Abstract
The biosynthesis of iron-sulfur clusters is a highly regulated process involving several proteins. Among them, so-called scaffold proteins play pivotal roles in both the assembly and delivery of iron-sulfur clusters. Here, we report the identification of two chloroplast-localized NifU-like proteins, AtCnfU-V and AtCnfU-IVb, from Arabidopsis (Arabidopsis thaliana) with high sequence similarity to a cyanobacterial NifU-like protein that was proposed to serve as a molecular scaffold. AtCnfU-V is constitutively expressed in several tissues of Arabidopsis, whereas the expression of AtCnfU-IVb is prominent in the aerial parts. Mutant Arabidopsis lacking AtCnfU-V exhibited a dwarf phenotype with faint pale-green leaves and had drastically impaired photosystem I accumulation. Chloroplasts in the mutants also showed a decrease in both the amount of ferredoxin, a major electron carrier of the stroma that contains a [2Fe-2S] cluster, and in the in vitro activity of iron-sulfur cluster insertion into apo-ferredoxin. When expressed in Escherichia coli cells, AtCnfU-V formed a homodimer carrying a [2Fe-2S]-like cluster, and this cluster could be transferred to apo-ferredoxin in vitro to form holo-ferredoxin. We propose that AtCnfU has an important function as a molecular scaffold for iron-sulfur cluster biosynthesis in chloroplasts and thereby is required for biogenesis of ferredoxin and photosystem I.
INTRODUCTION
Iron-sulfur proteins that contain iron-sulfur clusters play important roles in systems such as electron transport and regulation of gene expression and are ubiquitously expressed in various organisms from lower bacteria to higher eukaryotes. Although many iron-sulfur proteins have been identified and studied, the exact mechanisms by which iron-sulfur clusters are assembled into various iron-sulfur proteins in vivo and how these clusters are maintained in the given proteins remain to be elucidated.
Pioneering studies on nitrogenase assembly by Dean and colleagues and the recent identification of bacterial iron-sulfur cluster formation (Isc)/nitrogen fixation (Nif) proteins that are involved in iron-sulfur cluster biosynthesis have established remarkable progress in this field (Zheng et al., 1998; Takahashi and Nakamura, 1999; Frazzon et al., 2002; Nakai et al., 2002). Iron-sulfur cluster biosynthesis involving the Isc/Nif proteins has been proposed as a general mechanism acting in various organisms, including eukaryotes (Muhlenhoff and Lill, 2000; Frazzon and Dean, 2003). The proposed mechanism is as follows. (1) First, sulfur atoms necessary for iron-sulfur cluster formation are supplied by the IscS/NifS protein, which acts as a Cys desulfurase to catalyze desulfuration from Cys (Zheng et al., 1993, 1994). (2) The extracted sulfur atom bound to IscS/NifS is then transferred to so-called scaffold proteins, such as IscU/NifU-like protein/IscA (Urbina et al., 2001). (3) Iron atoms also are supplied to the scaffold protein by an as yet unknown mechanism, and then a transient iron-sulfur cluster is assembled on the scaffold. (4) Finally, the assembled cluster is delivered to various substrate apo-proteins to form the iron-sulfur protein (Agar et al., 2000a, 2000b; Nishio and Nakai, 2000; Yuvaniyama et al., 2000; Krebs et al., 2001; Ollagnier-de-Choudens et al., 2001; Wu et al., 2002; Tong et al., 2003).
Yeast (Saccharomyces cerevisiae) has traditionally been the model organism to investigate eukaryotic iron-sulfur cluster biosynthesis. Elegant studies by Lill and coworkers and by others have revealed that mitochondria contain a set of proteins homologous to the bacterial Isc proteins and that these proteins indeed play an important role in iron-sulfur cluster biosynthesis (Nakai et al., 1998, 2001; Lill et al., 1999). The mitochondrial Isc proteins were found to supply iron-sulfur clusters not only to the mitochondrial iron-sulfur proteins but also to the cytosolic iron-sulfur proteins (Kispal et al., 1999).
Plant cells possess two distinct organelles, mitochondria and chloroplasts, both of which contain several metabolically important iron-sulfur proteins. Because both organelles are believed to originate from their respective ancestral endosymbiotic bacteria, it was predicted that two distinct molecular mechanisms for iron-sulfur cluster biosynthesis might have derived from each ancestor. Indeed, biochemical studies have suggested the presence of iron-sulfur cluster biosynthetic activity also in chloroplasts (Takahashi et al., 1986; Merchant and Dreyfuss, 1998; Li et al., 1990; Suzuki et al., 1991; Nishio et al., 1999), although no likely components have so far been identified in this organelle. In addition to a mitochondrion-localized NifS homolog (Kushnir et al., 2001), another NifS homolog was recently localized to the chloroplast, and this protein was able to catalyze desulfuration from Cys (Leon et al., 2002; Pilon-Smits et al., 2002). These studies suggest that, in the plant cell, not only the mitochondrion but also the chloroplast has retained an Isc/Nif system for iron-sulfur cluster biosynthesis.
Here, we report the identification and characterization of chloroplastic NifU-like proteins from Arabidopsis (Arabidopsis thaliana), which can act as molecular scaffolds in iron-sulfur cluster biosynthesis. NifU was originally identified, along with NifS, as a protein involved in the assembly of nitrogenase in a nitrogen-fixing bacterium, Azotobacter vinelandii, in pioneering work by Dean and colleagues (Zheng et al., 1993; Zheng and Dean, 1994). Later, NifU was shown to provide a scaffold for NifS-mediated assembly of transient iron-sulfur clusters (Agar et al., 2000a; Yuvaniyama et al., 2000). Interestingly, various non-nitrogen-fixing bacteria and eukaryotic mitochondria possess an essential scaffold protein, IscU/Isu, for iron-sulfur cluster biosynthesis, and this protein shows a high sequence identity with the N-terminal domain of A. vinelandii NifU (Agar et al., 2000a, 2000b). By contrast, most extant non-nitrogen-fixing cyanobacteria whose genomic sequences are known do not possess any homologs of IscU. Instead, another NifU-like protein with sequence similarity to the C-terminal domain of A. vinelandii NifU was found to be highly conserved among different cyanobacteria. We previously demonstrated that the cyanobacterial NifU-like protein can function as a molecular scaffold for iron-sulfur cluster assembly and delivery (Nishio and Nakai, 2000), and we now term this protein CnfU (C-terminal domain of NifU). Our study reveals that Arabidopsis chloroplasts contain multiple proteins (named AtCnfU) that are highly homologous to the cyanobacterial CnfU protein. To elucidate their physiological significance, we characterized Arabidopsis mutants in which a nuclear gene for one of the AtCnfU proteins was knocked out. We also investigated whether the purified chloroplastic CnfU protein can act as a molecular scaffold for the delivery of iron-sulfur cluster to an apo-substrate protein in vitro.
RESULTS
The Arabidopsis Genome Contains Multiple Genes Whose Translation Products Show Sequence Similarity to the Cyanobacterial NifU-Like Protein CnfU
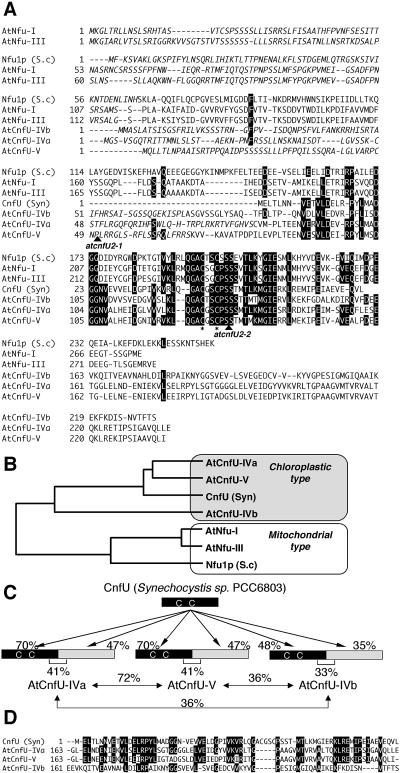
We previously demonstrated that CnfU from cyanobacterium Synechocystis PCC6803 functions as a scaffold for the assembly and delivery of iron-sulfur clusters (Nishio and Nakai, 2000). Recently, CnfU was found to be essential for cell viability, suggesting that it serves as a major scaffold for iron-sulfur cluster biosynthesis in this organism (K. Morimoto, T. Yabe, and M. Nakai, unpublished results). Because chloroplasts are believed to evolve from cyanobacterial-like ancestral endosymbionts, we hypothesized that higher plant chloroplasts also may have kept a CnfU homolog to act in iron-sulfur cluster biosynthesis in the organelle. Therefore, we searched the Arabidopsis EST database and genomic database using the TBLASTN program (http://www.ncbi.nlm.nih.gov/blast) for any protein homologous to the cyanobacterial CnfU and identified five different hypothetical proteins (Table 1). All of them had EST records, albeit in varying degrees, indicating that they are expressed in vivo. Next, we did a sequence alignment of the identified Arabidopsis homologs with the cyanobacterial CnfU and also with a yeast mitochondrial Nfu1p that also showed sequence similarity to the C-terminal domain of the A. vinelandii NifU (Figure 1A). Interestingly, all five Arabidopsis homologs contained N-terminal extensions, compared with the cyanobacterial CnfU. Analysis by TargetP (http://www.cbs.dtu.dk/services/TargetP/) and PSORT (http://psort.ims.u-tokyo.ac.jp/form.html) programs for the prediction of subcellular localization suggested that three of the proteins might localize to chloroplasts (named AtCnfU-IVa, AtCnfU-IVb, and AtCnfU-V, which are coded by At4g25910 [atCNFU3], At4g01940 [atCNFU1], and At5g49940 [atCNFU2], respectively) and that the other two (named AtNfu-I and AtNfu-III, which are coded by At1g51390 [atNFU1] and At3g20970 [atNFU2], respectively) might reside on mitochondria. According to the presumptive mitochondrial targeting sequences, the latter two contain an extra conserved domain (residues 113 to 183 of AtNfu-I and residues 118 to 188 of AtNfu-III) that is common to known mitochondrial Nfu proteins and α-proteobacterial homologs. Phylogenetic analysis (Figure 1B) also suggests that the Arabidopsis homologs can be divided into two evolutionarily distinct groups: a mitochondrial type, including AtNfu-I and AtNfu-III, which show high similarity to the yeast mitochondrial Nfu1p, and a chloroplastic type, including AtCnfU-IVa, AtCnfU-IVb, and AtCnfU-V, which are more strongly related to the cyanobacterial CnfU. This classification supports the predicted intracellular localizations as described above.
Table 1.
