Abstract
Histone acetylation and deacetylation are connected with transcriptional activation and silencing in many eukaryotic organisms. Gene families for enzymes that accomplish these modifications show a surprising multiplicity in sequence and expression levels, suggesting a high specificity for different targets. We show that mutations in Arabidopsis (Arabidopsis thaliana) HDA6, a putative class I histone deacetylase gene, result in loss of transcriptional silencing from several repetitive transgenic and endogenous templates. Surprisingly, total levels of histone H4 acetylation are only slightly affected, whereas significant hyperacetylation is restricted to the nucleolus organizer regions that contain the rDNA repeats. This switch coincides with an increase of histone 3 methylation at Lys residue 4, a modified DNA methylation pattern, and a concomitant decondensation of the chromatin. These results indicate that HDA6 might play a role in regulating activity of rRNA genes, and this control might be functionally linked to silencing of other repetitive templates and to its previously assigned role in RNA-directed DNA methylation.
INTRODUCTION
Nuclear DNA is organized in a higher order structure, which overcomes the space constraints in the nucleus and facilitates the spatio-temporal regulation of gene expression. The first level of compaction is achieved by nucleosomal packaging of DNA. Each nucleosome comprises 147 bp of DNA wrapped around a histone octamer that consists of two molecules each of histone proteins H2A, H2B, H3, and H4 (Luger et al., 1997). The histone proteins are subject to various covalent modifications, particularly within their N-terminal tails. These modifications include methylation, acetylation, phosphorylation, ubiquitination, and ADP-ribosylation (Jenuwein and Allis, 2001; Berger, 2002). In addition, DNA itself may be modified by methylation at cytosine residues. DNA methylation and histone tail modifications are believed to help organize chromatin into transcriptionally active (euchromatin) or transcriptionally inert (heterochromatin) regions by influencing the accessibility of DNA to the transcriptional machinery (Fischle et al., 2003).
Whereas heterochromatin is enriched in hypoacetylated histones, methylated DNA and histone H3 methylated at Lys residue 9 (H3K9met), euchromatin is characterized by highly acetylated histone H4, and histone H3 methylated at Lys residue 4 (H3K4met) (Nishioka et al., 2002; Peters et al., 2002).
Acetylation of Lys residues was one of the first histone modifications described to correlate with transcriptional activity (Allfrey et al., 1964). Acetylation was initially suggested to influence transcription by neutralizing the positive charge of the histone tails and decreasing their affinity for DNA; however, there is growing evidence that acetylation helps shape the binding surface for activators and repressors (Kurdistani and Grunstein, 2003).
Histone acetylation of a particular region of chromatin is regulated by a balance between the activities of histone acetyltransferases and histone deacetylases (HDACs). Histone acetyltransferases are transcriptional coactivators and components of large multisubunit complexes (e.g., SAGA, NuA4; Grant et al., 1998; Sterner and Berger, 2000), and HDACs are found associated with sequence-specific regulatory factors (Sin3, NuRD, and CoREST; Ahringer, 2000; You et al., 2001). HDACs also can be recruited by high DNA methylation levels, via association with methyl-DNA binding domain (MBD) containing proteins such as MeCP2 and MBD2 (Bird and Wolffe, 1999), or directly via recruitment by the maintenance DNA methyltransferase itself (Fuks et al., 2000). Evaluation across kingdoms indicates that HDAC families comprise conserved as well as highly divergent members (Pandey et al., 2002). The large number of different HDACs suggests that they have evolved to have specific and/or overlapping roles concerning their targets. In addition, HDACs are regulated in various ways, including by subcellular compartmentalization, posttranscriptional modification, and interacting proteins (Yang and Seto, 2003). Although a few HDACs are relatively well characterized for their role in transcriptional regulation, it will take an enormous effort to decipher the biological function for each family member in every experimental system. Defined loss-of-function mutations in genes for individual HDACs have helped to elucidate the role in some model organisms. For example, RPD3 from bakers' yeast is required to maintain histone hypoacetylation levels in vivo. Through its interaction with the transcriptional repressors SIN3 and UME6, RPD3 can be targeted to promoters and repress genes containing UME6 binding sites (Rundlett et al., 1998). Coupled chromatin immunoprecipitation (ChIP) and DNA microarray analyses indicated that RPD3 affects the acetylation of genes in virtually all cellular pathways (Robyr et al., 2002) but preferentially associates with promoters that direct high transcriptional activity such as ribosomal protein genes or rRNA genes (Kurdistani et al., 2002).
In the Arabidopsis (Arabidopsis thaliana) genome, 16 potentially functional HDACs have been identified, and these can be classified into three families (Pandey et al., 2002; see also http://chromdb.biosci.arizona.edu): the RPD3/HDA1-like histone deacetylases, the members of the SIR2-like family, and the plant-specific HD2-like HDACs originally identified as acidic nucleolar phosphoproteins from maize (Zea mays) (Lusser et al., 1997). Interference of HDAC functions in plants has been studied using inhibitors such as trichostatin A, SAHA, or butyrate and using transgenic plants containing antisense or overexpressing constructs. These approaches have provided evidence that HDACs are involved in regulation of histone acetylation and thereby gene expression, with consequences for plant morphology and development (Chen and Pikaard, 1997; Wu et al., 2000; Tian and Chen, 2001). Dissecting the function of individual HDAC members is problematic in these studies; therefore, the analysis of loss-of-function mutations of individual HDAC genes should add valuable information on specific roles.
Mutants in an Arabidopsis RPD3-like HDAC gene, AtHDA6, were found in two independent mutant screens based on their effects on specific transgene expression (Murfett et al., 2001; Aufsatz et al., 2002). The HDA6 mutant allele axe1 lead to higher expression from a marker gene with an auxin-responsive promoter element (Murfett et al., 2001), whereas the rts1 alleles of the locus interfere with double-stranded RNA-directed transcriptional silencing (Aufsatz et al., 2002).
The sil1 (modifiers of silencing 1, Furner et al., 1998) mutation was selected as a monogenic recessive trait reactivating silent and methylated transgenes (Furner et al., 1998). Here, we report the identification of the gene mutated by the sil1 mutation. The sil1 mutant is a new allele of AtHDA6. We show that sil1, as well as axe1-5, reactivate transcriptionally silent transgenes and endogenous repeats. We further provide evidence that mutations in the Arabidopsis HDA6 gene influence histone acetylation levels. Specifically, rDNA loci become enriched in acetylated histone H4, whereas total H4 acetylation levels are only slightly increased. The rDNA repeats in the mutant plants become locally hypermethylated at H3K4 and DNA hypomethylated, concomitant with significant changes in the structural organization of rDNA loci. The AtHDA6 gene product is therefore implicated in determining transcription, DNA methylation, and structural organization of multiple classes of repetitive DNA.
