. tabacum (Nt). Genes are grouped into 4 groups: A (green), B (red), C (blue) and D (orange) [4]. Potato genes are marked with dots. The numbers on the nodes are percentages from a bootstrap analysis of 1000 replicates. The scale bar indicates the branch length that corresponds to 0.06 substitutions per site.
Regulation of potato MKK family gene expression in developmental processes and stress
We first analysed the gene-expression metadata for the potato MKK family for developmental processes and after exposure to different biotic and abiotic stress [40]. The tissue specificities of the MKK family members differed between both of the potato varieties studied here, the potato Group Phureja Clone DM, and Group Tuberosum Clone RH, especially regarding the expression in tubers and stolons. In general, this analysis showed that in potato, StMKK1/2 and StMKK4/5 are relatively uniformly expressed, with the highest expression in tubers and stolons. StMKK3 transcripts had the highest abundance in carpels and petals, and StMKK7/9 in tubers and flowers, and also in roots for Clone RH. Under biotic and abiotic stress conditions, all of the potato MKKs except StMKK4/5, were down-regulated after wounding. None of the other stress conditions and hormonal treatments influenced the MKK family gene expression by more than two-fold compared to controls (File S2).
In a previous transcriptome analysis of potato HR responses to PVY infection [24], we checked the expression profile of identified potato MKKs [35]. According to the gene expression profiles obtained for the potato cv. Rywal, three out of five MKKs showed differential expression for the HR response to PVY: StMKK3, StMKK6 and StMKK4/5 (Figure 2). StMKK6 showed the strongest response of all, thus its expression data were also confirmed by quantitative real-time PCR (qPCR) (Table S2 in File S1). StMKK6 was strongly up-regulated before lesion formation (3 dpi). In contrast, at the same time, StMKK3 was down-regulated. StMKK4/5 showed two-fold induction at the later time (6 dpi) following inoculation. For the SA-deficient plants (NahG-Rywal), the regulation of these MKKs at the gene level was either attenuated (for StMKK3 and StMKK4/5) or delayed (for StMKK6). Only StMKK7/9 was induced earlier and stronger in the NahG-Rywal plants, compared to the non-transgenic plants.
Figure 2. Gene expression pattern of MKK family in the HR response against PVY.

Cultivar Rywal (HR response, conferred by Ny-1 gene) and NahG-Rywal (impaired accumulation of SA) were analysed for whole transcriptome response 1, 3 and 6 days after PVYN-Wi infection [24]. A. thaliana and S. tuberosum PGSC orthologues were assigned to each probe. Log2 fold changes of PVY in infected vs. mock-inoculated plants are indicated for each time point (1, 3 and 6 dpi). Statistically significant differences (FDR corrected p<0.05) are in bold. Up-regulated values are in blue and down-regulated values are in yellow.
Based on these data we focused on further investigations on the AtMKK6 orthologue in potato, StMKK6.
StMKK6 orthologues across the plant kingdom
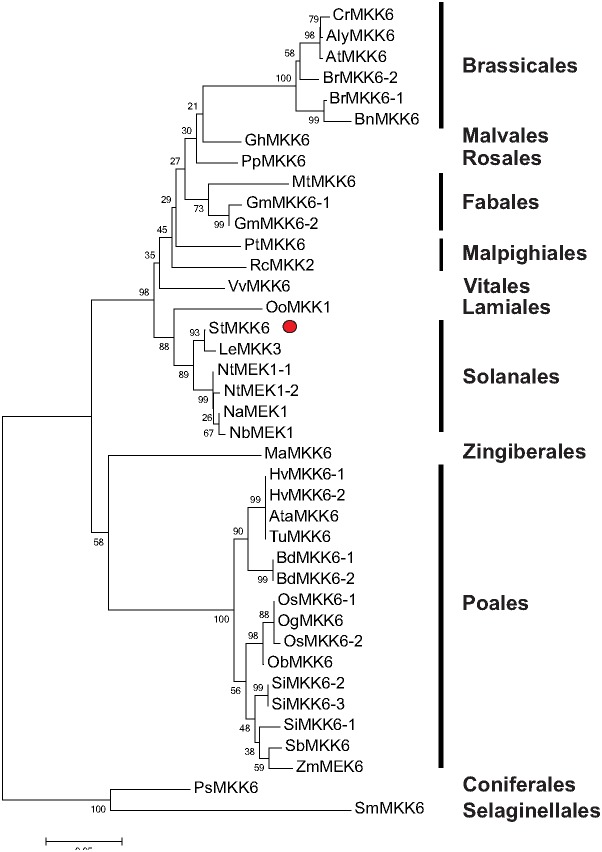
To study the sequence diversity and evolution of the MKK6 gene, we performed a phylogenetic analysis within the plant kingdom. We identified 39 plant MKK6 orthologues among higher plants. As well as for different families of Angiosperms, one MKK6 gene was also identified in Gymnosperms, and one in mosses (Figure 3 and File S3). MKK6 is mostly a single-copy gene, as these 39 MKK6 orthologues belong to 31 plant species (Figure 3).
Figure 3. Phylogenetic tree of potato MKK6 and its orthologues in different plant species, divided into classes.
The species of origin for each MKK is marked by species acronym before the protein name: Cr, Capsella rubella; Aly, Arabidopsis lyrata; At, Arabidopsis thaliana; Br, Brassica rapa; Bn, Brassica napus; Gh, Gossypium hirsutum; Pp, Prunus persica; Mt, Medicago trunculata; Gm, Glycine max; Pt, Populus trichocarpa; Rc, Ricinus communis; Vv, Vitis vinifera; Oo, Origanum onites; St, Solanum tuberosum; Le, Solanum lycopersicum (Lycopersicum esculetum); Nt, Nicotiana tabacum; Na, Nicotiana attenuata; Nb, Nicotiana benthamiana; Ma, Musa acuminata; Hv, Hordeum vulgare, Ata, Aegilops tauschii; Tu, Triticum urartu; Bd, Brachypodium distachyon; Os, Oryza sativa, Og, Oryza glaberrima; Ob, Oryza brachyantha; Si, Setaria italica; Sb, Sorghum bicolor; Zm, Zea mays, Ps, Picea sitchensis; Sm, Selaginella moellendorffii. Potato MKK6 is marked with red dot. The numbers on the nodes are percentages from a bootstrap analysis of 1000 replicates. The scale bar indicates the branch length that corresponds to 0.06 substitutions per site.
Comparison of the MKK6 sequence from potato cv. Rywal and the published potato genome sequence of S. tuberosum Group Phureja, Clone DM showed that the coding domain sequences are different for three nucleotides at different sites, which results in only one amino-acid difference; this is not part of the protein kinase domain.
Regulation of StMKK6 gene expression in development and stress
To better understand the function of the MKK6s, we examined the gene expression data that is available for A. thaliana MKK6 in Genevestigator. AtMKK6 is up-regulated in developmental processes (callus formation, germination) and after treatments with translation inhibitor cyclohexamide and hormones auxins. AtMKK6 is also up-regulated after flagellin 22 treatment, and under abiotic stress (cold, salt, acidic pH, hypoxia). AtMKK6 is down-regulated after treatments with abscisic acid and combinations of abscisic acid with jasmonate or SA.