Arabidopsis NifU-Like Proteins
| Protein Names | Chromosomea | AGI Accession No.b | Gene Names | Predicted Target Organellesc | Number of EST Recordsd |
|---|---|---|---|---|---|
| AtNfu-I | I | At1g51390 | atNFU1 | Mitochondria | 4 |
| AtNfu-III | III | At3g20970 | atNFU2 | Mitochondriae | 6 |
| AtCnfU-IVa | IV | At4g25910 | atCNFU3 | Chloroplast | 2 |
| AtCnfU-IVb | IV | At4g01940 | atCNFU1 | Chloroplaste | 11 |
| AtCnfU-V | V | At5g49940 | atCNFU2 | Chloroplaste | 14 |
The chromosomal location for the genes encoding each of the NifU-like proteins is indicated.
Accession number in the Arabidopsis Genome Initiative database.
Subcellular localization was predicted by sequence similarity and by TargetP and PSORT programs as described in the text.
Number of EST clones for each of the NifU-like proteins found by The Arabidopsis Information Resource BLAST search (http://www.arabidopsis.org/Blast/) as of December 9, 2003 is indicated.
Subcellular localization was determined in this study.
Figure 1.
Sequence Analysis of Arabidopsis NifU-Like Proteins.
(A) Sequence alignment of five Arabidopsis NifU-like proteins (see also Table 1) with cyanobacterial CnfU (GenBank accession number BAA18665) and yeast mitochondrial Nfu1p (GenBank accession number NP012884.1). Identical amino acid residues conserved in at least four of the seven homologs are full-tone-inverted. Conserved Cys residues are indicated by asterisks, and presumed presequences for intracellular targeting to organelles are indicated with italics. Positions of T-DNA insertions in the two atcnfU2 mutant alleles are denoted by arrowheads. S.c., S. cerevisiae; Syn, Synechocystis PCC6803.
(B) A phylogenetic tree was constructed by comparing predicted mature moieties of Arabidopsis and yeast mitochondrial homologs with cyanobacterial CnfU. S.c., S. cerevisiae; Syn, Synechocystis PCC6803.
(C) Schematic representation of tandem repeats of the CnfU unit found in three Arabidopsis CnfU proteins.
(D) Sequence alignment of the C-terminal repeats of three Arabidopsis CnfU proteins with cyanobacterial CnfU. Syn, Synechocystis PCC6803.
Further sequence analysis revealed that all three chloroplastic-type homologs are comprised of two repeated cyanobacterial CnfU domains (Figure 1C). In addition, the sequence similarity to the cyanobacterial CnfU was higher in the N-terminal domains, rather than at the C termini. Interestingly, whereas their N-terminal CnfU domains retain the highly conserved (Ala)-Cys-X-X-Cys motif, the C-terminal domains lack this motif (Figure 1D). The cyanobacterial CnfU is known to form a dimer, with one [2Fe-2S] cluster assembled intermoleculary between two Cys-X-X-Cys motifs; four Cys residues therefore function as the ligand for one [2Fe-2S] cluster (Nishio and Nakai, 2000). It is possible that chloroplastic CnfUs might therefore also form a dimer, which binds a [2Fe-2S] cluster between two Cys-X-X-Cys motifs of the two identical N-terminal domains. This possibility is addressed later in this report (Figures 10, 11, and 12).
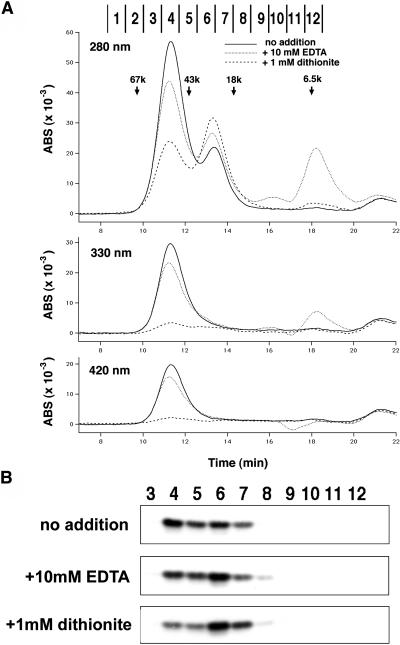
Figure 10.
Gel Filtration Analysis Revealed a Dimeric Holo-State and a Predominantly Monomeric Apo-State of AtCnfU-V.
(A) Gel filtration chromatograms of holo- and apo-AtCnfU-V. After incubation with (dotted lines) or without (solid lines) 10 mM EDTA or 1 mM dithionite (dashed lines) on ice for 1 h, purified AtCnfU-V (25 μg) was applied to a Superdex 75 column (Amersham Biosciences) and equilibrated with buffer containing 50 mM Hepes-KOH, pH 7.5, 150 mM KCl, and 5 mM DTT. Eluates were monitored simultaneously by absorbance at 280 nm (top), 330 nm (middle), and 420 nm (bottom) and divided into 12 fractions. Molecular mass marker proteins used were BSA (67 kD), ovalbumin (43 kD), myoglobin (18 kD), and aprotinin (6.5 kD). ABS, absorbance.
(B) Fractions (3 to 12) obtained from each gel filtration chromatography analysis shown in (A) were analyzed by protein gel blotting using an anti-AtCnfU-V antibody.
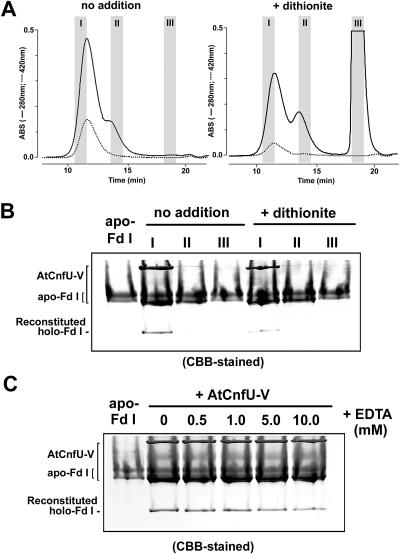
Figure 11.
Only the Dimeric Holo-State of AtCnfU-V Can Serve as an Iron-Sulfur Cluster Donor to Apo-Ferredoxin.
(A) Purified AtCnfU-V (200 μg) was directly applied to a Superdex 75 column (no addition) or incubated with 10 mM dithionite briefly (+ dithionite) just before gel filtration. To avoid cross-contamination between the AtCnfU-V dimeric and monomeric fractions, we saved the faster half the dimeric state peak fraction (I) and the slower half of the monomeric state peak fraction (II) as indicated. Salts fractions (III) also were saved to recover any low molecular weight impurities in the purified AtCnfU-V proteins or liberated from AtCnfU-V by the incubation with dithionite. ABS, absorbance.
(B) Twenty-microliter aliquots of the I, II, or III fractions were incubated with 20 μg of the purified apo-Fd I and analyzed as described in the legend to Figure 9D. Twenty micrograms of apo-Fd I also was loaded on the leftmost lane for comparison. Gels were stained with Coomassie blue (CBB).
(C) The purified AtCnfU-V (20 μg) and apo-Fd I (20 μg) were incubated in the presence of the indicated concentration of EDTA at 25°C for 60 min and analyzed as described above. CBB-stained, Coomassie blue staining.
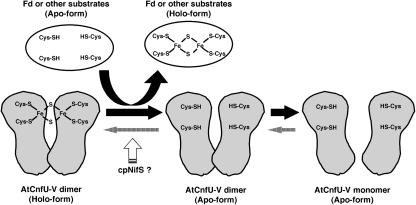
Figure 12.
Model of Scaffold Function of AtCnfU-V in Iron-Sulfur Cluster Transfer to Apo-Proteins.
AtCnfU-V exists as a [2Fe-2S]-containing dimer. The iron-sulfur cluster is assembled intermolecularly between the two identical N-terminal domains in the dimer. The cluster can be transferred to certain substrate apoproteins, including ferredoxin (Fd). Upon the transfer, the holo-AtCnfU-V dimer is converted to the apo-dimer or further to apo-monomers. The new [2Fe-2S] cluster is then recharged on the apo-dimeric form of AtCnfU-V, possibly with the aid of the chloroplast-localized NifS-like protein, cpNifS, as a sulfur donor.
AtCnfU Proteins Are Localized to the Stroma of the Chloroplast
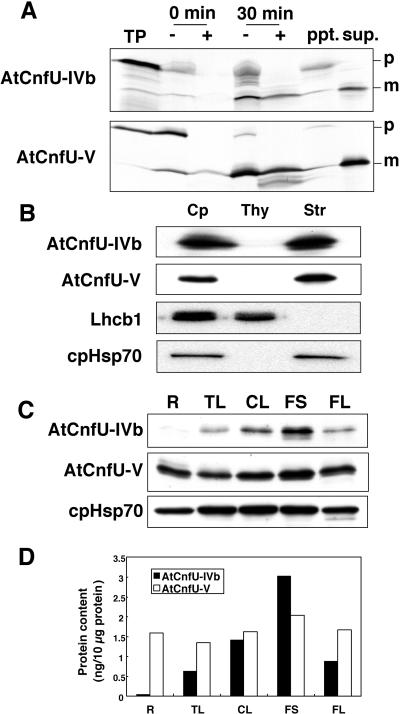
To identify the localization of the AtCnfU proteins on the chloroplast, an in vitro import study and protein gel blot analysis were performed as shown in Figures 2A and 2B, respectively. AtCnfU-IVa and AtCnfU-V are closely related to each other (72% identity between their presumptive mature sequences) and also show significant similarity in their predicted transit sequences. Based on the numbers of EST records and on our quantitative estimation (described below), AtCnfU-IVb and AtCnfU-V are predominantly expressed, whereas AtCnfU-IVa is a minor player. Therefore, AtCnfU-IVb and AtCnfU-V were analyzed as representatives of this protein type. Both precursor proteins could be imported into isolated Pisum sativum (pea) chloroplasts and processed to their mature forms, which were recovered in the stromal fraction (Figure 2A). The stromal localization of AtCnfU-IVb and AtCnfU-V was confirmed by protein gel blotting using specific antibodies as shown in Figure 2B. We also confirmed the localization of one of the two highly homologous AtNfu proteins (90% overall identity between their presumed mature sequences), AtNfu-III, as mitochondrial by in vitro import into isolated P. sativum mitochondria (T. Yabe and M. Nakai, unpublished results). Our results therefore showed that Arabidopsis possesses multiple chloroplast-localized CnfU homologs, AtCnfU-IVb, AtCnfU-V, and most likely AtCnfU-IVa.
Figure 2.
Chloroplast Localization and Expression Profiles of AtCnfU-IVb and AtCnfU-V.
(A) In vitro import of AtCnfU-IVb and AtCnfU-V precursors into chloroplasts. [35S]-labeled precursors were incubated with chloroplasts isolated from P. sativum leaves. After import reactions, imported proteins were analyzed by SDS-PAGE and fluorography. TP, translation products; −, without trypsin; +, with trypsin digestion; ppt, membrane pellets; sup, supernatant containing stromal fraction of chloroplast; p, precursor; m, mature protein.