RESULTS
The sil1 Mutation Is an Allele of HDA6
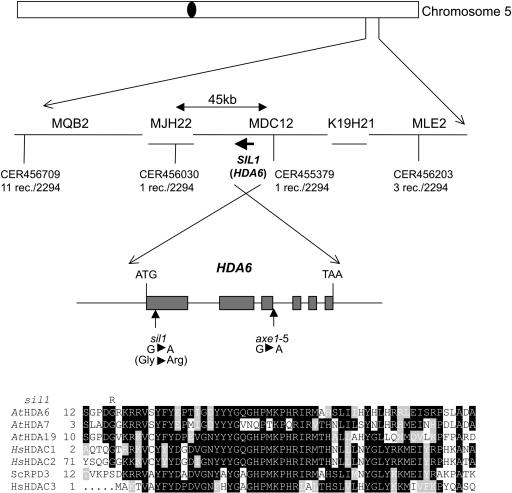
The mutant sil1 was identified in a screen for mutations releasing silencing of the complex, rearranged transgenic locus C containing the chalcone synthase gene (CHS) and the resistance marker genes neomycin phosphotransferase and hygromycin phosphotransferase. The sil1 mutation reactivates mainly the resistance marker genes, whereas the homology-dependent silencing of the endogenous and transgenic CHS copies are only weakly affected (Furner et al., 1998). In contrast with other mutations that alleviate silencing of the C locus (hog1 and ddm1), sil1 does not affect DNA methylation at the transgenes or at rDNA loci (Furner et al., 1998). The sil1 mutation has been mapped to chromosome V between markers CER456030 and CER455379 (Figure 1). This region encompasses the putative HDAC gene HDA6 (At5g63110), a homolog of yeast (Saccharomyces cerevisiae) RPD3 (Rundlett et al., 1996) and human HDAC1 (Taunton et al., 1996). Additional recessive mutant alleles of this gene (axe1-1, axe1-2, axe1-3, axe1-4, and axe1-5) were identified and shown to upregulate expression of other complex transgenic loci (produced by transformation with plasmids pDR5 and p2xD0) that contain marker genes under the control of an auxin-responsive promoter (Murfett et al., 2001). Crosses between wild-type plants and plants homozygous for sil1 and the C locus yielded hygromycin-sensitive hybrids, indicating that the C locus is quickly resilenced in a sil1-heterozygous background. Crosses between plants homozygous for different alleles of transgene-free axe1 mutant plants and homozygous sil1 mutants containing the C locus resulted exclusively in hygromycin-resistant hybrids, indicating a lack of resilencing of the C locus, failure of complementation, and allelism between the axe1 and sil1 mutations. Sequencing of the HDA6 coding region from sil1 genomic DNA revealed a G-to-A transition in the N-terminal part of the protein, resulting in replacement of Gly residue 16 by Arg (Figure 1). The allelism between axe1 and sil1 implies that the upregulation of DR5 and 2xD0 transgenes is based on alleviation of silencing because of an impaired function of HDA6 rather than an effect on auxin signaling (Murfett et al., 2001).
Figure 1.
sil1 Has a Mutation in the AtHDA6 Gene.
The sil1 mutation maps between markers CER456030 and CER455379 at the bottom of chromosome 5. Sequencing of the HDA6 gene in the sil1 mutant reveals a point mutation, 46 bases after the ATG initiation codon, leading to the replacement of Gly16 by Arg. The axe1-5 mutant has a base substitution at position 1635 downstream of the ATG at the third exon-intron junction. Alignment of AtHDA6 with Arabidopsis (At) and human (Hs) RPD3-like HDACs and yeast (Sc) RPD3 reveals a conservation of Gly16 in plant and human RPD3-like HDACs.
All HDA6 Mutations Release Transcriptional Gene Silencing
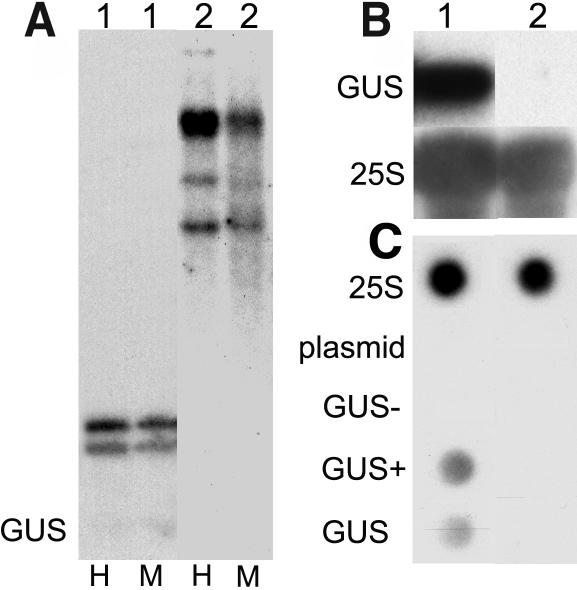
The locus C (reactivated by the mutant sil1) and genomic insertions formed by pDR5 and p2xD0 integration (reactivated by the axe1 mutants) are all very complex transgenic loci, consisting of multiple, rearranged, and methylated transgene copies. These features made it likely that they were transcriptionally inactivated and that the mutations interfered with transcriptional gene silencing (TGS). To verify this assumption, we crossed alleles of HDA6 mutant plants (sil1, axe1-1, axe1-3, axe1-4, and axe1-5) to a well-established TGS test line. This transgenic line, L5 (Morel et al., 2000), is homozygous for an insert carrying multiple and methylated copies (Figure 2A) of a transgene consisting of the 35S promoter of Cauliflower mosaic virus and the ß-glucuronidase (GUS) marker gene. The 35S:GUS transgene is silenced at the transcriptional level, as determined by RNA gel blot analysis (Figure 2B) and transcriptional run-on assays (Figure 2C). F2 seeds derived from the crosses were grown under axenic conditions, and seedlings were stained for GUS activity 1 week after germination. Approximately 19% of each F2 progeny expressed the GUS marker gene. This corresponds to the expected 3:16 ratio of F2 plants homozygous for an HDA6 mutation and carrying one or two copies of the L5 insert. Conversely, none of 150 F2 seedlings resulting from a cross between a wild-type plant and line L5 expressed GUS, indicating that the maintenance of TGS at the L5 insert requires the HDA6 gene product and is unaffected by crossing.
Figure 2.
The 35S:GUS Transgene at the L5 Locus Is Methylated and Transcriptionally Silenced.
The transcriptionally active transgenic line Hc1 (1) and the silenced line L5 (2) were characterized by a combination of DNA gel blot (A), RNA gel blot (B), and nuclear run-on analysis (C). The presence of high molecular weight fragments observed after digestion by the methylation-sensitive restriction enzymes HpaII (H) and MspI (M) and hybridization with radiolabeled 35S and GUS probes indicate that the entire insert in line L5 is strongly methylated (A). Hybridization of 10 μg of total RNA with a probe corresponding to the GUS coding region (top panel) or a 25S rDNA probe reveals the absence of GUS cytoplasmic transcript in line L5 (B). Run-on experiments using labeled RNA extracted from leaf nuclei of adult plants for hybridization of dot blots demonstrate the lack of nascent GUS transcript in line L5. Dots contain 2 μg DNA each of the 25S rDNA-containing plasmid (25S), single-stranded pBluescript KS+ (plasmid), and GUS-containing plasmids (GUS−, sense single-stranded; GUS+, antisense single-stranded; GUS, double-stranded) (C).
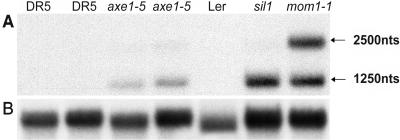
We also tested the effect of the HDA6 mutations on silencing of endogenous targets. A specific class of pericentromeric repeats termed TSI (transcriptionally silent information, not expressed in wild-type plants) is transcribed in plants homozygous for the sil1 allele (Steimer et al., 2000). We analyzed the reactivation of TSI repeats by RNA gel blots in the axe1-5 mutant, which does not express an HDA6 transcript of the expected size but shorter and longer mRNAs because of a splice-site mutation (Murfett et al., 2001). RNA gel blots show that axe1-5 plants express TSI (Figure 3). Interestingly, the point mutant sil1 results in similar if not higher TSI expression compared with the splice-site mutant axe1-5 (Figure 3), but both mutants have lower TSI levels than mom1, a TGS mutant that causes significant but moderate transcription from TSI templates without changing their DNA methylation (Amedeo et al., 2000). The possibly nuclear nonpolyadenylated, 1250-nucleotide TSI fragment and the cytoplasmic, polyadenylated 2500-nucleotide RNA (Steimer et al., 2000) accumulate to similar levels in the mutant mom1. By contrast, the HDA6 mutants predominantly express the shorter nonpolyadenylated fragment (Figure 3). As in mom1 mutant plants, the release of silencing does not result in obvious phenotypic alterations, with the exception of a significant delay in flowering in both sil1 and axe1-5 plants.
Figure 3.
sil1 and axe1-5 Alleles Release Silencing of an Endogenous TSI Sequence.