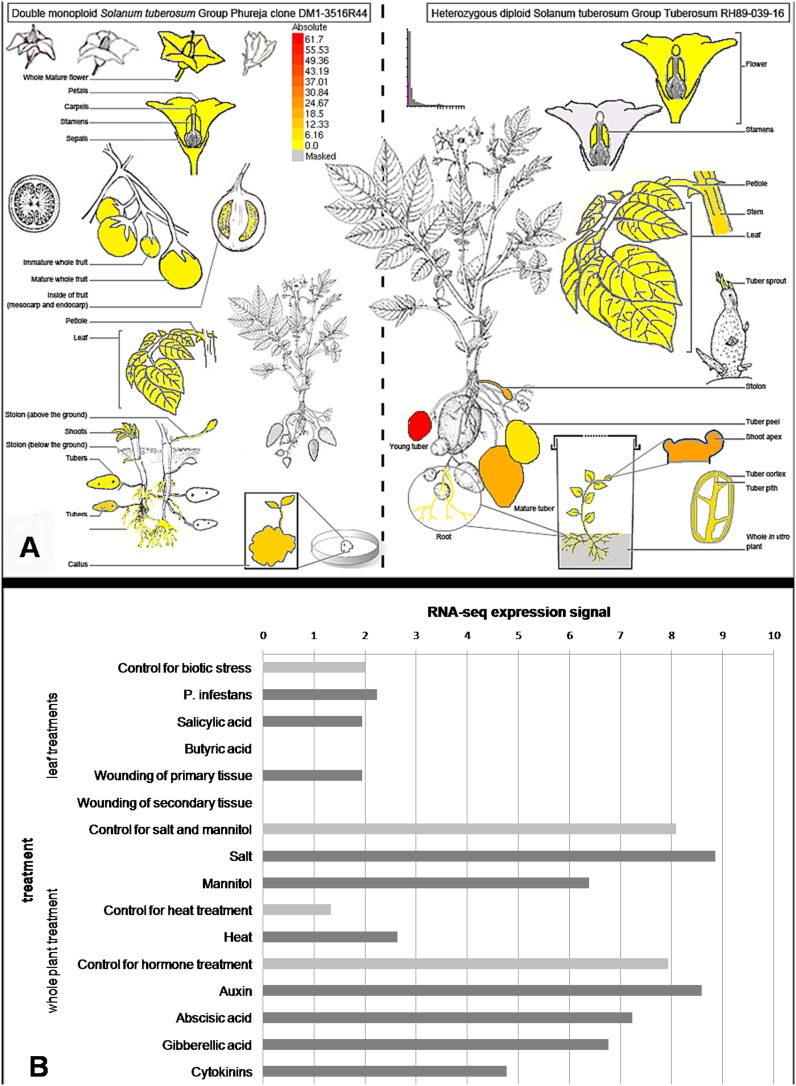
The expression profiles of StMKK6 obtained from the Potato eFP Browser database showed that in the potato Group Phureja Clone DM, StMKK6 tissue expression was high in the tubers and callus, and very low (below detection levels) in the flowers. In the Group Tuberosum Clone RH under non-stress conditions, StMKK6 gene expression was strong also in the shoot apex (Figure 4A; [40]). Among all of the stress-related conditions included, compared to the controls, StMKK6 was down-regulated after wounding of the secondary leaves and after treatment with the fungal elicitor DL-β-amino-n-butyric acid (Figure 4B).
Figure 4. StMKK6 gene expression in development and stress [40].
Data are obtained from the potato eFP browser [39]. A. Tissue and developmental gene expression pattern of StMKK6 in double monoploid S. tuberosum Group Phureja (left) and in heterozygous diploid S. tuberosum Group Tuberosum (right). B. Changes in StMKK6 gene expression under biotic stress conditions (treatments of leaves with P. infestans, SA analogues acibenzolar-S-methyl and fungal elicitor DL-β-amino-n-butyric acid and wounding) and abiotic stress conditions (treatments of whole plants with salt, heat and hormones cytokinins, 6 benzylaminopurine; gibberellins, GA3; abscisic acid; and auxin, IAA) and control treatments.
Analysis of the StMKK6 native promoter
In the Potato Genome Sequencing Consortium (PGSC) Potato Genome Browser, StMKK6 is assigned to PGSC gene ID PGSC0003DMG403005720, and it is localised to chromosome 3. Based on this information, we also cloned the StMKK6 promoter sequence from two cultivars of S. tuberosum Group Tuberosum, both of which are resistant to PVY infection: cv. Rywal (GenBank accession number KF837128) and cv. Santé (GenBank accession number KF837129). The 790 bp-long promoter isolated from cv. Rywal (Table S3 in File S1) is identical to the promoter of S. tuberosum Group Phureja, but different from that of cv. Santé (Table S4 in File S1), due to a 27-bp-long region that is localised 230 bp upstream of StMKK6. In general, the most common domains were the TATA-box, CAAT box and domains related to light responsiveness. Based on the development and stress-related domains found, StMKK6 appears to be involved in development of the endosperm and in responses to heat stress and wounding. StMKK6 is also predicted to be involved in gibberellic acid and salicylic acid signalling, and is under regulation by the circadian clock (Figure 5).
Figure 5. Predicted regulatory domains of StMKK6 native promoter from S. tuberosum cv. Rywal, 790-bp-long.
The crucial domains that regulate stress responses and development are CCAAT-box (MYBHv1 binding site; yellow), GCN4 motif (endosperm expression; pink), HSE (heat stress; brown), P-box (gibberellins; blue), Skn-1 motif (endosperm expression; purple), TCA element (salicylic acid; green), WUN motif (wounding; red) and circadian motif (control of circadian clock; grey). The arrow indicates the predicted beginning of the 5’ UTR region and the red shaded ATG indicates the start of StMKK6 coding region. The complete list of StMKK6 promoter domains from cv. Santé and cv. Rywal with their position, strand and sequence are specified in Tables S3 and S4 in File S1.
Regulation of StMKK6 by salicylic acid
Promoter analysis of StMKK6 has demonstrated potential involvement of StMKK6 in SA signalling. Therefore, we examined expression of StMKK6 under the SA treatment. The expression of StMKK6 was measured by qPCR 24 h after treatment with SA in Rywal and NahG-Rywal plants.
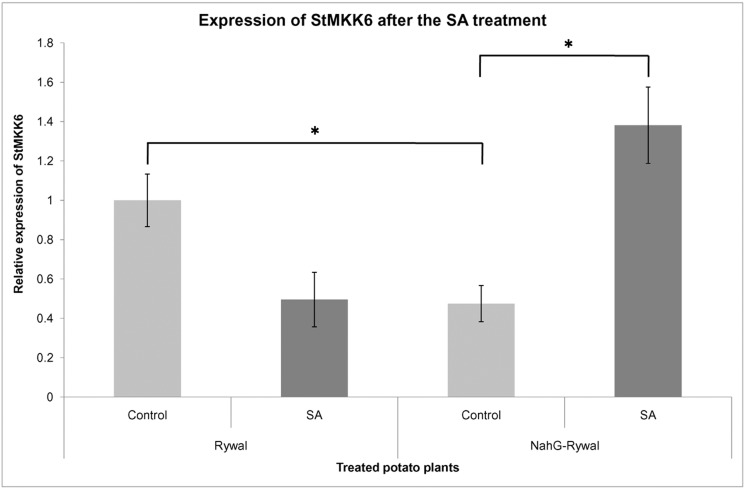
The treatment showed StMKK6 to be sensitive to the present active form of SA since significant changes of StMKK6 expression were observed in the SA-deficient genotype NahG-Rywal (Figure 6). There was a substantial, two-fold rise, of the StMKK6 RNA, compared to the non-treated plants. In the Rywal plants, that received the same SA treatment, the change in the StMKK6 expression was not significant (Table S5 in File S1).
Figure 6. Expression of StMKK6 in control and SA-treated Rywal and NahG-Rywal potato plants.
Expression of the endogenous StMKK6 in NahG-Rywal is at least two-fold lower than in Rywal. After the SA-treatment the expression of StMKK6 in NahG-Rywal increased for at least three times, comparing to the non-treated control. Difference in the endogenous StMKK6 expression between Rywal and NahG-Rywal as well as difference in the StMKK6 expression between SA-treated and non-treated NahG-Rywal plants are statistically significant (p<0.05).
We also observed significant difference in the RNA level of the endogenous StMKK6 in the non-treated Rywal, compared to the NahG-Rywal (Figure 6). In Rywal plants there was at least twice as much of expressed StMKK6 comparing to the level in the NahG-Rywal (Table S6 in File S1).