(B) Immunological detection of AtCnfU-IVb and AtCnfU-V in the chloroplastic stroma. Equivalent amounts of each fraction were analyzed by protein gel blotting. Cp, chloroplasts prepared from Arabidopsis leaves; Thy, thylakoids; Str, the stromal fraction.
(C) Expression profiles of AtCnfU-IVb and AtCnfU-V in various tissues, including roots (R), true leaves (TL), cauline leaves (CL), flower stalks (FS), and flowers (FL) of 23-d-old wild-type Arabidopsis plants. Proteins were extracted and analyzed by protein gel blotting.
(D) AtCnfU-IVb and AtCnfU-V protein bands detected in (C) were quantified densitometrically against their respective recombinant proteins as a standard. R, roots; TL, true leaves; CL, cauline leaves; FS, flower stalks; FL, flowers.
AtCnfU-V Is Constitutively Expressed in Various Plant Tissues
To investigate the expression pattern of chloroplastic CnfU proteins in various plant tissues, we performed protein gel blot analysis on protein extracts prepared from roots, true leaves, cauline leaves, flower stalks, and flowers of 23-d-old wild-type Arabidopsis. AtCnfU-IVb was predominantly expressed in aerial parts, whereas AtCnfU-V was constitutively expressed in various tissues, including roots (Figure 2C). This pattern coincided with that of the stromal molecular chaperone, cpHsp70, suggesting that AtCnfU-V has a widespread housekeeping function in the plant. Quantification using recombinant proteins as standards indicated that AtCnfU-IVb and AtCnfU-V each represent ∼0.03% of total cellular protein of Arabidopsis (Figure 2D).
Because of the high sequence similarity, anti-AtCnfU-V antibodies used in this study could cross-react with recombinant AtCnfU-IVa, a minor homolog (data not shown), although no additional immunoreactive bands were observed in the protein extracts prepared from AtCnfU-V–lacking plants, as described later (Figure 3B). Therefore, from the observed cross-reactivity of anti-AtCnfU-V antibodies to the recombinant AtCnfU-IVa, we could estimate the expression level of AtCnfU-IVa to be less than one-tenth of AtCnfU-V (data not shown).
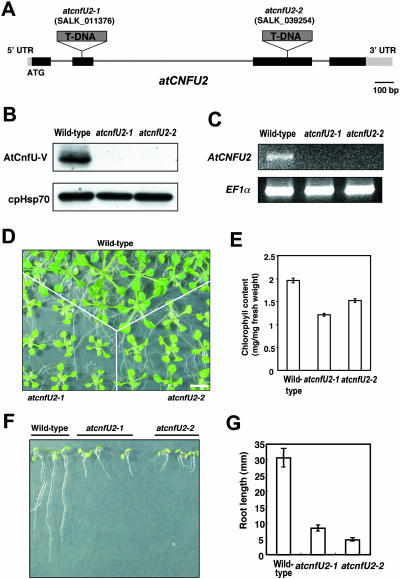
Figure 3.
Homozygous Mutants Lacking AtCnfU-V Showed a Dwarf Phenotype with Slightly Pale-Green Leaves.
(A) Positions of the T-DNA insertion in the atcnfU2-1 and atcnfU2-2 alleles. Solid boxes, exons; thin lines, introns; UTR, untranslated region.
(B) Protein gel blot analysis of AtCnfU-V in wild-type and atcnfU2 mutant plants. Protein extracts prepared from whole plants were analyzed by protein gel blotting using anti-AtNifU-V and anti-Hsp70 (as control) antibodies.
(C) RT-PCR analysis of AtCnfU-V transcripts. Total RNA was isolated from 18-d-old whole plants. Wild-type or atcnfU2 mutants were grown on MS medium containing 2% sucrose. RT-PCR was performed using gene-specific primers for AtCnfU-V or EF1α as a control. After agarose gel electrophoresis, amplified DNA bands were visualized with ethidium bromide.
(D) Dwarf phenotype of atcnfU2 mutants. Wild-type and atnifu2 mutant plants were geminated and grown for 14 d on MS medium containing 2% sucrose. Bar = 10 mm.
(E) Chlorophyll content of atcnfU2 mutant leaves. Chlorophyll was extracted from leaves of 14-d-old seedlings and quantified (n = 5).
(F) and (G) Root growth of atcnfU2 mutants. Wild-type and mutant plants were grown vertically on MS medium containing 2% sucrose for 7 d after germination. Average root lengths (n = 3) are presented in (G) with error bars.
Identification of T-DNA Insertion Mutants, atcnfU2-1 and atcnfU2-2
To investigate the physiological role of chloroplast-localized CnfU proteins, we characterized Arabidopsis mutants in which a nuclear gene (atCNFU2) encoding the constitutively expressed AtCnfU-V was disrupted. In the sequence-indexed Arabidopsis T-DNA insertion mutants provided by the Salk Institute Genomic Analysis Laboratory, several candidate mutant lines were identified. From these lines, two homozygous mutant lines, atcnfU2-1 and atcnfU2-2, each carrying a T-DNA insertion at a different site in the atCNFU2 gene were isolated (Figure 3A). Gene disruptions by the T-DNA insertion were confirmed by protein gel blot analysis and RT-PCR (Figures 3B and 3C). No immunoreactive bands were obvious by protein gel blot analysis, indicating that both alleles are null.
AtCnfU-V Deficiency Leads to a Marked Dwarf Phenotype
Both of the T-DNA insertion mutants described above were fertile and could be maintained as a homozygous line, suggesting that the atCNFU2 gene is not necessary for viability. However, both homozygous plants showed a marked dwarf phenotype in aerial parts and strikingly in the roots, even when they were germinated and grown on rich Murashige and Skoog (MS) medium supplemented with 2% sucrose (Figures 3D, 3F, and 3G). The leaves of the mutant plants were pale green and recorded a decrease in chlorophyll level (Figure 3E). Growth of heterozygous lines was indistinguishable from that of wild-type plants, indicating that both alleles are recessive. Because two independently isolated mutants (atcnfU2-1 and atcnfU2-2) showed similar dwarf and pale-green leaf phenotypes (as shown here) and because these phenotypes were clearly linked to the presence of a homozygous T-DNA insertion (i.e., for either allele, cosegregation was found in 9 plants out of 32), we conclude that the observed phenotypes were actually caused by the AtCnfU-V deficiency.
We then examined the growth defects of the homozygous atcnfU2-1 mutant in more detail. First, we compared the growth of wild-type and atcnfU2-1 plants under long-day or short-day conditions in MS medium with or without 2% sucrose (Figures 4a to 4h). Under all conditions analyzed, the mutants showed considerable dwarf and pale-green leaf phenotypes. The most marked growth inhibition and chlorosis were observed in plants grown under short-day conditions without sucrose (Figure 4h). These results suggest that photosynthetic growth is largely affected by the AtCnfU-V deficiency. We confirmed that photosynthetic oxygen evolution activity per unit of leaf area was indeed decreased in the mutants as compared with the wild type (see Supplemental Figure 1 online).
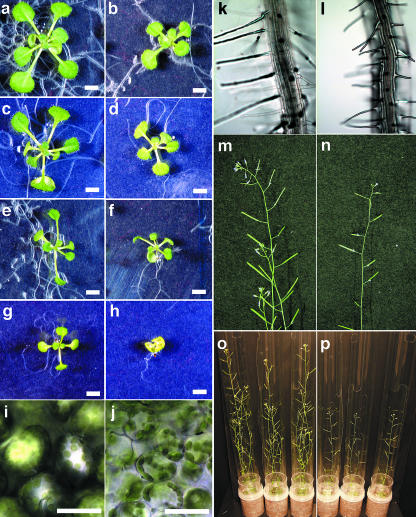
Figure 4.
Phenotypes of atcnfU2-1 Plants under Various Conditions and at Various Growth Stages.
(a) to (h) Wild-type ([a], [c], [e], and [g]) and atcnfU2-1 ([b], [d], [f], and [h]) plants were germinated and grown on MS media for 14 d with ([a], [b], [e], and [f]) or without ([c], [d], [g], and [h]) 2% sucrose under long-day ([a] to [d]) or short-day ([e] to [h]) conditions. Bars = 3 mm.
(i) to (l) Light micrographs of true leaf cells ([i] and [j]) and root cells ([k] and [l]) of wild-type ([i] and [k]) and atcnfU2-1 ([j] and [l]) plants grown on MS media containing 2% sucrose for 18 d after germination are shown. Bars = 50 μm.
(m) to (p) Photographs of wild-type ([m] and [o]) and atcnfU2-1 mutant ([n] and [p]) plants taken 39 d after germination.
Light microscopic observation of 18-d-old true leaves revealed that leaf cells were significantly smaller in the atcnfU2-1 mutant than in the wild type (Figures 4i and 4j). Although the apparent chloroplast size in the mutant was not obviously different from the wild type, mutant cells seemed to contain slightly fewer chloroplasts, which might be the reason for the pale-green appearance of the mutant leaves. Root hairs of the mutants were shorter than those of wild-type plants, and the longitudinal lengths of atcnfU2-1 root cells were markedly shorter, making the root hairs appear more densely distributed in the mutant roots. The dwarf phenotype of atcnfU2-1 also was prominent at the flower stalks, inflorescence, and siliques (Figures 4m to 4p), and the seed yield was severely diminished in the mutants. All phenotypes described here for the atcnfU2-1 mutants also were observed in the atcnfU2-2 mutants (data not shown).
Steady State Levels of Photosystem I Subunits and Stromal Ferredoxin Were Significantly Reduced in atcnfU2 Mutant Plants
To examine whether steady state levels of various chloroplast proteins would be altered in the mutants, protein gel blot analysis of protein extracts from 28-d-old wild-type, atcnfU2-1, and atcnfU2-2 mutant plants that were grown photoautotrophically without sucrose was performed. As shown in Figure 5A, the non-iron-sulfur proteins RbcL and RbcS were present at equivalent levels in all protein extracts. Among the stromal iron-sulfur proteins analyzed in this study, levels of ferredoxin, a multifunctional electron carrier containing [2Fe-2S] cluster, were severely diminished in the mutant plants (∼30% of the wild type), and sulfite reductase and nitrate reductase levels were slightly decreased (∼75% of the wild type) (Figure 5B). By contrast, the levels of the stromal chaperone proteins cpHsp70 and cpCpn60a were moderately higher in the mutants.
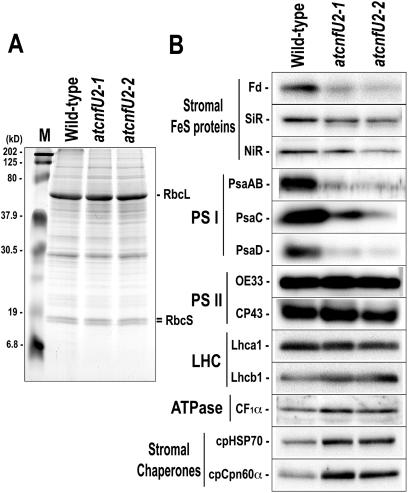
Figure 5.