(A) RNA gel blot analysis using the TSI pA2 fragment as probe reveals TSI transcripts in the two HDA6 mutant alleles axe1-5 and sil1. Lanes 1, 2, and 5 show silencing of the endogenous TSI repeats in the transgenic background of the axe1-5 mutants (DR5) and the Ler ecotype, whereas lanes 3, 4, 6, and 7 show reactivation of TSI in the two HDA6 mutant alleles and the mom1-1 mutant, respectively. Predominantly, two transcripts are expressed—a longer, polyadenylated one (Steimer et al., 2000) as well as a shorter transcript. nts, nucleotides.
(B) The blot was reprobed with RAN (small GTP binding protein) (Haizel et al., 1997) as a loading reference. Total RNA (20 μg per lane) was extracted from rosette leaves of adult plants.
rDNA Repeats Become Highly Acetylated in HDA6 Mutants
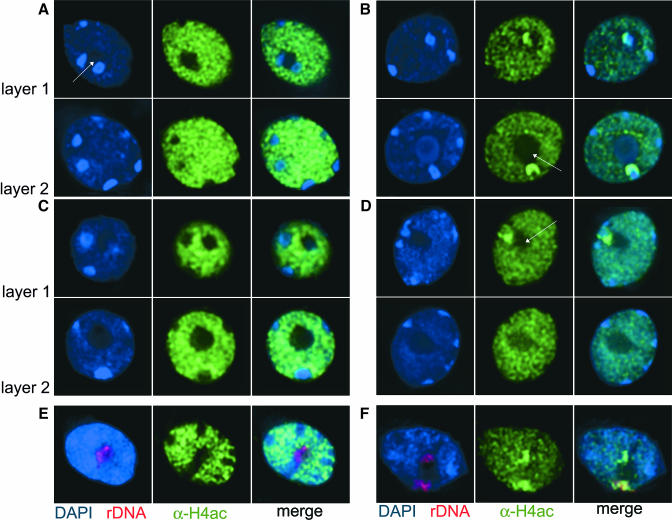
Because the HDA6 gene product has sequence homology with other nuclear proteins shown to have HDAC activity, we studied the effect of HDA6 mutations on the nuclear distribution of histone H4 acetylation. Mesophyll protoplasts from wild-type Landsberg erecta (Ler), the sil1 mutant, DR5 (the transgenic background of the axe1-5 mutant), and the axe1-5 mutant were fixed, stained with an antibody detecting tetra-acetylated histone H4 (α-H4ac), and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). DAPI staining of DNA in interphase nuclei of Arabidopsis distinguishes the nucleolus devoid of dye, the loosely packed euchromatin, and 8 to 10 condensed heterochromatic regions (Figure 4). The latter, known as chromocenters, contain centromeric and pericentromeric chromatin, and as many as four chromocenters also include rDNA repeats (Maluszynska and Heslop-Harrison, 1991). The DAPI-stained nuclei of mutants were indistinguishable from the wild type. Chromatin containing tetra-acetylated histones was found exclusively in euchromatin in all wild-type nuclei (Figures 4A and 4C). However, nuclei of axe1-5 and sil1 mutant plants contained chromocenters that were intensively stained with the H4ac antibody. This effect was more pronounced in the axe1-5 mutant than in the sil1 mutant (Figures 4B and 4D, Table 1). The labeled chromocenters were always in close association with the nucleolus (Figures 4B and 4D, arrow). Layer-by-layer analysis of mutant nuclei revealed that the number of highly acetylated heterochromatic regions never exceeds 4 in one nucleus and comprises only part of the chromocenter (Figures 4B and 4D). The tight association of the highly acetylated chromocenters with the nucleolus, together with the proposed role in rRNA gene repression described for the HDA6 homolog RPD3 in yeast (Sandmeier et al., 2002), suggested that these represent the rDNA loci. To examine a possible relationship between acetylated histones and rDNA, we combined immunodetection of modified histones with fluorescent in situ hybridization (FISH) for rDNA repeats. The rDNA loci of wild-type and mutant nuclei are localized close to the nucleolus. However, whereas all chromocenters, including those with the rDNA repeats, were free of any H4ac signal in DR5 (Figure 4E), the double labeling technique revealed an overlap between the bright H4ac immunosignals and rDNA FISH signals in mutant nuclei (Figure 4F). Therefore, the loss of functional HDA6 resulted in a drastic enrichment of histone acetylation specifically at rDNA repeats.
Figure 4.
rDNA Repeats Are Hyperacetylated in Nuclei of HDA6 Mutants.
(A) to (D) Distribution of histone H4 acetylation revealed by DAPI staining of DNA (blue, left panel) and immunodetection with an antibody specific for tetra-acetylated H4 (green, middle panel) in nuclei of control lines DR5 (A) and Ler (C) and in axe1-5 (B) and sil1 (D) mutant nuclei. Right panels show merged images. For each nucleus, two layers were selected from deconvoluted image stacks, arrows mark the nucleolus.
(E) and (F) FISH using rDNA repeats (red, left panel) after immunostaining with α-H4ac antibodies (green, middle panels) shows that the rDNA loci indeed are devoid of H4ac staining in the wild type (E) but become highly enriched with H4ac in mutant nuclei (F).
Table 1.
Number of HDA6 Mutant Nuclei with NOR-Specific Enrichment in Acetylated Histone H4 and Methylated H3K4
| Mutant | H4ac | H3K4met | ||
|---|---|---|---|---|
| axe1-5 | 98% | n = 100 | 62.4% | n = 303 |
| sil1 | 39.6% | n = 306 | 19.1% | n = 308 |
Total Histone H4 Acetylation Levels Are Not Increased in HDA6 Mutant Plants
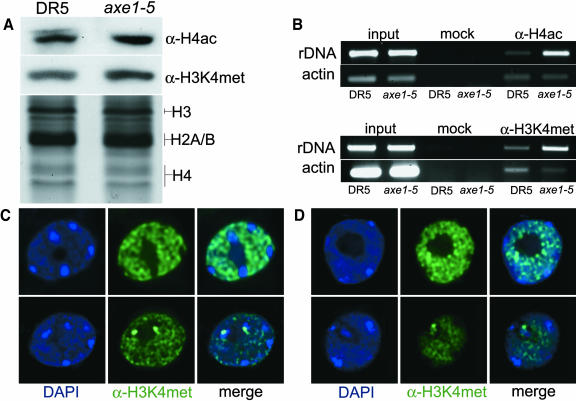
All HDA6 alleles were originally isolated as mutations affecting loci other than rRNA genes. Intense H4ac immunosignals could indicate a global increase in histone acetylation that would appear more prominent at rDNA loci because these include long stretches of silent rRNA genes that are highly condensed in comparison with euchromatic regions (Pontes et al., 2003). To test the possibility that reduced HDA6 activity affects histone acetylation levels globally, we isolated histones from DR5 and axe1-5 plants and performed protein gel blot analysis. There is no obvious increase in tetra-acetylated H4 in mutant plants (Figure 5A). Also, the amount of methylation at position 4 of histone H3, another epigenetic mark for actively transcribed genes, is not significantly increased in the mutants (Figure 5A). Although we cannot exclude the possibility that the HDA6 protein in axe1-5 is still partially functional and sufficient to maintain a basal level of hypoacetylation, it seems likely that HDA6 is not the major HDAC in Arabidopsis but may represent a type directed to selected targets, such as rDNA repeats or complex transgenes.
Figure 5.
Changes in Levels of H4ac and H3K4met Are Limited to Specific Loci in HDA6 Mutants.
(A) Protein gel blot analysis detecting H4ac (top panel) and H3K4met (middle panel) using α-H4ac and α-H3K4met antibodies, respectively, on protein extracts from wild-type (DR5) and axe1-5 mutant plants. Bottom panel, Coomassie staining shows equal protein loading.