Localisation of the StMKK6 protein in healthy and PVY-infected epidermal leaf cells
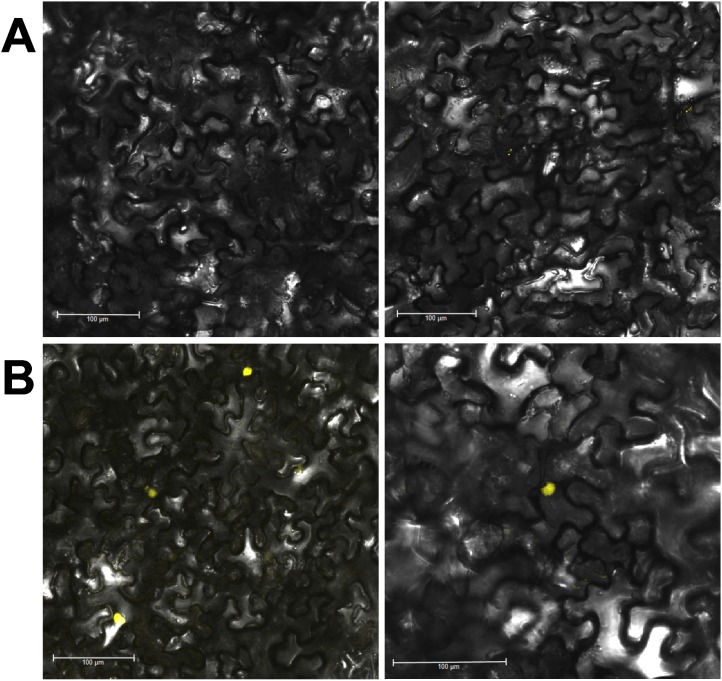
To learn more about the function of the StMKK6 protein in defence responses against the virus PVY, we analysed StMKK6 localisation in healthy and infected N. benthamiana plants. The infected leaves of N. benthamiana developed no necrotic lesions due to infection with PVY or overexpression of StMKK6. In-silico analysis predicted the localisation of StMKK6 as in the nucleus and cytoplasm. The same results were obtained also for AtMKK6 (Table S7 in File S1). An in-vivo study of the subcellular localisation of StMKK6 was performed under the native promoter and in C-terminal translational fusion with YFP in N. benthamiana leaves. In the mock-inoculated leaves, no fluorescence of the StMKK6-YFP fusion protein was observed (Figure 7A), while in PVY-infected leaves, there was strong StMKK6-YFP fluorescence in the nucleus and very weak StMKK6-YFP fluorescence in the cytoplasm (Figure 7B).
Figure 7. Epidermal cells of N. benthamiana expressing translational fusion of StMKK6 with YFP under native promoter.
Leaves were agroinfiltrated when the virus has spread uniformly through the inoculated leaves (8 days after inoculation) and observed after 72 h in two independent experiments. Examples from two plants (left and right panels) are shown. Control of transformation (fluorescent marker without StMKK6 fusion) is in the Figure S1A. A. Localisation of StMKK6 in mock-inoculated leaves. No fluorescence was observed. B. Localisation of StMKK6 in PVY-inoculated leaves, where the protein accumulates predominantly in nucleus. Additional images of StMKK6 localisation under native promoter are in Figures S1D and S1E.
The fluorescent protein itself did not affect the localisation of StMKK6, as fusions with different fluorescent proteins were observed in the same intracellular compartments. The overexpression of StMKK6, driven by the CaMV 35S promoter, resulted in localisation of the protein in the cytoplasm and nucleus in mock-inoculated as well as in PVY-infected leaves (Figures S1B and S1C).
Search for StMKK6 co-regulated genes and potential targets in the MAPK cascade
To further improve our knowledge of StMKK6 function, we searched for the other components of the MAPK module that is involved in the HR response in potato, the genes that are co-expressed with StMKK6, and the possible MAPK targets.
We found 22 co-expressed genes (File S4), for which we analysed the promoter regions. The most common biotic stress-related domains in their promoters were TC-rich repeat motifs (an element involved in defence and stress responsiveness; 13 promoters with the region), TGACG and CGTCA motifs (regulatory elements involved in methyl jasmonate responsiveness; 9 promoters with the region), Box-W1 (fungal elicitor responsive element; 8 promoters with the region), and TCA elements (SA responsiveness; 8 promoters with the region). The only region that was common to all 22 genes was the Skn-1 motif, which is related to endosperm expression.
In an A. thaliana interactome study [34], AtMKK6 was shown to interact with: AtMAPK4 (AT4G01370), AtMAPK6 (AT2G43790), AtMAPK11 (AT1G01560), AtMAPK12 (AT2G46070), AtMAPK13 (AT1G07880), and the calmodulin-like protein TCH3 (AT2G41100).
We first identified the orthologues of all of the potential interacting MAPKs in potato. AtMAPK4 together with AtMAPK11 and AtMAPK12 represent one orthologue cluster with StMAPK4_1 (PGSC003DMP400037535) and StMAPK4_2 (PGCS003DMP400000144). We also identified specific orthologues of AtMAPK6 in Sotub08g010260.1.1, and for AtMAPK13 in PGSC003DMP400044003 (Figure S2). We further cloned all of the potential MAPK interactors of StMKK6 from potato cv. Rywal. Comparisons of the four MAPKs cloned from potato cv. Rywal and those published in the potato genome sequence of S. tuberosum Group Phureja, Clone DM showed Rywal MAPK4_1 to be one-amino-acid different from PGSC0003DMP400037535, while Rywal MAPK 4_2 and MAPK13 are identical to their Phureja orthologues. Rywal MAPK6 had no corresponding proteins in Group Phureja, Clone DM. Yeast two-hybrid assays confirmed the interactions of StMKK6 with three MAPKs: StMAPK4_2, StMAPK6 and StMAPK13 (Figure 8). StMAPK4_1 could not be confirmed as a target of StMKK6 in potato cv. Rywal.
Figure 8. Yeast two-hybrid assays screening StMKK6 interaction partners.
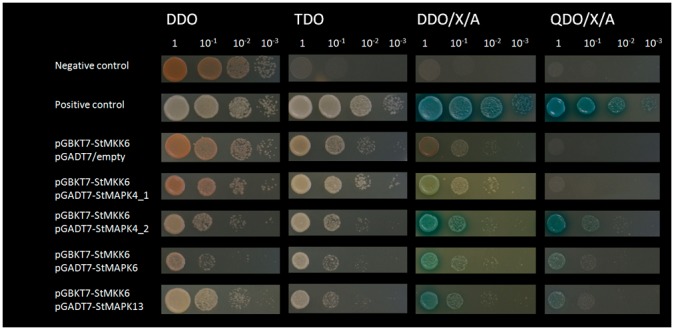
StMKK6 protein was fused with Gal DNA-binding domain as bait and StMAPK4_1, StMAPK4_2, StMAPK6 and StMAPK13 were used as prey fused with Gal DNA-activation domain. Interaction pairs P53/SV40 large T-antigen and Lam/SV40 large T-antigen were used as positive and negative controls respectively. StMKK6/empty vector pair was used as a control to discard auto-activation of bait protein. Serial dilutions of interaction pairs were plated on DDO media for co-transformation selection. Interactions of bait and prey proteins were examined by assessing growth on several selective media with different levels of restrictiveness i.e. SD/-Leu/-Trp/-His (TDO), SD/-Leu/-Trp/x-a-Gal/Aba (DDO/X/A) and QDO/X/A. Only co-transformed colonies growing on the most restrictive media (QDO/X/A) were considered as positive interaction transformants.
StMAPK4_2 and StMAPK13 show differential gene expression patterns after PVY inoculation (Figure 9), and have different expression patterns than StMKK6 (Figure 2).
Figure 9. Gene expression pattern of confirmed StMKK6 targets in the HR response against PVY.