Steady State Levels of Stromal Ferredoxin and PSI Subunits Were Severely Reduced in atcnfU2 Mutants.
Total protein extracts (10 μg proteins) of 28-d-old plants (wild type, atcnfU2-1, and atcnfU2-2) grown under continuous light without sucrose were separated by SDS-PAGE followed by Coomassie blue staining (A) or protein gel blot analysis (B). Fd, ferredoxin (2Fe-2S); SiR, sulfite reductase (4Fe-4S); NiR, nitrate reductase (4Fe-4S).
Surprisingly, the levels of all photosystem I (PSI) polypeptides that were analyzed, including PsaAB, PsaC, and PsaD, were markedly lower in the mutants (20 to 30% of the wild type), whereas levels of subunits of photosystem II (PSII) and other protein complexes, including light-harvesting complex (LHC), ATPase (Figure 5B), and cytochrome b6f (data not shown), were found to be normal.
Photosynthetic Electron Transport through PSI Was Impaired in atcnfU2 Mutant Plants
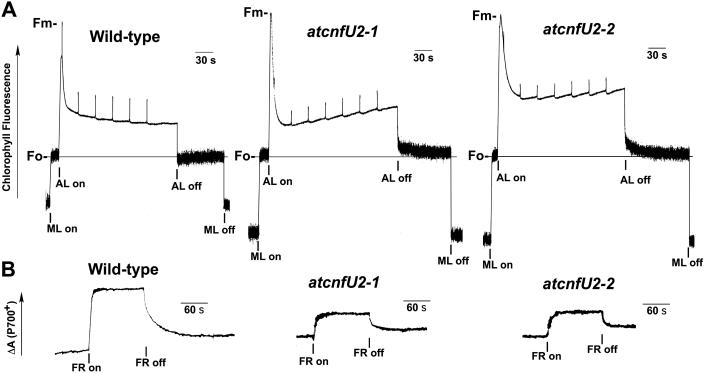
The electron flux through the photosynthetic electron transport chain can be assessed in vivo by monitoring chlorophyll fluorescence, which represents the redox state of PSII (Figure 6A), and by measurement of P700 absorbance changes, which represent the redox state of PSI (Figure 6B).
Figure 6.
Chlorophyll Fluorescence Induction and Redox States of P700 in Wild-Type and atcnfU2 Mutant Leaves.
(A) Fluorescence induction curve of 24-d-old wild-type, atcnfU2-1, and atcnfU2-2 mutant leaves grown under continuous light without sucrose. Representative traces of three to five independent experiments are shown. ML, measuring light beam; AL, white actinic light (520 μmol/m2/s); Fm, maximum fluorescence yield; Fo, minimum fluorescence yield when PSII centers are open.
(B) The oxidation of P700 in 25-d-old wild-type, atcnfU2-1, and atcnfU2-2 leaves (24 mm2 each) grown as above was induced by exposure to far-red light (FR; 30 W/m2) for 2 min. Representative traces of five to seven independent experiments are shown.
As shown in Figure 6A, when PSII is in the open state, the level of fluorescence is low (Fo) because the majority of light energy absorbed by PSII can be used to drive electron transfer. By contrast, when the PSII reaction center is closed (fully reduced) upon illumination, fluorescence reaches a maximum level (Fm). Fluorescence is then quenched by photochemical (qP) and nonphotochemical processes and finally when the actinic light is switched off, fluorescence returns to the Fo level. The Fv/Fm ratio, in which Fv represents Fm-Fo, was considerably diminished in the atcnfU2 mutant plants (Figure 6A, Table 2). However, mutant plants showed increased 1-qP values, indicating more reduced states of plastoquinone QA of PS II. Furthermore, even after the actinic light was switched off, chlorophyll fluorescence in the mutants did not return to the Fo level as quickly as in the wild-type. All these phenomena could be attributed to an impaired electron flow downstream of PSII.
Table 2.
Chlorophyll Fluorescence Parameters and the Redox State of P700 of Wild-Type and atcnfU2 Mutant Plants
| Fv/Fm | 1-qP | ΔA820/A820 (× 10−3) | |
|---|---|---|---|
| Wild type | 0.78 ± 0.02 | 0.60 ± 0.02 | 4.70 ± 0.31 |
| atcnfU2-1 | 0.65 ± 0.05 | 0.76 ± 0.05 | 1.71 ± 0.20 |
| atcnfU2-2 | 0.62 ± 0.06 | 0.80 ± 0.04 | 1.68 ± 0.27 |
The data represent mean values ± se from three to five independent measurements.
Determination of the absorbance changes of P700 of PSI under far-red light revealed that ΔA820/A820 was at least 2.7-fold lower in the atcnfU2 mutant plants than in the wild-type plants (Figure 6B, Table 2). Taken together, these data clearly indicate that electron transport through PSI is significantly affected in the mutant plants.
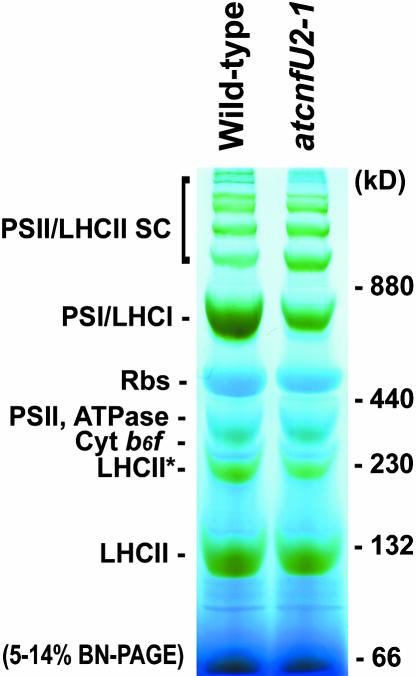
To directly assess the assembly of PSI and other photosynthetic complexes, we used blue native gel electrophoresis (Schägger et al., 1994) with isolated wild-type or atcnfU2-1 mutant chloroplasts solubilized with 0.5% n-dodecyl-β-d-maltoside (Figure 7). Although the mutant chloroplasts had normal or moderately increased levels of assembled PSII/LHCII supercomplexes, ribulose-1,5-bisphosphate carboxylase/oxygenase, ATPase, cytochrome b6f, and LHCII, they had markedly reduced levels of assembled PSI complex. This observation was consistent with results obtained from the protein gel blot analysis shown in Figure 5 and the measurements of photosynthetic electron transport activities shown in Figure 6 and Table 2. Semiquantitative RT-PCR analysis with representative genes for PSI subunits and ferredoxin demonstrated that the atcnfU2 mutations had no effect on the accumulation of their transcripts (see Supplemental Figure 2A online), suggesting that AtCnfU-V is crucial for posttranscriptional steps in biogenesis of PSI and ferredoxin.
Figure 7.
PSI Levels Were Dramatically Decreased in the atcnfU2-1 Mutant Chloroplasts.
Chloroplast protein (67 μg) prepared from wild-type and atcnfU2-1 leaves that had been grown under continuous light without sucrose were solubilized with 0.5% (w/v) n-dodecyl-β-d-maltoside and separated by 5 to 14% blue native gel electrophoresis (BN-PAGE). Bands corresponding to various photosynthetic complexes were identified by protein gel blot analysis (data not shown) and were indicated. Marker proteins used are described in Methods. PSII/LHCII SC, PSII/LHCII supercomplex; Rbs, ribulose-1,5-bisphosphate carboxylase/oxygenase.
The atcnfU2 Mutant Stroma Showed a Lowered Activity of Iron-Sulfur Cluster Insertion into Apo-Ferredoxin in Vitro
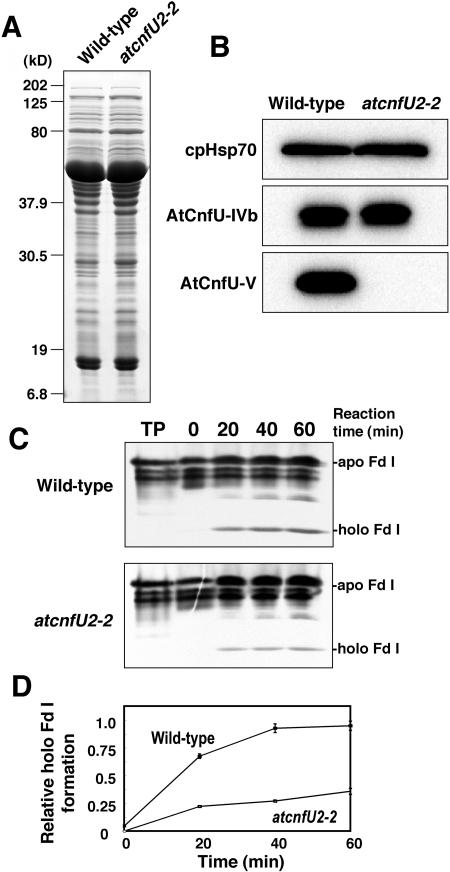
Cyanobacterial CnfU has been shown to function as an iron-sulfur cluster scaffold protein (Nishio and Nakai, 2000), and proper insertion of three [4Fe-4S] clusters (FA, FB, and FX) is known to be essential for overall assembly and accumulation of PSI subunits (Chitnis, 2001). Therefore, the severe reduction in the steady state levels of both PSI and ferredoxin might be attributable to decreased iron-sulfur cluster biosynthetic activity in the atcnfU2 mutant chloroplasts. We analyzed the iron-sulfur cluster biosynthetic activity using chloroplast extracts prepared from the wild-type and mutant plants. However, when the mutants were grown photoautotrophically without sucrose, plants showed a severe dwarf phenotype and chloroplasts were less developed; therefore, it was difficult to isolate sufficient mutant chloroplasts for such biochemical analysis. Consequently, chloroplasts were isolated from mutant and wild-type plants that were grown with sucrose. As shown in Supplemental Figure 3 online, under these growth conditions, levels of ferredoxin were severely diminished in the mutant plants compared with the wild type, whereas levels of other iron-sulfur proteins remained normal (sulfite reductase) or were only slightly decreased (PsaC) in the mutants. Levels of ferredoxin mRNA were not markedly reduced in the mutants (see Supplemental Figure 2B online), suggesting that the lowered steady state level of ferredoxin was probably because of a posttranscriptional defect in the mutants. Whereas other possibilities (e.g., decreased translation or increased protein turnover) seemed to be possible, we tested whether this posttranscriptional defect was inefficient conversion of apo-ferredoxin to holo-ferredoxin in the atcnfU2 chloroplasts by measuring the stromal activity of iron-sulfur cluster insertion into externally added apo-ferredoxin in vitro. Stromal extracts were prepared from either the wild-type or the atcnfU2-2 chloroplasts and showed similar profiles on SDS-PAGE (Figure 8A) and protein gel blotting, with the exception of the absence of AtCnfU-V in the mutant extracts (Figure 8B). AtCnfU-IVb levels remained unchanged in the mutant stroma. Stromal extracts were incubated at 25°C with the in vitro–synthesized apo-ferredoxin (a maize [Zea mays] photosynthetic ferredoxin, Fd I). Conversion of apo-ferredoxin to the holo-form was analyzed by nondenaturing PAGE and fluorography after various incubation periods (Figures 8C and 8D). Holo-ferredoxin formation was still observed in the atcnfU2-2 stromal extracts, but the rate was significantly lower than that in the wild type (∼35%). Similar results were obtained under more reducing conditions (i.e., in the presence of 1 mM DTT, see Supplemental Figure 4 online). To exclude the possibility that the observed lowered iron-sulfur cluster insertion activity might be a secondary effect of the reduced amount of ferredoxin residing in the mutant stroma, we added back a comparable amount of purified holo-ferredoxin to the mutant stroma and analyzed the activity. The addition of holo-ferredoxin could not rescue the mutant stroma (see Supplemental Figure 5 online). Thus, we concluded that the stromal iron-sulfur cluster insertion activity was indeed partially impaired in the absence of AtCnfU-V protein and that this lowered activity most likely causes the decreased accumulation of steady state levels of ferredoxin.