(B) ChIP performed in the control line DR5 and the mutant allele axe1-5 reveals an increase in H4ac and H3K4met at rDNA repeats. The Actin2/7 gene is equally present in mutant and control precipitates. If the antibodies are omitted during the procedure (mock), neither target is amplified, whereas the equal strength of bands after PCR with the input fraction indicates equal amounts of chromatin before immunoprecipitation.
(C) and (D) Distribution of histone H3 methylated at Lys 4 revealed by DAPI staining (blue, left panel) and immunodetection with an antibody specific for H3K4met (green, middle panel) in nuclei of control lines (top row) DR5 (C), Ler (D), and mutants (bottom row) axe1-5 (C) and sil1 (D). Right panels show merged images.
HDA6 Mutations Affect Histone H3K4 Methylation Patterns
An increase in histone acetylation is often correlated with another specific modification—methylation at Lys residues at position 4 of histone H3 (H3K4) (Strahl and Allis, 2000). To investigate this correlation for the hyperacetylated rDNA loci in HDA6 mutants, antibodies specific for H3K4 methylation were included in our immunostaining experiments. All chromocenters in the wild type were free of H3K4 methylation, but we observed an enrichment of H3K4 methylation in the axe1-5 and sil1 mutants at the chromocenters presumably containing the rDNA (Figures 5C and 5D, bottom panels). This change affects a significant number of nuclei in both mutants, although the proportion is lower in sil1 (Table 1). To confirm the local hyperacetylation at rDNA loci and the concomitant increase in H3K4 methylation at the molecular level, we performed ChIP on 3-week-old soil-grown plants of DR5 and the mutant allele axe1-5. Amplification with primers specific for a 280-bp region of the 25S rRNA gene showed that indeed rDNA repeats are enriched in both H4ac and H3K4met immunoprecipitates in the mutant compared with the control line DR5. These chromatin modifications at the Actin2/7 gene, serving as reference (Johnson et al., 2002; Tariq et al., 2003), remain unaffected by the HDA6 mutation (Figure 5B). Comparison of mutant and wild-type chromatin in the α-H3K4met–precipitated fraction by ChIP dot blot analysis confirmed the enrichment of rDNA to a similar extent as in the PCR-based assay (data not shown). Therefore, the irregular histone acetylation in the HDA6 mutants at rDNA-comprising chromocenters is correlated with an increase in H3K4 methylation.
Hyperacetylation at rDNA Is Not Accompanied by Increased rRNA Expression in the Mutants
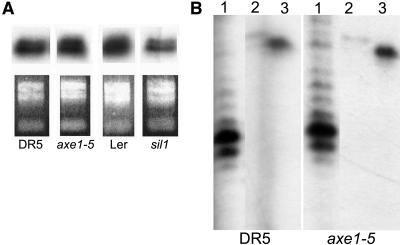
Only a subset of rDNA repeats in eukaryotic cells is transcribed at a given time (McKnight and Miller, 1976; Morgan et al., 1983; French et al., 2003). The hyperacetylation of rDNA repeats in the HDA6 mutants and the increased histone H3K4 methylation at the repeats suggested that these changes may reflect an increase in rRNA transcription. However, comparison of rRNA levels relative to total RNA using semiquantitative RT-PCR (data not shown) or using an S1 nucelase protection assay to detect pre-rRNAs initiated directly at the gene promoter (Figure 6A) did not reveal any differences between mutant and the wild type. However, potential upregulation of rRNAs might be masked when normalized to total RNA because rRNA represents the major species of RNA. Therefore, a subsequent S1 nuclease protection experiment compared rRNA transcript levels relative to the mRNA levels for ubiquitin and actin (Figure 6B). The results of this experiment reveal that axe1-5 mutant plants contain the same or even slightly reduced amounts of rRNA transcripts compared with the wild type (Figure 6B); thus, there is no indication for increased rRNA transcription concomitant with hyperacetylation at rDNA.
Figure 6.
rDNA Expression Is Not Increased in HDA6 Mutant Plants.
Total RNA from control lines DR5 and Ler and mutants axe1-5 and sil1 was subjected to S1 nuclease protection using probes specific for the 5′ end of pre-rRNA transcripts and compared with total RNA amounts, as seen from ethidium bromide staining (A). The signals obtained for rRNA (1) of DR5 and axe1-5 were normalized against signals obtained with probes specific for protein-coding genes ubiquitin (2) or actin (3) (B). All lanes are from the same exposure of the same autoradiogram.
rDNA Repeats Become Decondensed in HDA6 Mutants
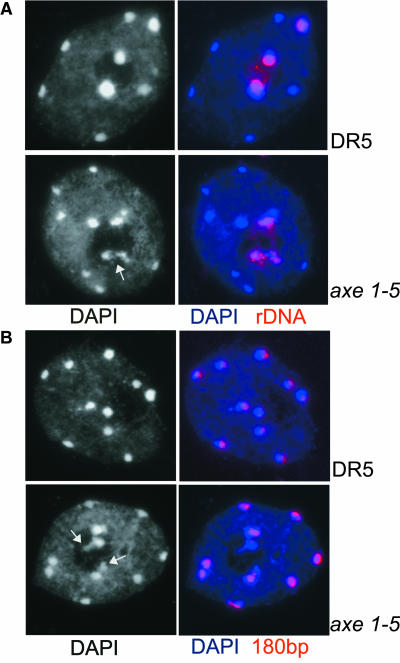
With the exception of the 5S RNA genes, rRNA genes of Arabidopsis are arranged in long tandem arrays comprising the two nucleolus organizer regions (NORs) on chromosomes II and IV (Maluszynska and Heslop-Harrison, 1991). Both NORs adjoin the telomeres (Copenhaver and Pikaard, 1996). FISH with rDNA probes on wild-type interphase nuclear spreads revealed the rDNA to be compactly organized in the chromocenter(s) close to the nucleolus, and only a few DNA repeats extend visibly into the nucleolus (Figure 7A). No obvious change in appearance occurred in the point mutation allele sil1 (data not shown). However, in the splice-site mutation axe1-5, the tight organization was abolished. The rDNA appears less condensed, and rDNA enters the nucleolus and overlaps with adjacent euchromatin (Figure 7A, bottom panel). By contrast, the core centromeric regions, represented by the 180-bp tandem repeats, do not become disorganized in either mutant (Figure 7B). This result appears distinct from the drastic decondensation of all chromocenters observed in ddm1 mutants (Mittelsten Scheid et al., 2002; Soppe et al., 2002; Probst et al., 2003) because of the general hypomethylation of heterochromatin. The decondensation of rDNA repeats is correlated with the high acetylation of histone H4 and possibly also with an increase in histone H3K4 methylation, suggesting a specific role for the HDA6 deacetylase in the regulation of chromatin structure at particular loci, such as the rDNA repeats.
Figure 7.
rDNA Loci, but Not Chromocenters in General, Are Decondensed in HDA6 Mutant Nuclei.
Interphase nuclear spreads of control lines DR5 and Ler and mutants axe1-5 and sil1 stained with DAPI (black and white in left panel, blue in merged images in the right panel) and FISH with biotin-labeled probes for rDNA repeats (A) and centromeric (180 bp) repeats (B). Arrows in the black and white images point to decondensed rDNA repeats in mutant nuclei in (A) and (B).