The experimental setup is as in Figure 2. To each probe an A. thaliana orthologue, potato orthologue and a PGSC gene ID were assigned. Log2 of gene expression differences between PVY-infected and mock-inoculated plants are indicated for each time point (1, 3 and 6 dpi). Statistically significant differences (FDR corrected p-value <0.05) are given in bold. The star (*) for StMAPK6 PGSC gene ID indicates that the gene was not predicted in PGSC gene model. Up-regulated values are in blue and down-regulated values are in yellow.
Discussion
To shed further light on the hypersensitive resistance signalling responses during PVY infection of potato, we analysed the involvement of potato MKKs in this process. The results revealed that the StMKK6 is regulated by the SA signalling pathway in the HR to PVY infection. The virus induces the expression of StMKK6 by the protein accumulating preferentially in nucleus.
Potato has five MKKs
To date, MKK genes have been analysed in several plants: in A. thaliana, 10 have been identified [4], in rice, 8 [48], in Brachypodium, 12 [49], in poplar, 10 [50], in apple, 9 [51] and in canola, 7 [52]. In the Solanaceae family, the MKK family is smaller than in other plant species. Our analysis showed that the potato and tomato genomes have five MKK genes, and that the N. benthamiana genome has eight. Five MKKs were identified also in N. attenuata, while in N. tabacum, an additional NbMKK1 orthologue was not found among the currently existing sequences (Figure 1, [53]). The phylogenetic tree of the different MKK genes shows that only N. benthamiana has three possible pairs of paralogues, with none found in potato or tomato. Higher numbers of MKK family members in other plant species might be the result of gene duplications in the MKK ancestors that occurred after divergence into different families. Consequently, it was not possible to assign a single A. thaliana orthologue to three out of five potato MKKs. This issue of assigning orthologues was previously discussed by Dóczi et al. 2012 [54], and was attributed to the high expansion and functional diversification of the MAPKs, and to the rapid evolution of MAPK signalling.
It has been speculated that all 10 AtMKKs are fully functional, although there have been doubts expressed for AtMKK8 and AtMKK10. Although AtMKK8 has all of the typical MKK motifs, there is no evidence of its expression, while AtMKK10 lacks a correctly constructed target site in the activation loop [7]. Thus, in plants with lower numbers of MKK genes, the plasticity of the MAPK signalling module is not necessarily also proportionally lower.
StMKK6 preferentially accumulates in the nucleus only after PVY infection
Transcriptomic analysis indicated that the StMKK6 gene is involved in the HR response against PVY, as the most intensively regulated of the MKKs (Figure 2). Thus, we focused on different aspects of StMKK6 function in this process. Here, meta-analysis of the publicly available datasets (Figure 4) and our in-planta localisation study (Figure 7) show that StMKK6 expression is very low in non-stressed potato leaves. Interestingly, the infection with PVY induces the expression of StMKK6, with the StMKK6 protein concentrated predominantly in nucleus.
The information on the intracellular localisation of StMKK6 and its orthologues is currently really limited. The only report on experimental results of MKKs intracellular localisation showed that AtMKK6 accumulated in the equatorial plane of the phragmoplast in dividing root epidermal cells, while all of the other AtMKKs (AtMKK1-5) did not [10]. In N. benthamiana, localisation of MKK1 (an orthologue of AtMKK7, -8 and -9) was studied for the HR to Phytophtora infestans infection [55], [56]. N. benthamiana MKK1 preferentially accumulated in the nucleus also under non-stress conditions. Liang et al. 2013 [52], studied the localisation of MKK2, -3 and -4 in Brassica napus epidermal leaf cells, and detected the accumulation of these MKKs both in the cytoplasm and the nucleus. On the other hand, other reports on the localisation of the MKK target MAPKs show preferential accumulation of those proteins in the cytoplasm [52], [55]. One however has to note that all above mentioned experiments were studying localisation of MKKs when expressed under strong (e.g. CaMV 35S) promoter, while we have studied localisation under its native promoter (Figure 7). Also in our hands expression of protein under 35S promoter caused ubiquitous cellular localisation (e.g. cytoplasm and nucleus, see Figures S1B and S1C) presumably due to excessive amounts of protein produced, which is known to generate localisation artefacts [57]. Anyhow, our results support the hypothesis that activation of MKK6 targets occurs predominately in the nucleus as indicated also by in silico localisation prediction of identified target MAPKs and that its activitiy is regulated on the level of MKK6 transcription.
Several studies have focused on the gene expression profiles of StMKK6 orthologues in other species. A. thaliana AtMKK6 and maize MKK6 (ZmMEK1) are involved in the regulation of cytokinesis [10], [58], [59] and are required for lateral root formation [60]. ZmMEK1 is also induced by polyethylene glycol, abscisic acid and SA, and it is down-regulated by NaCl [61]. In rice, OsMEK1 (referred to as OsMKK6 in Hamel et al. 2006 [7]) was reported to be involved in signalling of moderately low temperature stress [62], [63]. The present investigation into StMKK6 expression in different tissues of the S. tuberosum group Phureja shows that StMKK6 is highly expressed in tissues with intensive cell proliferation, similar to what has been reported for its orthologues in A. thaliana, tobacco and maize [10], [58], [59], [64].
After the SA treatment of the NahG-Rywal, the level of StMKK6 transcripts three-fold increased, while in non-transgenic Rywal we detected trend of decrease in transcription of StMKK6 gene (Figure 6). Higher basal SA levels in cv. Rywal could explain different regulation of StMKK6 after exogenous treatment. In fact, our results showed that basal levels of StMKK6 transcripts in Rywal are at least two-fold higher than in tha SA-deficient NahG-Rywal which indicates that SA is necessary for the maintenance of basal levels of the StMKK6. This effect was previously reported for other SA-dependent defence-related genes in cv. NahG-Rywal [24] and cv. NahG-Desirée [23]. The network of SA signalling is complex [23], [65]–[67] and expected to show some nonlinear behaviour. Consequently the response we see might be both time as well as concentration dependent and we will not be able to fully predict the outcome of SA signalling before its appropriate dynamic model exisits. In line with hypothesis that StMKK6 is regulated by SA, the analysis of the StMKK6 promoter revealed motifs that are responsive to SA (Figure 5). To the best of our knowledge, there has been only one study linking StMKK6 expression and SA signalling for any of the StMKK6 orthologues (ZmMEK1) [61]. In B. napus, MKK1, MKK2, MKK4 and MKK9 were induced by SA, while only MKK3 was not [52], which indicates that the majority of MKKs are targets of the SA signalling network.
The microarray results show that only MKK3 (group B MKKs) responded in the opposite direction; i.e., MKK3 was repressed after viral infection, contrary to all of the other potato MKKs, which were induced. Similarly, it has been shown also for A. thaliana and tobacco, that the MKK3 acts as a negative regulator in the JA signalling pathway and response to herbivores [11], [16], [68].
MAPK signalling network in hypersensitive resistance response to PVY
We identified here three downstream targets of StMKK6 in potato, StMAPK4_2 (orthologue of AtMAPK4, -11 and -12), StMAPK6 (orthologue of AtMAPK6 and tobacco SIPK) and StMAPK13 (orthologue of AtMAPK13 and tobacco NTF6/NRK1). All of these interactions had already been confirmed in A. thaliana [34], [69], [70]. In rice, OsMEK1 (or OsMKK6) interacts with OsMAPK1, -3, -5 and -6 (orthologues of AtMAPK6, -3 and -11/4, respectively [48], [63]). To date, only one MAPK interactor has been identified for the MKK6 orthologues in Solanaceae: in N. tabacum, NQK1/NtMEK1 was reported to interact with NTF6/NRK1 (orthologue of AtMAPK13 [64]). According to this, we can conclude that the MKK6 signalling module is evolutionarily relatively stable, as most of the interactions appear to be conserved between unrelated species. There are, however, some specificities. For example, out of two potato MAPK4/11/12 paralogues StMKK6 interacts with only one of them, while in A. thaliana it interacts with all three, and in rice, with two.