Figure 8.
atcnfU2 Stroma Exhibited Lowered Activity of Iron-Sulfur Cluster Insertion into Apo-Ferredoxin.
(A) Stromal extracts prepared either from wild-type or atcnfU2-2 chloroplasts (10 μg chlorophylls) were analyzed on 12.5% SDS-PAGE gels and stained with Coomassie blue.
(B) Content of AtCnfU-IVb, AtCnfU-V, and cpHsp70 in the stromal extracts shown in (A) were analyzed by protein gel blotting.
(C) In vitro holo-ferredoxin formation with the stromal extracts. [3H]-labeled apo-ferredoxin (TP, inputted translated products of maize Fd I) was incubated with the stromal extracts prepared from either wild-type or atcnfU2-2 chloroplasts. After the indicated incubation time, conversion of apo-ferredoxin to the holo-form was analyzed by nondenaturing PAGE followed by fluorography.
(D) Bands corresponding to holo-ferredoxin were quantified by densitometry. The average quantity of holo-ferredoxin formed in the wild-type stroma at 60 min was set at 1.0. Experiments were repeated three times independently, and average values (n = 3) are plotted with error bars.
Recombinant AtCnfU-V Protein Binds a [2Fe-2S]-Like Cluster That Can Be Transferred to Apo-Ferredoxin
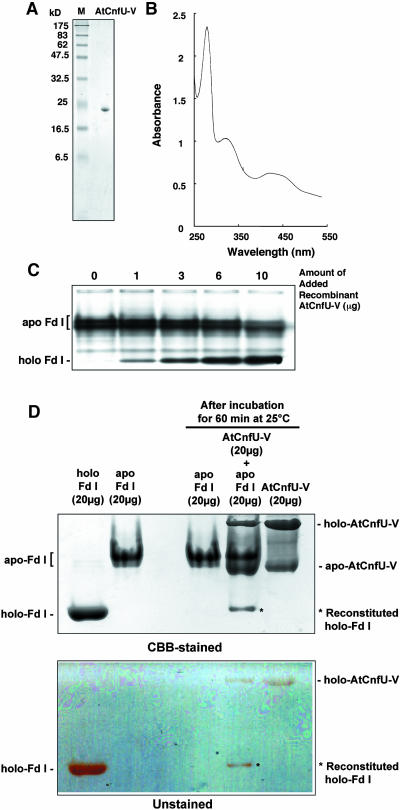
We previously showed that cyanobacterial CnfU binds a [2Fe-2S] cluster and can deliver the cluster to apo-ferredoxin without the aid of other factors (Nishio and Nakai, 2000). Therefore, we examined whether the chloroplast-localized CnfU homolog also binds a [2Fe-2S] cluster and can deliver the cluster to apo-ferredoxin. To do this, a predicted mature moiety of AtCnfU-V (corresponding to amino acid residues 83 to 235) was expressed in Escherichia coli and purified to homogeneity as a 23-kD protein on SDS-PAGE gels; this was slightly larger than the calculated molecular size of 16,821, probably because of its fairly acidic nature (pI = 4.1) (Figure 9A). The purified AtCnfU-V showed a characteristic UV/visible absorption spectrum with peak maxima at 330, 420, and 460 nm, which are typical for the [2Fe-2S]-type cluster (Figure 9B). The absorption spectrum of purified AtCnfU-V was remarkably similar to that of the cyanobacterial CnfU (Nishio and Nakai, 2000).
Figure 9.
Iron-Sulfur Cluster Assembled in AtCnfU-V Can Be Transferred to Apo-Ferredoxin in Vitro.
(A) Purified recombinant AtCnfU-V. One microgram of purified AtCnfU-V was analyzed by SDS-PAGE and Coomassie blue staining. M, molecular mass marker proteins.
(B) UV/visible absorption spectra of purified AtCnfU-V (2 mg/mL) were recorded on a UV-2500PC spectrophotometer (Shimadzu, Columbia, MD; light path = 1 cm).
(C) Iron-sulfur cluster transfer from AtCnfU-V to apo-ferredoxin. Recombinant AtCnfU-V was incubated with [3H]-labeled apo-ferredoxin (Fd I) for 30 min at 25°C in the presence of 1 mM DTT. After incubation, holo-ferredoxin formation was analyzed by nondenaturing PAGE as described in the legend to Figure 8.
(D) Twenty micrograms of purified AtCnfU-V was incubated with 20 μg of purified apo-Fd I at 25°C for 60 min in 50 μL of the buffer containing 50 mM Tris-HCl, pH7.5, 50 mM KCl, and 1 mM DTT and analyzed as above. Simultaneously, mock incubations including either one of two purified proteins also were performed. Purified holo- and apo-Fd I (20 μg) also were loaded on the nondenaturing PAGE for comparison. An unstained gel is shown in the lower panel and a Coomassie blue–stained gel in the upper panel. Positions of reconstituted holo-Fd I bands are indicated by asterisks.
Then we examined whether the [2Fe-2S]-like cluster-containing AtCnfU-V could deliver its iron-sulfur cluster to apo-ferredoxin. Upon incubation with the purified AtCnfU-V, the in vitro–synthesized apo-ferredoxin was efficiently converted to holo-form in a dose- and temperature-dependent manner (Figure 9C, see Supplemental Figure 6 online). To know if a stoichiometry between holo-ferredoxin formed and holo-AtCnfU-V consumed during the reaction, we conducted an iron-sulfur cluster transfer assay using a defined amount of purified apo-ferredoxin instead of the radiolabeled protein and purified AtCnfU-V, as already established in our previous study on the cyanobacterial CnfU protein (Nishio and Nakai, 2000). Apo-ferredoxin was prepared by boiling in the presence of EDTA and DTT followed by gel filtration to remove low molecular weight molecules such as sulfur, iron, or their derivatives liberated from the ferredoxin polypeptides by denaturation. As shown in Figure 9D, the holo-ferredoxin retained its characteristic pink-red color during electrophoresis (unstained gel), whereas the apo-form was completely colorless but could be visualized after Coomassie blue staining (CBB-stained gel). The purified AtCnfU-V separated into two major bands: a faster migrating band, which was colorless during electrophoresis and most likely corresponded to its monomeric apo-form, and a slower migrating band that was pink-red, characteristic of the iron-sulfur cluster, suggesting it corresponded to the dimeric holo-AtCnfU-V. This interpretation was consistent with the results of subsequent gel filtration analysis (Figure 10). Holo-ferredoxin (∼3 μg) was reconstituted when apo-ferredoxin (20 μg) was incubated with purified AtCnfU-V (20 μg) (Figure 9D), whereas approximately half of the added AtCnfU-V (∼10 μg) was converted to colorless apo-form. The loss of color intensity in the holo-AtCnfU-V band appeared to be well correlated with the gain of color in the reconstituted holo-ferredoxin (Figure 9D, unstained gel). Also, given one [2Fe-2S] cluster initially bound each holo-AtCnfU-V dimer as suggested later (Figure 10), 0.59 nmoles (= 0.295 nmoles of dimer) of AtCnfU-V (molecular weight = 16821; 10 μg equivalent) was consumed to reconstitute 0.29 nmoles of holo-ferredoxin (molecular weight = 10512; 3 μg equivalent), suggesting stoichiometric transfer of iron-sulfur cluster between these two molecules.
The [2Fe-2S]-Like Cluster-Containing AtCnfU-V Exists as a Dimer, and Loss of the Cluster Leads to Dissociation to Monomers
We next performed gel filtration to learn more about the molecular status of the purified AtCnfU-V. Absorbance at 280 nm (Trp and tyrosine residues) of the gel filtrate revealed two peaks: a major peak (around fraction 4) and a minor peak (around fraction 6) whose apparent molecular sizes were estimated as ∼48 kD and 23 kD, respectively (Figure 10A, top, solid line). Only the major peak exhibited absorption at both 330 nm and 420 nm, which is characteristic for the presence of a [2Fe-2S] cluster (Figure 10A, middle and bottom, solid lines). Protein gel blotting confirmed that both peaks contained AtCnfU-V protein (Figure 10B, top). Because the purified AtCnfU-V fraction used for the gel filtration was essentially pure, as shown in Figure 9A, we concluded that the purified AtCnfU-V existed as an iron-sulfur cluster-containing dimer and also as an apo-monomer.
The [2Fe-2S] cluster assembled in the cyanobacterial CnfU became unstable upon incubation with EDTA (Nishio and Nakai, 2000). Furthermore, the cluster was extremely labile upon reduction by dithionite (Na2S2O4) (K. Morimoto, T. Yabe, and M. Nakai, unpublished results). Such a reductively labile property is known to be common among the iron-sulfur clusters assembled in other scaffold proteins (IscU, IscA, and NifU) involved in iron-sulfur cluster biosynthesis (Morimoto et al., 2002, 2003). Therefore, we analyzed the effect of iron chelation and reduction on the stability of the [2Fe-2S]-like cluster retained in the AtCnfU-V dimer. As shown in Figure 10A, whereas EDTA caused a slight decrease in the absorption at 280, 330, and 420 nm of the peak around fraction 4 corresponding to the AtCnfU-V dimer, dithionite resulted in almost a complete loss of iron-sulfur cluster from the AtCnfU-V dimer as judged by the decrease in absorbance at 330 nm and 420 nm. In both cases, corresponding amounts of the apo-form of AtCnfU-V monomer accumulated, which were apparent as an increase in absorbance at 280 nm of the smaller peak around fraction 6 (Figure 10A). Conversion of dimeric AtCnfU-V to a monomeric form upon incubation with EDTA or dithionite was confirmed by protein gel blotting, as shown in Figure 10B. Although dithionite seemed to remove almost all of the iron-sulfur cluster retained in the holo-AtCnfU-V dimer and most of the dimer was converted to the apo-monomer, a significant amount of dimeric AtCnfU-V still remained (Figure 10B, bottom), suggesting that a fraction of AtCnfU-V was in an apo-dimeric form.