HDA6 Mutations Affect DNA Methylation Levels Specifically at rDNA Loci
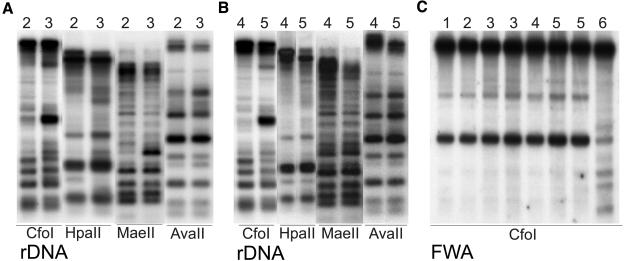
Inhibition of HDACs by trichostatin A (TSA) in Neurospora crassa results in reduced DNA methylation at specific transgenic loci (Selker, 1998). No significant changes in DNA methylation levels of either transgene or rDNA was reported in the initial study of the sil1 mutation (Furner et al., 1998) or the axe1 mutations (Murfett et al., 2001). By contrast, the rts1 and rts2 alleles caused limited demethylation at the target site analyzed (Aufsatz et al., 2002). We investigated whether the increased H4 acetylation at the rDNA loci in HDA6 mutants was accompanied by changes in DNA methylation at these targets. We incubated DNA from wild-type plants (Columbia and Ler), the transgenic line DR5, and the mutants axe1-5 and sil1 (devoid of the C locus) with different methylation-sensitive restriction enzymes and performed DNA gel blot analysis with an rDNA probe. Clear changes in the methylation pattern between the wild type and mutants were detected using CfoI (Gm5CGC; Figures 8A and 8B). The hypomethylation in the axe1-5 background was slightly stronger, and this line also showed more pronounced hyperacetylation at rDNA repeats (Figure 4B, Table 1). Other enzymes, which also are specifically inhibited by CG methylation, HpaII (Cm5CGG) and MaeII (Am5CGT), confirmed the rDNA hypomethylation in both mutants (Figures 8A and 8B). The digest with the enzyme AvaII, which is inhibited by either CG, CNG, or CNN methylation (GGW m5CC and GGWCm5C; Figures 8A and 8B), showed only minor reductions in cytosine methylation at the rDNA repeats. Though the changes in CG methylation are significant and distinct, the demethylation is much less pronounced than in DNA of ddm1-5 (Jeddeloh et al., 1999) used as demethylation control and strongly affected in methylation patterns at many repetitive sequences. The changes caused by the two HDA6 mutant alleles are relatively subtle and may have been missed in our earlier studies (Furner et al., 1998).
Figure 8.
DNA Methylation Patterns at rDNA Repeats Are Affected in HDA6 Mutants.
Genomic DNA samples from wild-type Columbia (1) and Ler (4), the transgenic line DR5 (2), the HDA6 mutant alleles axe1-5 (3) and sil1 (5), and from the DNA methylation mutant ddm1-5 (6) (Jeddeloh et al., 1999) were analyzed. The DNA was digested with methylation-sensitive restriction enzymes CfoI, HpaII, MaeII, and AvaII ([A] and [B]), subjected to DNA gel blot analysis, and probed with rDNA (Vongs et al., 1993). A CfoI digest hybridized with a FWA probe (Saze et al., 2003) is shown in (C).
To examine whether the mutations induce specifically rDNA demethylation or a more general genome-wide demethylation, DNA gel blots were reprobed for other potential candidate genes containing appropriate restriction sites. Suggested by the delay in flowering time in the HDA6 mutants, the membrane with the CfoI digest was hybridized with the promoter of the FWA gene (Saze et al., 2003), a positive regulator of flowering. We also analyzed the HpaII digest for methylation changes at the (weakly expressed) TSI genes and at the 180-bp centromeric repeats. Only very subtle changes could be detected with the FWA and the TSI probes, whereas the methylation at the 180-bp repeats appeared unaffected by the HDA6 mutations (Figure 8C; data not shown).
DISCUSSION
The Arabidopsis gene HDA6 is a putative HDAC based on its close homology to yeast RPD3 and mouse HDAC1 and previously has been identified by seven different mutated alleles; the first five were isolated as recessive mutations increasing expression of transgenes with auxin-responsive promoters. A further two alleles were recovered in a screen for interference with transcriptional silencing acting in trans via RNA-directed DNA methylation (Aufsatz et al., 2002). The sil1 mutant was isolated as a modifier of transgene silencing (Furner et al., 1998). Here, we report that sil1 is another allele of HDA6. By crossing five of these alleles to a line having a well-characterized cis-transcriptionally silenced locus, we observed a release of silencing, indicating that HDA6 is involved in epigenetic regulation. The HDA6 mutants also express noncoding RNA from endogenous repetitive templates (Steimer et al., 2000). This indicates that functional HDA6 is required to maintain TGS at certain, probably repetitive target sequences. All of the HDA6 mutants analyzed accumulate less TSI transcripts than other TGS mutants that affect global DNA methylation, such as met1 (Saze et al., 2003) or ddm1 (Steimer et al., 2000). This could either be because of the fact that none of the sil or axe alleles are true null mutations or that there might be redundancy with other members of the HDAC family (Pandey et al., 2002). An interesting peculiarity of the HDA6 mutations is the predominant accumulation of the smaller nonpolyadenylated TSI transcript (cf. with mom1, ddm1, and met1 mutants). The origin and/or the processing of the TSI transcript family are not yet well understood; however, this might reflect release of expression from particular transcriptional initiation sites or from templates that lack appropriate polyadenylation signals. The changes of chromatin modifications at rDNA loci that also produce nonpolyadenylated transcripts suggest further studies to investigate whether HDA6 has a specific role for regulation of transcripts lacking this 3′ end modification.
The known HDA6 mutations occur throughout the coding region of the gene. With the exception of the Gly mutated in sil1 that is not conserved in the yeast counterpart RPD3, all amino acid exchange mutations affect residues that are highly conserved between plant, animal, and yeast HDACs (Murfett et al., 2001). Although the mutations cause different degrees of transcriptional reactivation, histone H4 acetylation, and histone H3K4 methylation at rDNA loci, these differences are not directly correlated with either single amino acid exchanges or splice-site mutations. HDA6 activity might be very sensitive to any structural changes of the protein.
Even though it cannot be excluded that HDA6 has an effect on acetylation of other, yet unidentified, target proteins, the significant increase of histone acetylation at the rDNA loci in HDA6 mutants strongly suggests that HDA6 is indeed a functional HDAC. Because the protein gel blot analysis indicated no significant increase in the total level of acetylated H4, HDA6 might remove acetyl residues only from specific targets, whereas other related family members are responsible for a more general control of histone deacetylation. HDAC genes form a large family, and many members were already shown to be responsible for the reversible and dynamic acetylation changes of histone tails (Kurdistani and Grunstein, 2003). Both yeast RPD3 (Rundlett et al., 1998) and mouse HDAC1 (Doetzlhofer et al., 1999) are required for transcriptional repression of reporter genes, and RPD3 is involved in the deacetylation of large chromosomal domains throughout the yeast genome (Vogelauer et al., 2000; Kurdistani et al., 2002; Robyr et al., 2002). There are 10 members of the RPD3/HDA1 gene family with complete HDAC domains in Arabidopsis falling into subgroups (Pandey et al., 2002) with varying and tissue-specific expression levels. Evidence for their role in histone modification and gene regulation so far was limited to HDA19 (synonyms AtRPD3A and AtHDA1), the closest homolog of HDA6 with detectable expression levels (Tian and Chen, 2001). Other members are weakly expressed or have yet to be analyzed for their transcript levels (http://www.chromdb.org/). Transcripts from HDA19 are highly abundant in leaves, stem, and flowers, and expression as a GAL4 fusion protein was shown to downregulate a reporter gene (Wu et al., 2000). Antisense-based downregulation of HDA19 resulted in a 10-fold increase in tetra-acetylated histone 4 (Tian and Chen, 2001). Therefore, HDA6 shares sequence homology and very likely enzymatic activity with its homolog HDA19 but seems to be responsible more for specific rather than for general deacetylation.