Among the StMKK6 downstream targets, the present study shows that only StMAPK13 expression (Figure 9) is significantly regulated in the HR response to PVY infection, although while StMKK6 is strongly up-regulated, StMAPK13 is strongly down-regulated. Interestingly StMAPK4_2 and StMAPK13 are significantly induced in SA deficient plants at the same time points as StMKK6. All of the target MAPKs are, however, expressed at (substantially) higher levels in both the mock-infected and PVY-infected leaves, compared to StMKK6 (Figure S3). Therefore, we can hypothesise that the crucial regulation of StMAPK4_2, 6 and 13 is not at the gene expression level, but at a later step; e.g., phosphorylation by newly synthesised StMKK6, or any other post-translational process (e.g. protein translocation or degradation).
As well as StMKK6, we showed that StMKK4/5 (also named as StMEK1 in some studies) is also up-regulated in the HR response of potato to PVY, albeit to a lower extent (slightly below the strict significance cut-off), and showed a delay comparable to StMKK6 in the NahG plants (Figure 2). In N. tabacum, both NtMEK2 (orthologue of StMKK4/5) and NQK1/NtMEK1 (orthologue of StMKK6) are required for N-mediated resistance against tobacco mosaic virus [71]–[73]. Similarly, the silencing of AtMEK1 and AtMEK2 allowed for higher amplification of Cucumber mosaic virus in A. thaliana [74]. This indicates the close connectivity and interdependency between the StMEK1 and StMEK2 signalling modules in plant defence.
Since an effective HR takes place in cv. Rywal in response to PVY, it is possible that some of the regulated potato MKKs are part of this response. A comprehensive analysis of the different MKK functions was performed in Pto-mediated resistance in tomato by Ekengren et al., 2003 [75], and Pedley and Martin, 2004 [76]. Ekengren et al. [75] showed that silencing of either MEK1 or MEK2 breaks the resistance against Pseudomonas syringae. However, Pedley and Martin [76] showed that LeMKK2 (orthologue of AtMKK4/5) and LeMKK4 (orthologue of AtMKKK7/8/9) caused programmed cell death when overexpressed, while LeMKK1 (orthologue of AtMKK1/2) and LeMKK3 (orthologue of MKK6) did not. In A. thaliana, two studies have shown that as well as the MKK4/MKK5 (MEK2) module, another branch of the group A MAPKKs, AtMKK1 and AtMKK2, function together as the second MAPK module involved in the HR response [68], [77]. In N. benthamiana MKK1 (orthologue of AtMKK7/8/9) is a potent inducer of HR-like cell death in response to P. infestans [11], [55], [78] and the same was shown also for AtMKK7 and MKK9 in A. thaliana [79]. In A. thaliana, MKK5 was shown to have a role in the cascade that triggers the HR response [80]. In the present study, there was strong repression of potato MKK3, while this dysregulation was diminished in the NahG-Rywal plants (Figure 2).
The interconnectivity between the NbMEK1 (orthologue of AtMKK6) and NbMEK2 (orthologue of AtMKK4/5) kinase signalling modules was studied mechanistically by del Pozo et al., 2004 [81]. They showed that the silencing of NbMEK1 abolished the cell death caused by constitutively active NbMEK2. In N. attenuata, MEK1 (orthologue of StMKK6) and SIPKK (orthologue of AtMKK1/2) are involved in the regulation of the accumulation of 12-oxo-phytodienoic acid and JA [53], which from another point, supports the role of SA in MKK6 signalling, due to the known antagonistic cross-talk between JA and SA in defence pathways [82].
Conclusions
Our results show that StMKK6 is an important player in potato HR response against PVY infection, as shown on the gene-expression level and protein localisation. We identified potential StMKK6 downstream targets and have shown that albeit the signalling network seems to be evolutionary relatively stable the fine-tuned interdependency within the broader MAPK signalling network might be different in different plants, and when a plant is exposed to different pathogens.
Supporting Information
Localisation of StMKK6, under 35S promoter and native promoter, in epidermal cells of N. benthamiana . A. Control of transformation. Epidermal cells, transformed with plasmids containing 35S::pH7CWG2-CFP (left) and 35S::pH7YWG2-YFP (right) fusion. The fluorescence of the CFP or YFP alone (without the fusion with StMKK6) is observed only in cytoplasm. B. Localisation of StMKK6 fused with CFP (upper panel) or YFP (lower panel) with expression under the CaMV 35S promoter in mock-inoculated epidermal cells. The protein is localised in cytoplasm and nucleus. C. Localisation of StMKK6 fused with CFP (upper panel) or YFP (lower panel) with expression under the CaMV 35S promoter in PVY-inoculated epidermal cells. The protein is localised in cytoplasm and nucleus. D. Localisation of StMKK6 fused with YFP with expression under native promoter in mock-inoculated epidermal cells, where no fluorescence is observed. E. Localisation of StMKK6 fused with YFP with expression under native promoter in PVY-inoculated epidermal cells, where the protein accumulates predominantly in nucleus.
(TIF)
Phylogenetic tree of potato and A. thaliana MAPKs from group A and group B. Potato (Sotub, PGSC and St) and A. thaliana (At) MAPKs from group A (MAPK3, 6 and 10) and group B (MAPK4, 5, 11, 12 and 13) as was already proposed by Ichimura et al. 2002 [4]. Besides the potato sequences from the PGSC Browser (Sotub and PGSC) the tree also includes four MAPKs from cv. Rywal: StMAPK4_1, StMAPK4_2, StMAPK6 and StMAPK13. The sequences from Arabidopsis are AtMAPK3 (AT3G45640.1), AtMAPK4 (AT4G01370.1), AtMAPK5 (AT4G11330.1), AtMAPK6 (AT2G43790.1), AtMAPK10 (AT3G59790.1), AtMAPK11 (AT1G01560.1), AtMAPK12 (AT2G46070.1) and AtMAPK13 (AT1G07880.2). The scale bar indicates the branch length that corresponds to 0.06 substitutions per site.
(TIF)
Expression pattern of St MKK6 and its confirmed targets St MAPK4_2 , St MAPK6 and St MAPK13 in mock-inoculated plants. Log2 of normalized signals for mock-inoculated plants 1 day post inoculation are shown. The expression is shown for four mock treated Rywal (R) and NahG-Rywal (nah) plants. In both sets of plants the expression of the MKK6 interacting MAPKs is higher than of StMKK6. Calculated are differences in the expression between Rywal and NahG-Rywal plants.
(TIF)
Table S1 in File S1. Oligonucleotides used for fragment isolation, qPCR and cloning. In primers for cloning, the underlined parts of sequences are complementary to the destination plasmids. Table S2 in File S1. Validation of St MKK6 microarray (µarray) results by real-time PCR (qPCR). To validate the microarray results for StMKK6, its expression was analysed by qPCR in the same RNA samples as for the microarray analysis. Log2 of ratio between virus- and mock-inoculated plants in cv. Rywal and NahG-Rywal 1, 3 and 6 days after PVY inoculation for StMKK6 obtained by both methods are shown. Statistically significant values (p<0.05) are marked with bold. Table S3 in File S1. Predicted regulatory domains of St MKK6 native promoter from S. tuberosum , cv. Rywal. The promoter sequence is 899 bp-long and analysed with PlantCare software [36]. Table S4 in File S1. Predicted regulatory domains of St MKK6 native promoter from S. tuberosum , cv. Santé. The promoter sequence is 247 bp-long and analysed with PlantCare software [36]. Table S5 in File S1. Expression values of StMKK6 in control and SA-treated potatoes cv. Rywal and NahG-Rywal. Two biological replicates per treatment were analysed. Relative expression values and fold-changes (compared to Rywal control) are shown in the table. Differences between control and SA-treated plants were statistically evaluated by t-test. Table S6 in File S1. Comparison of StMKK6 basal expression values in Rywal and NahG-Rywal plants. Two biological replicates per treatment were analysed. Relative expression values and fold-changes (compared to Rywal control) are shown in the table. Differences between Rywal and NahG-Rywal plants were statistically evaluated by t-test. Table S7 in File S1. Subcellular localisation prediction of StMKK6 and AtMKK6. Amino acid sequence of AtMKK6 (GenBank accession number NM_125041.2) and StMKK6 (GenBank accession number KF837129.1) proteins were used as query in PredictProtein service. The localisation for each was predicted by three different prediction algorithms: PROSITE, LOCkey and LOCtree. For each subcellular prediction a confidence to the prediction is given.