Evidence for Direct Transfer of Iron-Sulfur Cluster from Holo-AtCnfU-V Dimer to Apo-Ferredoxin
To demonstrate that it is specifically the iron-sulfur cluster-containing AtCnfU-V dimer that serves as the iron-sulfur cluster donor for reconstitution of holo-ferredoxin and not its apo-monomer or any low molecular weight impurities in the purified AtCnfU-V fraction used in the reconstitution assay shown in Figure 9, we performed a similar reconstitution assay with purified apo-ferredoxin and fractions that were obtained after reseparation of the purified AtCnfU-V proteins on gel filtration. Only the dimeric AtCnfU-V (fraction I), and not the apo-monomer (fraction II) or any salts (fraction III), led to formation of holo-ferredoxin (Figures 11A and 11B). This reconstitution activity decreased moderately when the AtCnfU-V was pretreated briefly with dithionite to liberate some bound iron and inorganic sulfur atoms before gel filtration separation (Figures 11A and 11B, + dithionite). This further discounted the possibility that any liberated low molecular weight compounds were responsible for holo-ferredoxin formation.
In addition, holo-ferredoxin formation could be observed even in the presence of millimolar levels of EDTA (Figure 11C), which caused only slight disruption of the iron-sulfur cluster assembled in the AtCnfU (Figure 10). By contrast, chemical iron-sulfur cluster formation with ferrous ion and sulfide was completely blocked by the addition of EDTA (see Supplemental Figure 9A online). We also found that the stromal activity of iron-sulfur cluster insertion could not be blocked by the addition of EDTA (see Supplemental Figure 9B online). Taken together, these data suggest that the formation of holo-ferredoxin by AtCnfU-V was the result of the direct transfer of the iron-sulfur cluster from the holo-AtCnfU-V dimer to apo-ferredoxin. Therefore, AtCnfU-V can serve as a scaffold for iron-sulfur cluster assembly and delivery to apo-ferredoxin.
DISCUSSION
We have demonstrated here that Arabidopsis chloroplast stroma contains multiple NifU-like CnfU proteins of cyanobacterial origin. We have further demonstrated that an Arabidopsis mutant lacking one of the chloroplast-localized CnfU proteins, AtCnfU-V, exhibited a significant dwarf phenotype along with reduction not only in the steady state levels of PSI and stromal ferredoxin but also the stromal iron-sulfur cluster insertion activity. In addition, we provide evidence that recombinant AtCnfU-V exists as a reductively labile, [2Fe-2S]-like cluster-containing homo-dimer, which can function as a direct [2Fe-2S] cluster donor to apo-ferredoxin in vitro. From these observations, we propose that the chloroplast-localized AtCnfU proteins function as scaffolds for iron-sulfur cluster assembly and delivery in the organelle.
The observed pale-green and dwarf phenotypes and photosynthetic growth defect of the atcnfU2 mutants lacking the constitutively expressed AtCnfU-V is most likely caused by an insufficient supply of iron-sulfur clusters for transfer to certain apoproteins in chloroplasts and probably also to other substrates in other plastids of various tissues. Notably, the significant decrease in stromal holo-ferredoxin, a well-known multifunctional electron carrier that plays a critical role in distributing reducing power to various assimilatory processes in both photosynthetic and nonphotosynthetic plastids, might be a major reason for such an overall growth defect.
Although several proteins are known to be involved in PSI biogenesis of higher plants (Stöckel and Oelmüller, 2004), this is an example of a mutation that affects both PSI and stromal ferredoxin biogenesis. It is possible that the impaired PSI biogenesis is some secondary or acclimation effect of the decreased levels of functional ferredoxin, an electron acceptor for PSI. However, this is unlikely because a dramatic decrease in PSI biogenesis such as was shown in this study was not observed in transgenic Solanum tuberosum (potato) plants in which ferredoxin levels were reduced because of expression of antisense ferredoxin RNA (Holtgrefe et al., 2003). Therefore, the observed defect in overall PSI biogenesis in the atcnfU2 mutant plants is most likely to be attributable to inefficient supply of iron-sulfur cluster to central components of PSI. Further study is required to elucidate whether AtCnfU actually serves as a direct or indirect iron-sulfur cluster donor to PSI iron-sulfur cluster-containing subunits.
The Arabidopsis genome encodes three chloroplast-localized CnfU proteins that all exhibit quite different levels and patterns of expression. AtCnfU-V is ubiquitously distributed among all tissues analyzed, whereas expression of AtCnfU-IVb was predominantly found in the aerial parts, and the expression level of AtCnfU-IVa was low. An interesting question that arises is whether these chloroplast-localized multiple CnfU proteins have complementary or distinct functions. To this end, we found that the atcnfU2 plants lacking the constitutively expressed AtCnfU-V were still viable. The mutant plants contained decreased but still marked levels of holo-ferredoxin as well as other iron-sulfur proteins (Figure 5B, see Supplemental Figure 2 online). PSI biogenesis was less severely disrupted in mutants when they were grown in the presence of sucrose (see Supplemental Figure 2 online). Moreover, even when grown in the absence of sucrose, atcnfU2 mutants had normal levels of cytochrome b6f complex containing Rieske iron-sulfur protein (Figure 7). Although it remains unknown whether the other iron-sulfur proteins retained their normal iron-sulfur clusters in the mutants, we also found residual iron-sulfur cluster insertion activity in the chloroplast stroma lacking AtCnfU-V. These data suggest that distinct CnfU proteins might have complementary or partially overlapping roles in the delivery of iron-sulfur clusters to various substrate apo-proteins in different plastids and/or under different growth conditions. Inhibition of root growth was the most obvious gross defect in the atcnfU2 mutant plants (Figure 3F), and this phenotype might be correlated with the low expression of AtCnfU-IVb in roots as shown in Figure 2C. Characterization of other single or double CnfU-knockout Arabidopsis mutants would help to answer the question.
Iron-sulfur cluster assembly (ISC) machinery, including IscU and IscS, is involved in iron-sulfur cluster biosynthesis in most bacteria (other than cyanobacteria) and eukaryotic mitochondria, and IscU is regarded as a major and essential scaffold protein for the assembly of iron-sulfur clusters. Interestingly, the Arabidopsis genome contains three genes that potentially encode IscU homologs, in addition to the NifU-like proteins described in this study, and all of them possess a putative mitochondrial-targeting sequence. Our recent experimental data indicate that these IscU homologs are localized to the mitochondrial matrix (T. Yabe and M. Nakai, unpublished results). This suggests that the Arabidopsis chloroplast does not express an IscU homolog like cyanobacteria and supports our view that chloroplastic CnfU proteins of cyanobacterial origin play an essential role in iron-sulfur cluster biosynthesis in the organelle as the major alternative scaffold protein to IscU. NifS (IscS) homologs also have been identified in both the chloroplast and mitochondrion; these proteins are necessary for the production of elemental sulfur atoms for iron-sulfur cluster biosynthesis (Kushnir et al., 2001; Leon et al., 2002; Pilon-Smits et al., 2002; T. Yabe and M. Nakai, unpublished results). Therefore, plant cells seem to contain distinct iron-sulfur cluster biosynthetic machineries in the mitochondrion and chloroplast, both of which are of bacterial origin; the former is thought to be derived from the bacterial ISC machinery, whereas the latter is proposed to be derived from an ancestral cyanobacteria-like endosymbiont. In yeast, the mitochondrial ISC machinery provides iron-sulfur clusters to mitochondrial as well as cytosolic proteins. It would be interesting to analyze whether chloroplasts, in addition to mitochondria, supply iron-sulfur clusters to cytosolic proteins in plant cells.
Recently, mobilization of sulfur (SUF) machinery was implicated in iron-sulfur cluster biosynthesis (Ellis et al., 2001; Takahashi and Tokumoto, 2002; Nachin et al., 2003), although direct involvement of this machinery for iron-sulfur cluster assembly and delivery remains to be demonstrated. The Arabidopsis genome indeed encodes SUF component homologs, which are potentially localized to chloroplasts. Therefore, an intriguing question is whether the chloroplast CnfU proteins identified in this study function in iron-sulfur cluster biosynthesis in concert, or in parallel, with the SUF machinery.
We demonstrated here that AtCnfU-V exists in the iron-sulfur cluster-containing holo-dimeric state. Because the UV/visible spectra of purified AtCnfU-V are consistent with the presence of a [2Fe-2S]-type cluster and because AtCnfU retains the highly conserved Cys-X-X-Cys motif in its N-terminal domain, it is possible to speculate that one [2Fe-2S] cluster is formed intermolecularly between the N-terminal domains of two identical monomers as illustrated in Figure 12. Interestingly, the C-terminal domains of AtCnfU proteins themselves show a significant degree of sequence similarity with the cyanobacterial CnfU. However, the function of this domain and why this domain lacks the Cys-X-X-Cys motif remain unclear. Upon loss of the [2Fe-2S]-like cluster, apo-AtCnfU-V dimers dissociate readily to monomers, indicating that the dimer does not form a stable complex. We speculate that, in vivo, AtCnfU cycles between a holo-dimer and apo-dimer (or apo-monomers). After transfer of the [2Fe-2S] cluster to a substrate protein, another [2Fe-2S] cluster is reassembled on the apo-AtCnfU-V dimer. It is possible that a chloroplast-localized NifS homolog (Leon et al., 2002; Pilon-Smits et al., 2002; T. Yabe and M. Nakai, unpublished results) might be involved in this recycling step as a sulfur donor. Although transfer of the iron-sulfur cluster from AtCnfU-V to apo-ferredoxin was found to occur in vitro without the aid of other factor(s), the observed transfer reaction seemed not to be very efficient, as shown in Figure 9D. Therefore, it might be possible that any additional stromal factor(s) could assist AtCnfU for more efficient iron-sulfur cluster transfer in vivo. Further biochemical experiments are required to elucidate the mechanistic picture of AtCnfU-mediated iron-sulfur cluster biosynthesis in chloroplasts.