A knockout of mouse HDAC1 leads to embryonic lethality because of severe proliferation defects (Lagger et al., 2002). Clr3 (for cryptic loci regulator; RPD3-like) and Clr6 (HDAC1-like) HDACs in fission yeast (Schizosaccharomyces cerevisiae) are involved in maintenance of silent mating type and centromeric heterochromatin (Grewal et al., 1998), and mutants for clr3 and clr6 show defective mitotic segregation (Grewal et al., 1998). No Arabidopsis mutant affected in any other HDA gene has been described, but downregulation of the HDA19 by antisense RNA expression resulted in strong pleiotropic effects in transgenic plants, including some that are attributable to secondary deregulation of genes controlling development (Tian and Chen, 2001). By contrast, HDA6 mutants do not have any drastic phenotype even after several generations of inbreeding (Furner et al., 1998; Murfett et al., 2001; Aufsatz et al., 2002), except a significant delay in flowering time. The fact that morphology is largely unaffected in all plants with a mutated HDA6 is further evidence that the regulatory role of this protein is restricted to very specific target genes.
Our FISH analysis on interphase chromosome spreads indicated that the organization of rRNA genes into chromocenters is affected in the HDA6 mutants. Decondensation of chromocenters also has been observed in plants mutated in a chromatin remodeling factor that shapes heterochromatin more generally and is required for proper DNA methylation and histone modification (Mittelsten Scheid et al., 2002; Soppe et al., 2002). Interestingly, in ddm1-5 mutant nuclei, mainly centromeric and pericentromeric repeats undergo decondensation, whereas in axe1-5 nuclei, specifically rDNA repeats are affected (Probst et al., 2003). Therefore, histone deacetylation by HDA6 may be required to establish a heterochromatin-like structure at the rDNA repeats, whereas downregulation of transcription may well be achieved also by other mechanisms, such as modulation of the initiation frequency at active decondensed rRNA genes (Sandmeier et al., 2002; Grummt and Pikaard, 2003). The number of active rDNA repeats can be variable, as evident by ultrastructural analysis in Drosophila melanogaster, Xenopus laevis, and yeast (McKnight and Miller, 1976; Morgan et al., 1983; French et al., 2003). A very drastic specific regulation of rRNA gene activity in numerous eukaryotes is known as nucleolar dominance (Pikaard, 2002a, 2002b), a phenomenon observed upon the formation of genetic hybrids between related but different species when one set of parental rDNA is suppressed while the other is active. Nucleolar dominance in interspecific hybrids of Brassica and Arabidopsis can be overcome by treatment with TSA, a general inhibitor of histone deacetylation, or by the DNA methyltransferase inhibitor 5-azacytidine (Chen and Pikaard, 1997). Suppressed rDNA in allotetraploid hybrids between Arabidopsis and A. arenosa also is characterized by DNA and histone modifications characteristic for heterochromatin, and nucleolar dominance is released by the same inhibitors as in Brassica (Lawrence et al., 2004). rRNA silencing was further shown to depend on HDAC HDT1, a member of the plant-specific class II HDAC family with nucleolar localization. Interference with HDT1 expression by RNA interference technology caused expression of the otherwise suppressed Arabidopsis rRNA, an increase in histone H3K4 methylation, and loss of cytosine methylation at rDNA (Lawrence et al., 2004). By contrast, the changes of chromatin features at rDNA in the HDA6 mutants seem to occur without major changes in transcription rates. This suggests several layers of regulation: a general control of transcription potential via accessibility of the templates and a secondary control of actual transcription by polymerase loading or activity of the polymerase complex. The existence of additional rDNA loci in the allopolyploid hybrids might feed back on both regulatory systems, whereas transcriptional activity in an inbred diploid background is unaffected even if functional HDA6 is missing. This assumption is further supported by the observation that RNA interference downregulation of HDA6 in allopolyploid hybrids does indeed interfere with the selective uniparental transcription of rDNA repeats (R.L. Lawrence and C.S. Pikaard, personal communication).
The similar effects of chemical or genetic interference with DNA methyltransferase or HDAC in nucleolar dominance suggest that DNA methylation and hypoacetylation collaborate in gene silencing mechanisms at rDNA loci. TSA treatment also resulted in derepression of two silenced loci in N. crassa (Selker, 1998) and induced a specific reduction of DNA methylation at these two silenced loci without affecting overall methylation levels. This also indicates a reinforcing relationship between acetylation and DNA methylation, although an actual histone hyperacetylation at the affected loci was not demonstrated. It has been well established that DNA methylation can lead to the recruitment of HDACs (Feng and Zhang, 2001), but our data suggest that histone H4 hyperacetylation also can affect DNA methylation levels. We observed clear differences in CG methylation of rDNA genes between wild-type and sil1/axe1-5 mutants upon digestion with several methylation-sensitive enzymes. These differences were not observed previously in the sil1 or rts1 mutants (Furner et al., 1998; Aufsatz et al., 2002), possibly because they are most obvious with restriction enzymes CfoI and MaeII that were not used in earlier studies. The reductions in rDNA methylation levels in sil1/axe1-5 mutants, compared with wild-type plants, were much less than those observed in other DNA methylation mutants, such as ddm1 and hog1 (Furner et al., 1998; Jeddeloh et al., 1999). Furthermore, we observed a more significant effect on cytosines followed by G residues than on cytosines in other contexts. This is in accordance with the results of Aufsatz and coworkers (2002), who used bisulfite sequencing to measure cytosine methylation levels in the promoter region silenced by RNA-directed TGS. The highest reductions in methylation levels between mutant rts1 and wild-type plants were observed in CG sites, and a lesser effect was observed in CNG sites. Nonsymmetrical CNN sites showed no significant decrease in cytosine methylation in the mutants. These results led Aufsatz and coworkers to propose a model for HDA6 function, in which HDA6 plays a role in reinforcing CG methylation after primary and intermediate de novo C(N)G methylation by other components, thus helping to lock in the silent state of the target gene. Our results are consistent with this model, which might explain why the HDA6 mutations discovered to date show only moderate reactivation of silenced target genes. Because methylation of rDNA and centromeric repeats was not affected in rts1 mutants and the HDA6 gene had not been identified in other screens for DNA hypomethylation or TGS mutants, it was suggested that HDA6 might be specifically involved in RNA-directed pathways of gene silencing (Aufsatz et al., 2002). However, we have now shown that the sil1/axe1 mutants not only alleviate silencing of a well-characterized TGS locus and endogenous transcriptionally silenced repeats, but also affect acetylation of histones and maintenance of chromatin structure at rDNA loci. These observations indicate that HDA6 is not restricted to its role in an RNA-dependent epigenetic regulation, but acts with a certain level of specificity on other selective targets.
The recent identification of a nucleolar remodeling complex (NoRC) in mouse (Santoro et al., 2002) might allow connecting DNA methylation and histone acetylation activities at the rDNA locus on a biochemical basis. NoRC, consisting of the large nucleolar proteins Tip5 and SNF2, can induce nucleosomal movement on chromatin templates in vitro that depends not only on ATP, but also specifically on the presence of the histone H4 tail (Strohner et al., 2001). Tip5 was shown to interact in vitro with the DNA methyltransferases DNMT1 and DNMT3b as well as with the deacetylase HDAC1. Being recruited to acetylated histone H4 tails via the bromodomain of Tip5, the complex might establish a repressive state by means of histone and DNA modifications. Interestingly, the failure to deacetylate histones also abolished DNA methylation of transfected rRNA gene templates, therefore supporting our observation that histone deacetylation can be required to maintain wild-type DNA methylation levels. It remains to be seen whether a NoRC-like complex with HDA6 (and/or HDT1) as a component exists in plants, or whether some other mechanism controls the equilibrium between decondensed active rDNA and condensed inactive rDNA repeats.
Hypoacetylation of rDNA repeats and DNA methylation also can be reinforced by methylation of histone H3K9, another hallmark of heterochromatin and gene silencing in eukaryotes (Zhang and Reinberg, 2001; Lawrence et al., 2004). Interestingly, recent studies in fission yeast have revealed that a mutation in the clr3 HDAC impairs methylation of histone H3K9 (Nakayama et al., 2001). It will be interesting to investigate if HDA6 mutants in Arabidopsis affect H3K9met on particular heterochromatic targets or have additional effects on other histone modifications. In our immunostaining experiments, we could not observe a clear reduction of H3K9 methylation at chromocenters close to the nucleolus (data not shown).