(DOC)
Gene expression of potato MKK family in different tissues and after several stress treatments. For each MKK, the RNA expression signals (FPKM values), based on the RNA sequencing of double monoploid Solanum tuberosum Group Phureja clone DM1-3 (DM) [40] and Genome sequence and analysis of the tuber crop potato [30] were collected in the Potato eFP browser [39]. A. Tissue expression of MKK gene family in Clone DM and Clone RH. In bold are the expression values that are more than 2-times different from the average expression in all the organs. B. Expression of MKK gene family after several stress treatments. In bold are the expression values that are more than 2-times different from the controls.
(XLSX)
St MKK6 orthologs with corresponding UniProt IDs. List of StMKK6 orthologues across plant kingdom. Listed are the Uniprot IDs for the proteins, names of the proteins as in the phylogenetic tree (Figure 3) and the organism the gene was originally isolated from.
(XLS)
Expression of genes co-regulated with St MKK6 and their regulatory domains in promoter regions. Cultivar Rywal (HR response, conferred by Ny-1 gene) and NahG-Rywal (impaired accumulation of SA) were analysed for whole transcriptome response 1, 3 and 6 dpi after PVY infection [24]. Log2 fold changes of PVY in infected vs. mock inoculated plants are indicated for each time point (1, 3 and 6 dpi). Statistically significant differences (FDR corrected p<0.05) are in bold. Higher expression values are blue and lower are yellow. The promoter sequences were obtained from the Solanum_tuberosum-PGSC_DM_v34_superscaffolds database. 1000 bp-long promoter sequences upstream of the gene were analysed by PlantCARE web service [36]. Biotic stress-related domains are coloured and the colours are explained in the legend on the right.
(XLS)
Acknowledgments
The authors would like to acknowledge Tina Demšar, Katja Stare, Tanja Guček and Klavdija Mohorič for all of their help and Chris Berrie for language revision. For the use of the confocal microscope, we thank the Laboratory of Biotechnology of the National Institute of Chemistry, Ljubljana, Slovenia.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
The work was supported by the Slovenian Research Agency (https://www.arrs.gov.si/en/dobrodoslica.asp, grant nos.: P4-0165, J1-4268), The European Cooperation in Science and Technology, COST action FA0806 (http://www.cost.eu/domains_actions/fa/Actions/FA0806) and the Slovenian-Polish bilateral project (2010-2011). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Meng X, Zhang S (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51: 245–266. [DOI] [PubMed] [Google Scholar]
- 2. Rodriguez MCS, Petersen M, Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61: 621–649. [DOI] [PubMed] [Google Scholar]
- 3. Pitzschke A, Hirt H (2009) Disentangling the complexity of mitogen-activated protein kinases and reactive oxygen species signaling. Plant Physiol 149: 606–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ichimura K, Shinozaki K, Tena G, Sheen J (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7: 301–308. [DOI] [PubMed] [Google Scholar]
- 5. Andreasson E, Ellis B (2010) Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci 15: 106–113. [DOI] [PubMed] [Google Scholar]
- 6. Rasmussen MW, Roux M, Petersen M, Mundy J (2012) MAP kinase cascades in Arabidopsis innate immunity. Front Plant Sci 3: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hamel L-P, Nicole M-C, Sritubtim S, Morency M-J, Ellis M, et al. (2006) Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci 11: 192–198. [DOI] [PubMed] [Google Scholar]
- 8. Qiu J-L, Zhou L, Yun BW, Nielsen HB, Fiil BK, et al. (2008) Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol 148: 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Teige M, Scheikl E, Eulgem T, Dóczi R, Ichimura K, et al. (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 15: 141–152. [DOI] [PubMed] [Google Scholar]
- 10. Takahashi Y, Soyano T, Kosetsu K, Sasabe M, Machida Y (2010) HINKEL kinesin, ANP MAPKKKs and MKK6/ANQ MAPKK, which phosphorylates and activates MPK4 MAPK, constitute a pathway that is required for cytokinesis in Arabidopsis thaliana. Plant Cell Physiol 51: 1766–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takahashi F, Yoshida R, Ichimura K, Mizoguchi T, Seo S, et al. (2007) The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 19: 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dóczi R, Brader G, Pettkó-Szandtner A, Rajh I, Djamei A, et al. (2007) The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell 19: 3266–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hwa CM, Yang XC (2008) The AtMKK3 pathway mediates ABA and salt signaling in Arabidopsis. Acta Physiol Plant 30: 277–286. [Google Scholar]
- 14. Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, et al. (2008) Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci U S A 105: 15629–15634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, et al. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. [DOI] [PubMed] [Google Scholar]
- 16. Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoo S-D, Cho YH, Tena G, Xiong Y, Sheen J (2008) Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou C, Cai Z, Guo Y, Gan S (2009) An arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol 150: 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang X, Dai Y, Xiong Y, DeFraia C, Li J, et al. (2007) Overexpression of Arabidopsis MAP kinase kinase 7 leads to activation of plant basal and systemic acquired resistance. Plant J 52: 1066–1079. [DOI] [PubMed] [Google Scholar]
- 20. Naseem M, Dandekar T (2012) The role of auxin-cytokinin antagonism in plant-pathogen interactions. PLoS Pathog 8: e1003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lewsey M, Palukaitis P, Carr JP (2009) Plant-virus interactions: defence and counter-defence. In: Parker J, editor. Annual Plant Reviews. Oxford: Wiley-Blackwell, Vol. 34: 134–176. [Google Scholar]
- 22. Vlot a C, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206. [DOI] [PubMed] [Google Scholar]
- 23. Baebler Š, Stare K, Kovač M, Blejec A, Prezelj N, et al. (2011) Dynamics of responses in compatible potato-Potato virus Y interaction are modulated by salicylic acid. PLoS One 6: e29009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baebler Š, Witek K, Petek M, Stare K, TuŠek-Žnidarič M, et al. (2014) Salicylic acid is an indispensable component of the Ny-1 resistance-gene-mediated response against Potato virus Y infection in potato. J Exp Bot 65: 1095–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Singh DP, Moore CA, Gilliland A, Carr JP (2004) Activation of multiple antiviral defence mechanisms by salicylic acid. Mol Plant Pathol 5: 57–63. [DOI] [PubMed] [Google Scholar]
- 26. Mur LAJ, Kenton P, Lloyd AJ, Ougham H, Prats E (2008) The hypersensitive response; the centenary is upon us but how much do we know? J Exp Bot 59: 501–520. [DOI] [PubMed] [Google Scholar]
- 27. Kogovšek P, Ravnikar M (2013) Physiology of the potato-Potato virus Y interaction. In: Lüttge U, Beyschlag W, Francis D, Cushman J, editors. Progress in Botany. Berlin, Heidelberg: Springer Berlin Heidelberg, Vol. 74: 101–133. [Google Scholar]
- 28. Szajko K, Chrzanowska M, Witek K, Strzelczyk-Zyta D, Zagórska H, et al. (2008) The novel gene Ny-1 on potato chromosome IX confers hypersensitive resistance to Potato virus Y and is an alternative to Ry genes in potato breeding for PVY resistance. Theor Appl Genet 116: 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 30. Potato Genome Sequencing Consortium (2011) Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195. [DOI] [PubMed] [Google Scholar]
- 31. Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saito N, Nei M (1987) The neighbour-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 34. Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramšak Ž, Baebler Š, Rotter A, Korbar M, Mozetič I, et al. (2014) GoMapMan: integration, consolidation and visualization of plant gene annotations within the MapMan ontology. Nucleic Acids Res 42: D1167–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, et al. (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rost B, Yachdav G, Liu J (2004) The PredictProtein server. Nucleic Acids Res 32: W321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tanz SK, Castleden I, Hooper CM, Vacher M, Small I, et al. (2013) SUBA3: a database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res 41: D1185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Winter D, Vinegar B, Nahal H, Ammar R, Wilson G V, et al. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Massa AN, Childs KL, Lin H, Bryan GJ, Giuliano G, et al. (2011) The transcriptome of the reference potato genome Solanum tuberosum Group Phureja clone DM1-3 516R44. PLoS One 6: e26801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, et al. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Theocharidis A, van Dongen S, Enright AJ, Freeman TC (2009) Network visualization and analysis of gene expression data using BioLayout Express(3D). Nat Protoc 4: 1535–1550. [DOI] [PubMed] [Google Scholar]
- 43.Rasmussen R (2001) Quantification on the LightCycler. In: Meuer S, Wittwer C, Nakagawara K, editors. Rapid Cycle Real-Time PCR: Methods and Applications, Heidelberg: Springer Berlin Heidelberg, 21–34.