METHODS
Plant Material and Culture Conditions
Arabidopsis (ecotype Columbia) was grown on MS medium (1× MS salts [Sigma, St. Louis, MO], 0.1% Gamborg's vitamin solution [Sigma], and 0.3% Phytagel [Sigma]) with or without 2% sucrose, or on vermiculite as soil, with a 16-h-light at 23°C/8-h-dark at 21°C cycle for a long-day condition or with continuous light at 23°C; for a short-day condition, the plants were grown with an 8-h-light/16-h-dark cycle. T-DNA insertion mutant lines of AtCnfU-V, SALK_011376 and SALK_039254, were provided by the Salk Institute Genomic Analysis Laboratory. The nucleotide sequences adjacent to the T-DNA insertion sites were confirmed by genomic PCR and DNA sequencing; in atcnfU2-1, the T-DNA left-border sequence starts downstream of base 268 (amino acid residue 50), if the translation initiation site is counted as base 1, and in atcnfU2-2, the T-DNA left-border sequence starts upstream of base 1355 (amino acid residue 132).
Preparation of First Strand cDNA and RT-PCR Analysis
Preparation of mRNA was performed using a Quick Prep mRNA purification kit (Pharmacia, Piscataway, NJ) from total RNAs, which were isolated from 8-d-old whole plants or from 23-d-old leaves of Arabidopsis using an RNeasy plant mini kit (Qiagen, Valencia, CA). Reverse transcription was performed to synthesize the first strand cDNA with an AMV reverse transcriptase first strand cDNA synthesis kit (Life Sciences, St. Petersburg, FL). RT-PCR was performed using an RT-PCR high-plus-kit (Toyobo, Osaka, Japan) for amplification of the AtCnfU-V, PsaA, PsaB, PsaC, PsaD, and EF1α (elongation factor 1α) cDNAs (Henrique et al., 2002).
Plasmid Constructions
Plasmids used for in vitro translation were constructed as follows: plasmid pTYC13 encoding AtCnfU-V precursor protein was constructed by inserting the full-length of AtCnfU-V cDNA into the EcoRI-HindIII site of pGEM-4Z (Promega, Madison, WI), and plasmid pGEM/pAtCnfU-IVb encoding the AtCnfU-IVb precursor protein was constructed by inserting a KpnI-SphI fragment of cDNA, provided by ABRC, into the KpnI-SphI site of pGEM-4Z.
To overexpress mature AtCnfU-IVb in E. coli, pTYC21 was constructed by inserting the AtCnfU-IVb cDNA fragment corresponding to amino acid residues 84 to 231 into the NcoI-XhoI site of pET-21d (Novagen, Madison, WI). To overexpress AtCnfU-IVa in E. coli, pTYC24 was constructed by inserting the AtCnfU-IVa cDNA fragment corresponding to amino acid residues 82 to 236 into the NdeI-XhoI site of pET-21d. To overexpress AtCnfU-V without the predicted transit peptide, pTYC22 was constructed by inserting the AtCnfU-V cDNA fragment corresponding to amino acid residues 83 to 235 into the NcoI-XhoI site of pET-21d. The plasmid used in the in vitro translation of maize mature Fd I was constructed as described previously (Hirohashi et al., 2001).
Preparation of Protein Extracts from Arabidopsis, Isolation of Chloroplasts, and in Vitro Protein Import Assays
Whole plant extracts were prepared from Arabidopsis plants as described previously (Martinez-Garcia et al., 1999). Preparation of chloroplasts was performed as described previously (Kogata et al., 1999). For the import study, chloroplasts were isolated from 12- to 14-d-old P. sativum leaves as described (Nakai et al., 1994). Import reactions contained the isolated chloroplasts (50 μg chlorophyll) in a 200-μL reaction mixture. Determination of chlorophyll content was performed as described previously (Lichtenthaler, 1987). The import assay was performed essentially as described previously (Hirohashi et al., 2001). The TNT coupled reticulocyte lysate system (Promega) was used for the in vitro translation with PRO-Mix l-[35S]-Met/Cys (Amersham Biosciences, Buckinghamshire, UK). The import buffer contained 5 mM DTT, 5 mM MgCl2, 5 mM Met, 5 mM Cys, and 5 mM ATP in HS buffer (50 mM Hepes-KOH, pH 7.8, and 330 mM sorbitol). The mixture was incubated at 25°C for 30 min under continuous illumination. At the end of the import reaction, chloroplasts were reisolated and divided into two aliquots, one of which was resuspended in 100 μL of 10 mM Hepes-KOH, pH 7.5. The hypotonically ruptured chloroplasts were centrifuged at 14,000 rpm for 10 min. The supernatants were removed as the stromal fractions and the membrane pellets were washed twice with the same buffer. Finally, all fractions were analyzed by SDS-PAGE followed by fluorography.
Purification of Recombinant Proteins
Plasmids pTYC21 and pTYC22 were introduced into E. coli strain BL21 (DE3) (Novagen). Plasmid-bearing cells were initially grown aerobically at 37°C on L medium (Silhavy et al., 1984) containing 150 μg/mL of ampicillin. Expression of recombinant proteins was then induced by the addition of 0.2 mM isopropylthio-β-galactoside at 20°C for 15 h. Cells were broken by sonication in buffer containing 50 mM Tris-HCl, pH 7.5, 100 mM KCl, and 1 mM DTT. After removal of membranes by centrifugation, the supernatants were used to purify AtCnfU-V and AtCnfU-IVb. They were first applied to a DE52 column (Whatman, Clifton, NJ) from which eluted fractions were collected and precipitated with 40 to 70% saturated ammonium sulfate. The precipitates were resuspended and applied to a HiPrep butyl FF chromatography column (Amersham Biosciences). Eluted protein fractions also were desalted through a HiPrep desalting column (Amersham Biosciences) and further purified over HiPrep Q XL and HiPrep Sephacryl S-100 HR columns (Amersham Biosciences). Finally, fractions containing purified proteins were concentrated with Centriprep YM-30 spin columns (Millipore, Bedford, MA) and stored at −80°C until use. Antibodies against the purified AtCnfU-IVb or AtCnfU-V were raised in rabbits and affinity purified using an antigen-bound HiTrap NHS-activated column (Amersham Biosciences). Protein concentrations were determined using the Bradford method (Bio-Rad, Hercules, CA) with BSA (Sigma) as a standard.
Assay of Iron-Sulfur Cluster Transfer Activity
Chloroplasts isolated from 18-d-old Arabidopsis plants as described above were stored at −80°C as pellets until use. The stromal extracts were prepared by hypotonic treatment of the chloroplasts with 10 mM Hepes-KOH, pH7.5 (at a concentration of 2 mg chlorophyll/mL), for 30 min on ice followed by centrifugation at 10,000g for 15 min at 4°C. [3H]-labeled ferredoxin (maize Fd I), which was synthesized in vitro using TNT coupled reticulocyte lysate systems (Promega), was incubated with the stromal extracts (obtained from 50 μg chlorophyll of chloroplasts) for 0 to 60 min at 25°C in 50 μL of 50 mM Hepes-KOH, pH 7.5, and 50 mM KCl with or without 1 mM dithionite. For assay using chemical amount of apo-ferredoxin as a substrate, apo-ferredoxin was prepared by boiling purified Fd I in the presence of 100 mM DTT and 100 mM EDTA and further purified by gel filtration column chromatography using a fast-desalting column (Amersham Pharmacia Biotech, Uppsala, Sweden) that had been equilibrated with 50 mM Tris-HCl, pH7.5, 50 mM KCl, and 1 mM DTT. Recombinant AtCnfU-V (0 to 20 μg) also was used in the reaction with 1 mM DTT instead of the stromal extacts. All solutions used in the assay were degassed thoroughly before use. Holo-ferredoxin formation was assessed using nondenaturing PAGE (Nishio and Nakai, 2000) followed by Coomassie Brilliant Blue staining or by fluorography.
Gel Filtration of the Purified AtCnfU-V
Purified AtCnfU-V (25 μg) dissolved in a buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM KCl, and 5 mM DTT were further treated either with 10 mM EDTA or 1 mM DTT on ice for 1 h. After treatment, samples were loaded onto Superdex 75 columns (Amersham Biosciences) and eluted with the same buffer. Protein elution was monitored by simultaneous absorbance measurements at 280, 330, and 420 nm (Figure 10A). Twelve 100-μL fractions were collected and analyzed by protein gel blotting as described above (Figure 10B).
To perform the subsequent ferredoxin iron-sulfur cluster reconstitution assay, purified AtCnfU-V (200 μg) dissolved in a buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM KCl, and 1 mM DTT was further treated with or without 10 mM dithionite on ice briefly and subjected to gel filtration column chromatography as above. Each 20-μL fraction obtained after gel filtration was incubated with 20 μg apo-ferredoxin for 60 min at 25°C and analyzed as described above.
Measurements of Oxygen Evolution, Chlorophyll Fluorescence, and P700 Redox State
Rates of oxygen evolution were measured with a leaf disc oxygen-electrode system (Hansatech, Kings Lynn, UK) at a saturating concentration of CO2 (∼10%) at 25°C. In vivo chlorophyll fluorescence of detached leaves was excited and detected with a pulse amplitude modulated fluorometer (PAM101/102/103; Walz, Effeltrich, Germany). A modulated nonactinic measuring beam was used to measure fluorescence. Pulses (1 s) of white light (4000 μmol/m2/s) spaced every 30 s were used to determine the maximum fluorescence (Fm) and the ratio (Fm-Fo)/Fm = Fv/Fm. Actinic light with an intensity of 520 μmol/m2/s was used to drive photosynthesis. The redox state of P700 was monitored by absorbance changes at 820 nm using an ED-P700 detector unit attached to the pulse amplitude modulated fluorometer. The oxidation of P700 was induced by illumination with a 2-min period of far-red light (30 W/m2) to detached leaves (24 mm2).
Blue Native Gel Electrophoresis
Blue native gel electrophoresis was performed as described (Schägger et al., 1994; Asakura et al., 2004). Chloroplast proteins (67 μg) were solubilized with 0.5% (w/v) n-dodecy-β-d-maltoside and separated by 5 to 14% blue native gel electrophoresis. Ferritin (880 kD and 440 kD), catalase (230 kD), and BSA (132 kD and 66 kD) were used as molecular mass marker proteins.
Supplementary Material
Acknowledgments
We thank all members of our laboratory and S. Itoh and H. Mino for helpful suggestions, A. Barkan, H. Oh-oka, T. Hase, and T. Hirohashi for various antibodies, and the ABRC and the Salk Institute Genomic Analysis Laboratory for the atcnfU2 mutants and EST clones. This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas and a Grant-in-Aid for Encouragement of Young Scientists from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to M.N.).
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Masato Nakai (nakai@protein.osaka-u.ac.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.020511.