In spite of the general correlation between hyperacetylation and active transcription, recent studies in barley (Hordeum vulgare) and Vicia faba indicated that histone acetylation correlates more with timing of replication during the S phase of the cell cycle than with transcriptional activity (Jasencakova et al., 2000, 2001). The question of whether a shift in replication timing because of nonremoval of acetyl groups in HDA6 mutants also occurs and under what conditions it is coupled with transcriptional activity remains for future analysis.
METHODS
Plant Material
Plants were grown in soil in a growth chamber under short-day conditions (12 h light, 21°C, 12 h dark, 16°C) or on MS medium (Ducheta Biochemie, Haarlem, The Netherlands) with or without appropriate selection.
Line L5 was obtained by transformation of wild-type Arabidopsis plants of the Columbia ecotype (Col-0) with a T-DNA composed of a GUS reporter gene driven by the 35S promoter of the Cauliflower mosaic virus and an NptII gene conferring resistance to kanamycin (Elmayan et al., 1998). GUS expression was detected in the hemizygous L5 primary transformant and in its homozygous progeny. Silencing took place between the second and third generation and was stably maintained to subsequent generations, whereas nonsilenced lines, such as the Hc1, continued to express the GUS transgene. Line L5 was previously cited as line 6b5 (Amedeo et al., 2000; Morel et al., 2000) and was renamed L5 to avoid confusion with a widely distributed posttranscriptionally silenced tobacco line named 6b5 (Elmayan and Vaucheret, 1996).
Transgene Expression Analysis
The sil1 and axe1 mutants were crossed to line L5, and the F1 progenies were allowed to self-fertilize. F2 plants homozygous for the TGS mutations and carrying the L5 insert were selected by staining with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid.
Map-Based Cloning of the SIL1 Gene
Mapping of the sil1 mutation was performed by crossing the sil1 mutant homozygous for the C insert in the Ler background with line L5 in the Col-0 background. The hygromycin-resistant F2 individuals that were homozygous for the sil1 mutation and carry the C insert were selected for mapping. The sil1 mutation was mapped at the bottom of chromosome 5. The insertion/deletion markers CER456709, CER456030, CER455379, and CER456203 were derived from the Cereon database (http://www.Arabidopsis.org/cereon/). The analysis of 2294 recombinant chromosomes allowed us to map the sil1 mutation between markers CER456030 and CER455379. Sequencing of the HDA6 gene was performed on PCR products and repeated several times to confirm the point mutation in the sil1 mutant.
RNA Gel Blot, Run-On, and DNA Gel Blot Analyses
Run-on and RNA gel blot analyses of the nonsilenced Hc1 control line and the silenced line L5 were performed as described (Mourrain et al., 2000). Genomic DNA of Hc1 and L5 was extracted by standard cetyl-trimethyl-ammonium bromide method, and their analysis by DNA gel blots was performed as described (Mourrain et al., 2000). RNA isolation and RNA gel blot analysis for TSI expression in sil1 and axe1-5 mutants and isolation of genomic DNA from sil1 and axe1 mutants for DNA gel blot analysis were performed as described (Mittelsten Scheid et al., 2002). The pA2 probe (Steimer et al., 2000) was used for TSI detection in RNA gel blot and DNA gel blots, an EcoRI fragment of the rRNA gene (Vongs et al., 1993) was used for hybridization to rDNA repeats, and the FWA promoter region (amplified with primers 5′-CAGCGTCTACCAAATCTACACT-3′ and 5′-TAGTGTCTCGACAACGAACAAG-3′) was used for the methylation analysis (Saze et al., 2003). The small GTP binding protein RAN was used as a loading control for RNA gel blots.
FISH
FISH was performed as described previously (Fransz et al., 1998; Probst et al., 2003). Young rosette leaves (1 to 1.5 cm) were fixed in 3:1 ethanol-acetic acid and stored at −20°C. After digestion with a combination of cellulase, pectolyase, and cytohelicase in citrate buffer, the suspension was stirred for 1 min at 45°C in 60% acetic acid, and the nuclei were then spread on glass slides and fixed in ethanol-acetic acid. After a postfixation in 2% paraformaldehyde in PBS, the slides were air-dried. Subsequently, the slides were baked at 60°C, treated with RNase (100 μg/mL in 2× SSC [1× SSC is 0.15 M NaCl and 0.015 M sodium citrate]) at 37°C for 1 h, then with pepsin (10 μg/mL in water, pH 2) at 37°C for 20 min. The pAL1 (180 bp) repeat was cloned into pBluescript KS+ vector, and labeled probes were generated by PCR with 0.1 mM dATP, dCTP, and dGTP, 0.065 mM dTTP, and 0.035 mM biotin-dUTP (Roche, Indianapolis, IN). rDNA probes were obtained with the biotin nick translation kit (Roche) using 18S- and 25S-rDNA–containing plasmids. Next, 1 μL of the PCR reaction or 3 μL of the nick translation mix were added to 20 μL of hybridization mix. After hybridization for ∼15 h in a wet chamber, slides were washed for 5 min in 2× SSC, 5 min in 0.1× SSC, 3 min in 2× SSC at 42°C, and 5 min in 2× SSC/0.1% Tween 20 at room temperature. The biotin-labeled probe was detected with Texas Red conjugated avidin (5 μg/mL, Vector Laboratories, Burlingame, CA), followed by a biotinylated goat-anti-avidin antibody (5 μg/mL; Vector Laboratories) and once more Texas Red avidin. DNA was counterstained with DAPI (2 μg/mL) in Vectashield mounting medium (Vector Laboratories). Images were analyzed with a Leitz DMR fluorescence microscope and documented with a SPOT RT camera (Diagnostic Instruments, Sterling Heights, MI). Images were merged and processed using Adobe Photoshop 7.0 (Mountain View, CA).
Immunostaining
Immunostaining experiments were performed as described (Probst et al., 2003). Protoplasts were isolated from young leaves of DR5, axe1-5, Ler, and sil1 plants by digestion with 1% cellulase and 0.25% macerozyme in Mes buffer (10 mM Mes, pH 5.7, 0.4 M mannitol, 30 mM CaCl2, 5 mM β-mercaptoethanol, and 0.1% BSA), washed in wash solution (4 mM Mes, pH 5.7, 2 mM KCl, and 0.5 M mannitol), attached to poly-Lys coated slides, fixed in 2% paraformaldehyde in Phem buffer (6 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 2 mM MgCl2, pH 6.9) for 10 min, permeabilized in 0.5% Nonidet P-40 in PHEM buffer, and postfixed in methanol:acetone 1:1 at −20°C. After rehydration in PBS, slides were blocked in 2% BSA in PBS (30 min, 37°C) and incubated with antibodies (Upstate, Charlottesville, VA) against tetra-acetylated H4 (dilution 1:100) or Lys4-dimethylated H3 (1:500) in blocking solution or 1% BSA in PBS (1 h, 37°C or overnight at 4°C). Detection was performed with an anti-rabbit fluorescein isothiocyanate–coupled antibody (1:100, 37°C, 45 min; Molecular Probes, Eugene, OR) in 0.5% BSA in PBS. DNA was counterstained with DAPI in Vectashield mounting medium.
For the combination of immunostaining and FISH, the slides were first processed as for immunostaining experiments. After incubation with the secondary antibody, the slides were dehydrated in an ethanol series (2 min in 70%, 2 min in 90%, and 2 min in 100%), air-dried, and baked at 60°C for 30 min. After an RNase treatment (100 μg/mL in 2× SSC) for 1 h at 37°C, the slides were washed in PBS, postfixed in 4% paraformaldehyde in PBS for 20 min at 4°C, washed again in PBS, dehydrated in an ethanol series, and air-dried. Hybridization, washing, and detection of the labeled probe were performed as for FISH on spread nuclei.