- 44. Weller SA, Elphinstone JG, Smith NC, Boonham N, Stead DE (2000) Detection of Ralstonia solanacearum strains with a Quantitative, Multiplex, Real-Time, Fluorogenic PCR (TaqMan) assay. Appl Environ Microbiol 66: 2853–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baebler Š, Krečič-Stres H, Rotter A, Kogovšek P, Cankar K, et al. (2009) PVY NTN elicits a diverse gene expression response in different potato genotypes in the first 12 h after inoculation. Mol Plant Pathol 10: 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Karimi M, De Meyer B, Hilson P (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105. [DOI] [PubMed] [Google Scholar]
- 47. Pohleven J, Obermajer N, Sabotič J, Anžlovar S, Sepcić K, et al. (2009) Purification, characterization and cloning of a ricin B-like lectin from mushroom Clitocybe nebularis with antiproliferative activity against human leukemic T cells. Biochim Biophys Acta 1790: 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Singh R, Jwa NS (2013) The rice MAPKK-MAPK interactome: the biological significance of MAPK components in hormone signal transduction. Plant Cell Rep 32: 923–931. [DOI] [PubMed] [Google Scholar]
- 49. Chen L, Hu W, Tan S, Wang M, Ma Z, et al. (2012) Genome-wide identification and analysis of MAPK and MAPKK gene families in Brachypodium distachyon. PLoS One 7: e46744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nicole MC, Hamel LP, Morency MJ, Beaudoin N, Ellis BE, et al. (2006) MAP-ping genomic organization and organ-specific expression profiles of poplar MAP kinases and MAP kinase kinases. BMC Genomics 7: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang S, Xu R, Luo X, Jiang Z, Shu H (2013) Genome-wide identification and expression analysis of MAPK and MAPKK gene family in Malus domestica. Gene 531: 377–387. [DOI] [PubMed] [Google Scholar]
- 52. Liang W, Yang B, Yu BJ, Zhou Z, Li C, et al. (2013) Identification and analysis of MKK and MPK gene families in canola (Brassica napus L.). BMC Genomics 14: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Heinrich M, Baldwin IT, Wu J (2011) Three MAPK kinases, MEK1, SIPKK, and NPK2, are not involved in activation of SIPK after wounding and herbivore feeding but important for accumulation of trypsin proteinase inhibitors. Plant Mol Biol Report 30: 731–740. [Google Scholar]
- 54. Dóczi R, Okrész L, Romero AE, Paccanaro A, Bögre L (2012) Exploring the evolutionary path of plant MAPK networks. Trends Plant Sci 17: 518–525. [DOI] [PubMed] [Google Scholar]
- 55. Takahashi Y, Nasir KH Bin, Ito A, Kanzaki H, Matsumura H, et al. (2007) A high-throughput screen of cell-death-inducing factors in Nicotiana benthamiana identifies a novel MAPKK that mediates INF1-induced cell death signaling and non-host resistance to Pseudomonas cichorii. Plant J 49: 1030–1040. [DOI] [PubMed] [Google Scholar]
- 56. Yoshihiro T, Nasir KH Bin, Ito A, Kanzaki H, Matsumura H, et al. (2007) A novel MAPKK involved in cell death and defense dignaling. Plant Signal Behav 2: 396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piston DW, Patterson, George H. Lippincott-Schwartz J, Claxton, Nathan S. Davidson MW (n.d.) Imaging parameters for fluorescent proteins.
- 58. Hardin SC, Wolniak SM (1998) Molecular cloning and characterization of maize ZmMEK1, a protein kinase with a catalytic domain homologous to mitogen- and stress-activated protein kinase kinases. Planta 206: 577–584. [DOI] [PubMed] [Google Scholar]
- 59. Hardin SC, Wolniak SM (2001) Expression of the mitogen-activated protein kinase kinase ZmMEK1 in the primary root of maize. Planta 213: 916–926. [DOI] [PubMed] [Google Scholar]
- 60. Zeng Q, Sritubtim S, Ellis BE (2011) AtMKK6 and AtMPK13 are required for lateral root formation in Arabidopsis. Plant Signal Behav 6: 1436–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu Y, Zhou Y, Liu L, Sun L, Zhang M, et al. (2012) Maize ZmMEK1 is a single-copy gene. Mol Biol Rep 39: 2957–2966. [DOI] [PubMed] [Google Scholar]
- 62. Wen J, Oono K, Imai R, Osmek R (2002) Two novel mitogen-activated protein signaling components, OsMEK1 and OsMAP1, are involved in a moderate low-temperature signaling pathway in rice. Plant Physiol 129: 1880–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xie G, Kato H, Imai R (2012) Biochemical identification of the OsMKK6-OsMPK3 signalling pathway for chilling stress tolerance in rice. Biochem J 443: 95–102. [DOI] [PubMed] [Google Scholar]
- 64. Soyano T, Nishihama R, Morikiyo K, Ishikawa M, Machida Y (2003) NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK-mediated MAPK cascade and is required for plant cytokinesis. Genes Dev 17: 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Naseem M, Philippi N, Hussain A, Wangorsch G, Ahmed N, et al. (2012) Integrated systems view on networking by hormones in Arabidopsis immunity reveals multiple crosstalk for cytokinin. Plant Cell 24: 1793–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Glazebrook J, Chen W, Estes B, Chang H-S, Nawrath C, et al. (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J 34: 217–228. [DOI] [PubMed] [Google Scholar]
- 67. Miljkovic D, Stare T, Mozetič I, Podpečan V, Petek M, et al. (2012) Signalling network construction for modelling plant defence response. PLoS One 7: e51822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Meng X, Xu J, He Y, Yang K-Y, Mordorski B, et al. (2013) Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25: 1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Melikant B, Giuliani C, Halbmayer-Watzina S, Limmongkon A, Heberle-Bors E, et al. (2004) The Arabidopsis thaliana MEK AtMKK6 activates the MAP kinase AtMPK13. FEBS Lett 576: 5–8 Available: http://www.ncbi.nlm.nih.gov/pubmed/15474000. [DOI] [PubMed] [Google Scholar]
- 70. Lin W-Y, Matsuoka D, Sasayama D, Nanmori T (2010) A splice variant of Arabidopsis mitogen-activated protein kinase and its regulatory function in the MKK6–MPK13 pathway. Plant Sci 178: 245–250. [Google Scholar]
- 71. Liu Y, Schiff M, Dinesh-Kumar SP (2004) Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J 38: 800–809. [DOI] [PubMed] [Google Scholar]
- 72. Liu Y, Ren D, Pike S, Pallardy S, Gassmann W, et al. (2007) Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J 51: 941–954. [DOI] [PubMed] [Google Scholar]
- 73. Jin H, Liu Y, Yang K-Y, Kim C, Baker B, et al. (2003) Function of a mitogen-activated protein kinase pathway in N gene-mediated resistance in tobacco. Plant J 33: 719–731. [DOI] [PubMed] [Google Scholar]
- 74. Shang J, Xi D-H, Xu F, Wang S-D, Cao S, et al. (2011) A broad-spectrum, efficient and nontransgenic approach to control plant viruses by application of salicylic acid and jasmonic acid. Planta 233: 299–308. [DOI] [PubMed] [Google Scholar]
- 75. Ekengren SK, Liu Y, Schiff M, Dinesh-Kumar SP, Martin GB (2003) Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J 36: 905–917. [DOI] [PubMed] [Google Scholar]
- 76. Pedley KF, Martin GB (2004) Identification of MAPKs and their possible MAPK kinase activators involved in the Pto-mediated defense response of tomato. J Biol Chem 279: 49229–49235. [DOI] [PubMed] [Google Scholar]
- 77. Gao M, Liu J, Bi D, Zhang Z, Cheng F, et al. (2008) MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res 18: 1190–1198. [DOI] [PubMed] [Google Scholar]