References
- Agar, J.N., Krebs, C., Frazzon, J., Huynh, B.H., Dean, D.R., and Johnson, M.K. (2000. b). IscU as a scaffold for iron-sulfur cluster biosynthesis: Sequential assembly of [2Fe-2S] and [4Fe-4S] clusters in IscU. Biochemistry 39, 7856–7862. [DOI] [PubMed] [Google Scholar]
- Agar, J.N., Yuvaniyama, P., Jack, R.F., Cash, V.L., Smith, A.D., Dean, D.R., and Johnson, M.K. (2000. a). Modular organization and identification of a mononuclear iron-binding site within the NifU protein. J. Biol. Inorg. Chem. 5, 167–177. [DOI] [PubMed] [Google Scholar]
- Asakura, Y., Hirohashi, T., Kikuchi, S., Belcher, S., Osborne, E., Yano, S., Terashima, I., Barkan, A., and Nakai, M. (2004). Maize mutants lacking chloroplast FtsY exhibit pleiotropic defects in the biogenesis of thylakoid membranes. Plant Cell 16, 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis, P.R. (2001). Photosystem I: Function and physiology. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 593–626. [DOI] [PubMed] [Google Scholar]
- Ellis, K.E., Clough, B., Saldanha, J.W., and Wilson, R.J. (2001). Nifs and Sufs in malaria. Mol. Microbiol. 41, 973–981. [DOI] [PubMed] [Google Scholar]
- Frazzon, J., and Dean, D.R. (2003). Formation of iron-sulfur clusters in bacteria: An emerging field in bioinorganic chemistry. Curr. Opin. Chem. Biol. 7, 166–173. [DOI] [PubMed] [Google Scholar]
- Frazzon, J., Fick, J.R., and Dean, D.R. (2002). Biosynthesis of iron-sulphur clusters is a complex and highly conserved process. Biochem. Soc. Trans. 30, 680–685. [DOI] [PubMed] [Google Scholar]
- Henrique, R., Jasik, J., Klein, M., Martinoia, E., Feller, U., Schell, J., Pais, M.S., and Koncz, C. (2002). Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Mol. Biol. 50, 587–597. [DOI] [PubMed] [Google Scholar]
- Hirohashi, T., Hase, T., and Nakai, M. (2001). Maize non-photosynthetic ferredoxin precursor is mis-sorted to the intermembrane space of chloroplasts in the presence of light. Plant Physiol. 125, 2154–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtgrefe, S., Bader, K.P., Horton, P., Scheibe, R., Schaewen, A.V., and Backhausen, J.E. (2003). Decreased content of leaf ferredoxin changes electron distribution and limits photosynthesis in transgenic potato plants. Plant Physiol. 133, 1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal, G., Csere, P., Prohl, C., and Lill, R. (1999). The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18, 3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogata, N., Nishio, K., Hirohashi, T., Kikuchi, S., and Nakai, M. (1999). Involvement of a chloroplast homolog of the signal recognition particle receptor protein, FtsY, in protein targeting to thylakoids. FEBS Lett. 447, 329–333. [DOI] [PubMed] [Google Scholar]
- Krebs, C., Agar, J.N., Smith, A.D., Frazzon, J., Dean, D.R., Huynh, B.H., and Johnson, M.K. (2001). IscA, an alternate scaffold for Fe-S cluster biosynthesis. Biochemistry 40, 14069–14080. [DOI] [PubMed] [Google Scholar]
- Kushnir, S., Babiychuk, E., Storozhenko, S., Davey, M., Papenbrock, J., De Rycke, R.R., Engler, G., Stephan, U., Lange, H., Kispal, G., Lill, R., and Van Montagu, M.M. (2001). A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell 13, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon, S., Touraine, B., Briat, J.F., and Lobreaux, S. (2002). The AtNFS2 gene from Arabidopsis thaliana encodes a NifS-like plastidial cysteine desulphurase. Biochem. J. 366, 557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.M., Theg, S.M., Bauerle, C.M., and Keegstra, K. (1990). Metal-ion-center assembly of ferredoxin and plastocyanin. Proc. Natl. Acad. Sci. USA 87, 6748–6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler, H.K. (1987). Chlorophylls and Carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382. [Google Scholar]
- Lill, R., Diekert, K., Kaut, A., Lange, H., Pelzer, W., Prohl, C., and Kispal, G. (1999). The essential role of mitochondria in the biogenesis of cellular iron-sulfur proteins. Biol. Chem. 380, 1157–1166. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Monte, E., and Quail, P.H. (1999). A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20, 251–257. [DOI] [PubMed] [Google Scholar]
- Merchant, S., and Dreyfuss, D.W. (1998). Posttranslational assembly of photosynthetic metalloproteins. Annu. Rev. Plant Physiol. 49, 25–51. [DOI] [PubMed] [Google Scholar]
- Morimoto, K., Nishio, K., and Nakai, M. (2002). Identification of a novel prokaryotic HEAT-repeats-containing protein which interacts with a cyanobacterial IscA homolog. FEBS Lett. 519, 123–127. [DOI] [PubMed] [Google Scholar]
- Morimoto, K., Sato, S., Tabata, S., and Nakai, M. (2003). HEAT-repeats containing protein, IaiH, stabilizes the iron-sulfur cluster bound to the cyanobacterial IscA homologue, IscA2. J. Biochem. (Tokyo) 134, 211–217. [DOI] [PubMed] [Google Scholar]
- Muhlenhoff, U., and Lill, R. (2000). Biogenesis of iron-sulfur proteins in eukaryotes: A novel task of mitochondria that is inherited from bacteria. Biochim. Biophys. Acta 1459, 370–382. [DOI] [PubMed] [Google Scholar]
- Nachin, L., Loiseau, L., Expert, D., and Barras, F. (2003). SufC: An unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 22, 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, M., Goto, A., Nohara, T., Sugita, D., and Endo, T. (1994). Identification of the SecA protein homolog in pea chloroplasts and its possible involvement in thylakoidal protein transport. J. Biol. Chem. 269, 31338–31341. [PubMed] [Google Scholar]
- Nakai, M., Nishio, K., Morimoto, K., Yabe, T., and Kikuchi, S. (2002). Molecular scaffolds involved in iron-sulfur cluster biosynthesis. In Recent Research Developments in Proteins, S.G. Pandalai, ed (Trivandrum, India: Transworld Research Network), pp. 1–11.
- Nakai, Y., Nakai, M., Hayashi, H., and Kagamiyama, H. (2001). Nuclear localization of yeast Nfs1p is required for cell survival. J. Biol. Chem. 276, 8314–8320. [DOI] [PubMed] [Google Scholar]
- Nakai, Y., Yoshihara, Y., Hayashi, H., and Kagamiyama, H. (1998). cDNA cloning and characterization of mouse nifS-like protein, m-Nfs1: Mitochondrial localization of eukaryotic NifS-like proteins. FEBS Lett. 433, 143–148. [DOI] [PubMed] [Google Scholar]
- Nishio, K., and Nakai, M. (2000). Transfer of iron-sulfur cluster from NifU to apoferredoxin. J. Biol. Chem. 275, 22615–22618. [DOI] [PubMed] [Google Scholar]
- Nishio, K., Nakai, M., and Hase, T. (1999). Fe-S cluster formation of ferredoxin in chloroplast stroma. In Photosynthesis: Mechanisms and Effects, G. Garab, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 3155–3158.
- Ollagnier-de-Choudens, S., Mattioli, T., Takahashi, Y., and Fontecave, M. (2001). Iron-sulfur cluster assembly: Characterization of IscA and evidence for a specific and functional complex with ferredoxin. J. Biol. Chem. 276, 22604–22607. [DOI] [PubMed] [Google Scholar]
- Pilon-Smits, E.A., Garifullina, G.F., Abdel-Ghany, S., Kato, S., Mihara, H., Hale, K.L., Burkhead, J.L., Esaki, N., Kurihara, T., and Pilon, M. (2002). Characterization of a NifS-like chloroplast protein from Arabidopsis. Implications for its role in sulfur and selenium metabolism. Plant Physiol. 130, 1309–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger, H., Cramer, W.A., and von Jagow, G. (1994). Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217, 220–230. [DOI] [PubMed] [Google Scholar]
- Silhavy, T.J., Berman, M.L., and Enquist, L.W. (1984). Experiments with Gene Fusions. (Cold Spring Habor, NY: Cold Spring Harbor Laboratory Press).
- Stöckel, J., and Oelmüller, R. (2004). A novel protein for photosystem I biogenesis. J. Biol. Chem. 279, 10243–10251. [DOI] [PubMed] [Google Scholar]
- Suzuki, S., Izumihara, K., and Hase, T. (1991). Plastid import and iron-sulfur cluster assembly of photosynthetic and non-photosynthetic ferredoxin isoproteins in maize. Plant Physiol. 97, 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., Mitsui, A., Hase, T., and Matsubara, H. (1986). Formation of iron-sulfur cluster of ferredoxin in isolated chloroplasts. Proc. Natl. Acad. Sci. USA 83, 2434–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., and Nakamura, M. (1999). Functional assignment of the ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster involved in the assembly of Fe-S clusters in Escherichia coli. J. Biochem. (Tokyo) 126, 917–926. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., and Tokumoto, U. (2002). A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 277, 28380–28383. [DOI] [PubMed] [Google Scholar]
- Tong, W.H., Jameson, G.N.L., Huynh, B.H., and Rouault, T.A. (2003). Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc. Natl. Acad. Sci. USA 100, 9762–9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina, H.D., Silberg, J.J., Hoff, K.G., and Vickery, L.E. (2001). Transfer of sulfur from IscS to IscU during Fe/S cluster assembly. J. Biol. Chem. 276, 44521–44526. [DOI] [PubMed] [Google Scholar]
- Wu, G., Mansy, S.S., Wu Sp, S.P., Surerus, K.K., Foster, M.W., and Cowan, J.A. (2002). Characterization of an iron-sulfur cluster assembly protein (ISU1) from Schizosaccharomyces pombe. Biochemistry 41, 5024–5032. [DOI] [PubMed] [Google Scholar]
- Yuvaniyama, P., Agar, J.N., Cash, V.L., Johnson, M.K., and Dean, D.R. (2000). NifS-directed assembly of a transient [2Fe-2S] cluster within the NifU protein. Proc. Natl. Acad. Sci. USA 97, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, L., Cash, V.L., Flint, D.H., and Dean, D.R. (1998). Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273, 13264–13272. [DOI] [PubMed] [Google Scholar]
- Zheng, L., and Dean, D.R. (1994). Catalytic formation of a nitrogenase iron-sulfur cluster. J. Biol. Chem. 269, 18723–18726. [PubMed] [Google Scholar]
- Zheng, L., White, R.H., Cash, V.L., and Dean, D.R. (1994). Mechanism for the desulfurization of L-cysteine catalyzed by the nifS gene product. Biochemistry 33, 4714–4720. [DOI] [PubMed] [Google Scholar]
- Zheng, L., White, R.H., Cash, V.L., Jack, R.F., and Dean, D.R. (1993). Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. USA 90, 2754–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.