- 78. Asai S, Ohta K, Yoshioka H (2008) MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 20: 1390–1406 doi:10.1105/tpc.107.055855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Popescu SC, Popescu G V, Bachan S, Zhang Z, Gerstein M, et al. (2009) MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev 23: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Liu H, Wang Y, Xu J, Su T, Liu G, et al. (2008) Ethylene signaling is required for the acceleration of cell death induced by the activation of AtMEK5 in Arabidopsis. Cell Res 18: 422–432. [DOI] [PubMed] [Google Scholar]
- 81. Del Pozo O, Pedley KF, Martin GB (2004) MAPKKKalpha is a positive regulator of cell death associated with both plant immunity and disease. EMBO J 23: 3072–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gimenez-Ibanez S, Solano R (2013) Nuclear jasmonate and salicylate signaling and crosstalk in defense against pathogens. Front Plant Sci 4: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Localisation of StMKK6, under 35S promoter and native promoter, in epidermal cells of N. benthamiana . A. Control of transformation. Epidermal cells, transformed with plasmids containing 35S::pH7CWG2-CFP (left) and 35S::pH7YWG2-YFP (right) fusion. The fluorescence of the CFP or YFP alone (without the fusion with StMKK6) is observed only in cytoplasm. B. Localisation of StMKK6 fused with CFP (upper panel) or YFP (lower panel) with expression under the CaMV 35S promoter in mock-inoculated epidermal cells. The protein is localised in cytoplasm and nucleus. C. Localisation of StMKK6 fused with CFP (upper panel) or YFP (lower panel) with expression under the CaMV 35S promoter in PVY-inoculated epidermal cells. The protein is localised in cytoplasm and nucleus. D. Localisation of StMKK6 fused with YFP with expression under native promoter in mock-inoculated epidermal cells, where no fluorescence is observed. E. Localisation of StMKK6 fused with YFP with expression under native promoter in PVY-inoculated epidermal cells, where the protein accumulates predominantly in nucleus.
(TIF)
Phylogenetic tree of potato and A. thaliana MAPKs from group A and group B. Potato (Sotub, PGSC and St) and A. thaliana (At) MAPKs from group A (MAPK3, 6 and 10) and group B (MAPK4, 5, 11, 12 and 13) as was already proposed by Ichimura et al. 2002 [4]. Besides the potato sequences from the PGSC Browser (Sotub and PGSC) the tree also includes four MAPKs from cv. Rywal: StMAPK4_1, StMAPK4_2, StMAPK6 and StMAPK13. The sequences from Arabidopsis are AtMAPK3 (AT3G45640.1), AtMAPK4 (AT4G01370.1), AtMAPK5 (AT4G11330.1), AtMAPK6 (AT2G43790.1), AtMAPK10 (AT3G59790.1), AtMAPK11 (AT1G01560.1), AtMAPK12 (AT2G46070.1) and AtMAPK13 (AT1G07880.2). The scale bar indicates the branch length that corresponds to 0.06 substitutions per site.
(TIF)
Expression pattern of St MKK6 and its confirmed targets St MAPK4_2 , St MAPK6 and St MAPK13 in mock-inoculated plants. Log2 of normalized signals for mock-inoculated plants 1 day post inoculation are shown. The expression is shown for four mock treated Rywal (R) and NahG-Rywal (nah) plants. In both sets of plants the expression of the MKK6 interacting MAPKs is higher than of StMKK6. Calculated are differences in the expression between Rywal and NahG-Rywal plants.
(TIF)
Table S1 in File S1. Oligonucleotides used for fragment isolation, qPCR and cloning. In primers for cloning, the underlined parts of sequences are complementary to the destination plasmids. Table S2 in File S1. Validation of St MKK6 microarray (µarray) results by real-time PCR (qPCR). To validate the microarray results for StMKK6, its expression was analysed by qPCR in the same RNA samples as for the microarray analysis. Log2 of ratio between virus- and mock-inoculated plants in cv. Rywal and NahG-Rywal 1, 3 and 6 days after PVY inoculation for StMKK6 obtained by both methods are shown. Statistically significant values (p<0.05) are marked with bold. Table S3 in File S1. Predicted regulatory domains of St MKK6 native promoter from S. tuberosum , cv. Rywal. The promoter sequence is 899 bp-long and analysed with PlantCare software [36]. Table S4 in File S1. Predicted regulatory domains of St MKK6 native promoter from S. tuberosum , cv. Santé. The promoter sequence is 247 bp-long and analysed with PlantCare software [36]. Table S5 in File S1. Expression values of StMKK6 in control and SA-treated potatoes cv. Rywal and NahG-Rywal. Two biological replicates per treatment were analysed. Relative expression values and fold-changes (compared to Rywal control) are shown in the table. Differences between control and SA-treated plants were statistically evaluated by t-test. Table S6 in File S1. Comparison of StMKK6 basal expression values in Rywal and NahG-Rywal plants. Two biological replicates per treatment were analysed. Relative expression values and fold-changes (compared to Rywal control) are shown in the table. Differences between Rywal and NahG-Rywal plants were statistically evaluated by t-test. Table S7 in File S1. Subcellular localisation prediction of StMKK6 and AtMKK6. Amino acid sequence of AtMKK6 (GenBank accession number NM_125041.2) and StMKK6 (GenBank accession number KF837129.1) proteins were used as query in PredictProtein service. The localisation for each was predicted by three different prediction algorithms: PROSITE, LOCkey and LOCtree. For each subcellular prediction a confidence to the prediction is given.
(DOC)
Gene expression of potato MKK family in different tissues and after several stress treatments. For each MKK, the RNA expression signals (FPKM values), based on the RNA sequencing of double monoploid Solanum tuberosum Group Phureja clone DM1-3 (DM) [40] and Genome sequence and analysis of the tuber crop potato [30] were collected in the Potato eFP browser [39]. A. Tissue expression of MKK gene family in Clone DM and Clone RH. In bold are the expression values that are more than 2-times different from the average expression in all the organs. B. Expression of MKK gene family after several stress treatments. In bold are the expression values that are more than 2-times different from the controls.
(XLSX)
St MKK6 orthologs with corresponding UniProt IDs. List of StMKK6 orthologues across plant kingdom. Listed are the Uniprot IDs for the proteins, names of the proteins as in the phylogenetic tree (Figure 3) and the organism the gene was originally isolated from.
(XLS)
Expression of genes co-regulated with St MKK6 and their regulatory domains in promoter regions. Cultivar Rywal (HR response, conferred by Ny-1 gene) and NahG-Rywal (impaired accumulation of SA) were analysed for whole transcriptome response 1, 3 and 6 dpi after PVY infection [24]. Log2 fold changes of PVY in infected vs. mock inoculated plants are indicated for each time point (1, 3 and 6 dpi). Statistically significant differences (FDR corrected p<0.05) are in bold. Higher expression values are blue and lower are yellow. The promoter sequences were obtained from the Solanum_tuberosum-PGSC_DM_v34_superscaffolds database. 1000 bp-long promoter sequences upstream of the gene were analysed by PlantCARE web service [36]. Biotic stress-related domains are coloured and the colours are explained in the legend on the right.
(XLS)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.