Images were analyzed with a Deltavision deconvolution microscope (Applied Precision, Issaquah, WA). The WoRx software (Applied Precision) was applied for deconvolution of the image stacks, and single layers were chosen for illustration.
Protein Gel Blot Analysis
Fresh leaf tissue (5 g) was ground in liquid nitrogen, transferred to extraction buffer (0.25 N HCl, 10 mM Tris-HCl, pH 7.5, 2 mM EDTA, 20 mM β-mercaptoethanol, and 0.2 mM phenylmethylsulfonyl fluoride [PMSF]) and treated with ultrasound. After centrifugation (10 min, 10,000 rpm), the supernatant was precipitated with TCA (25% final concentration). After centrifugation at 17000g for 20 min, the pellet was washed two times in acetone, dried, and resuspended in 1× SDS loading buffer (75 mM Tris-HCl, pH 6.8, 0.6% SDS, 15% glycerol, and 1.075 M β-mercaptoethanol). The proteins were separated on a 14% SDS page and blotted to a Hybond ECL membrane (Amersham, Buckinghamshire, UK). The membrane was blocked with 3% dry milk in protein gel blot basic buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20) and incubated overnight at 4°C with α-tetra-acetylated H4 or α-H3K4met (Upstate; 1:2000 in protein gel blot basic buffer supplemented with 1% BSA). After washing, the primary antibody was detected with secondary anti-rabbit horseradish peroxidase coupled antibody (1:7500; Amersham) at room temperature for 45 min. Visualization was achieved using the ECL system (Amersham).
ChIP
ChIP and PCR analysis was performed as described (Gendrel et al., 2002; Johnson et al., 2002) with minor modifications. In brief, leaves of 3-week-old plants of DR5 and axe1-5 mutant plants grown in soil were vacuum-infiltrated with 1% formaldehyde in buffer 1 (0.4 M sucrose and 10 mM Tris-HCl, pH 8), ground to powder in liquid nitrogen, resuspended in buffer 1 supplemented with 5 mM β-mercaptoethanol, 1 mM PMSF, and protein inhibitors, filtered, and centrifuged. The pellet was dissolved in buffer 2 (0.25 M sucrose, 10 mM Tris-HCl, pH 8, 10 mM MgCl2, 1% Triton X-100, and 5 mM β-mercaptoethanol), centrifuged again, resuspended in buffer 3 (1.7 M sucrose, 10 mM Tris-HCl, pH 8, 2 mM MgCl2, 0.15% Triton X-100, 5 mM β-mercaptoethanol, and 1 mM PMSF), and layered on top of an equal amount of buffer 3. The pellet was finally resuspended in nuclei lysis buffer (50 mM Tris-HCl, pH 8, 10 mM EDTA, and 1% SDS), diluted with ChIP dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8, and 167 mM NaCl), and sonicated. The sheared chromatin was precleared with salmon sperm DNA/protein-A agarose (Upstate), and the histone-DNA complexes were immunoprecipitated with α-H4ac and α-H3K4met antibodies (Upstate). After a wash in wash buffer 1 (20 mM Tris-HCl, pH 8, 150 mM NaCl, 1% Triton X-100, and 2 mM EDTA), wash buffer 2 (20 mM Tris-HCl, pH 8, 500 mM NaCl, 1% Triton X-100, and 2 mM EDTA), wash buffer 3 (10 mM Tris-HCl, pH 8, 250 mM LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, and 1 mM EDTA), and two washes in Tris-EDTA (TE) buffer, the chromatin was eluted with 0.1 M NaHCO3 and 1% SDS. Cross-linking was reversed overnight at 65°C in the presence of 0.2 M NaCl, and samples were treated with proteinase K for 3 h. After phenol-chloroform extraction, the DNA was resuspended in TE supplemented with RNase A to 10 μg/mL. The immunoprecipitated DNA was analyzed by PCR as described (Johnson et al., 2002). Amplification of the Actin2/7 gene (Tariq et al., 2003) was performed for 40 cycles, the rDNA repeats for 25 cycles using primers 5′-GATTCCCTTAGTAACGGCG-3′ and 5′-CGGTACTTGTTCGCTATCGG-3′.
S1 Nuclease Protection
Ten micrograms of total RNA was hybridized to oligonucleotide probes corresponding to the non-RNA (antisense) strands of Arabidopsis genes. Probes were 5′ end–labeled with T4 polynucleotide kinase (New England Biolabs, Beverly, MA) and γ-32P-ATP. The rRNA probe, which spans the transcription start site, was 5′-GGGTTCCCCACGGACTGCCCAGACTCCCTCAACACCCACCCCCCTATATAGCTGCC-3′; the ubiquitin probe, matching internal UBQ10 transcript sequences, was 5′-GGAACGGAAACATAGTAGAACACTTATTCATCAGGGATTATACAAGGCCCCCCGG-3′; and the actin probe, matching internal ACT2 sequences, was 5′-GCTCGTTGTAGAAAGTGTGATGCCATATCTTTTCCATGTCATGGGCCCCCC-3′. Three to six nucleotides at the 3′ termini of the oligonucleotides probes were purposely designed to be noncomplementary to the target RNAs so that undigested probe could be discriminated from S1 digested products. Hybridization, nuclease treatment, and anaylsis of products was done essentially as described by Lawrence et al. (2004).
Acknowledgments
We thank J. Lichota for excellent help with the protein gel blot analysis, Z. Jasencakova for technical advice with the immunostaining-FISH protocol, K. Afsar for technical assistance, and D. Schübeler, M. Tariq, and J. Paszkowski for critical reading of the manuscript. J.M. was supported by the National Science Foundation Grant IBN 0090996. Work of K.E., R.J.L., and C.S.P. was supported by National Institutes of Health Grant R01-GM60380. O.M.S. was supported by the Swiss Federal Office for Education and Science Grant 00.0224-2 and by the European Communion Grant QLRT-2000-00078.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Aline V. Probst (aline.probst@bioveg.unige.ch).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.018754.
References
- Ahringer, J. (2000). NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 16, 351–356. [DOI] [PubMed] [Google Scholar]
- Allfrey, V.G., Faulkner, R.M., and Mirsky, A.E. (1964). Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA 51, 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amedeo, P., Habu, Y., Afsar, K., Mittelsten Scheid, O., and Paszkowski, J. (2000). Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature 405, 203–206. [DOI] [PubMed] [Google Scholar]
- Aufsatz, W., Mette, M.F., Van Der Winden, J., Matzke, M., and Matzke, A.J. (2002). HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 21, 6832–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, S.L. (2002). Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12, 142–148. [DOI] [PubMed] [Google Scholar]
- Bird, A.P., and Wolffe, A.P. (1999). Methylation-induced repression—Belts, braces, and chromatin. Cell 99, 451–454. [DOI] [PubMed] [Google Scholar]
- Chen, Z.J., and Pikaard, C.S. (1997). Epigenetic silencing of RNA polymerase I transcription: A role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 11, 2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver, G.P., and Pikaard, C.S. (1996). RFLP and physical mapping with an rDNA-specific endonuclease reveals that nucleolus organizer regions of Arabidopsis thaliana adjoin the telomeres on chromosomes 2 and 4. Plant J. 9, 259–272. [DOI] [PubMed] [Google Scholar]
- Doetzlhofer, A., Rotheneder, H., Lagger, G., Koranda, M., Kurtev, V., Brosch, G., Wintersberger, E., and Seiser, C. (1999). Histone deacetylase 1 can repress transcription by binding to Sp1. Mol. Cell. Biol. 19, 5504–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan, T., Balzergue, S., Beon, F., Bourdon, V., Daubremet, J., Guenet, Y., Mourrain, P., Palauqui, J.C., Vernhettes, S., Vialle, T., Wostrikoff, K., and Vaucheret, H. (1998). Arabidopsis mutants impaired in cosuppression. Plant Cell 10, 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan, T., and Vaucheret, H. (1996). Expression of single copies of a strongly expressed 35S transgene can be silenced post-transcriptionally. Plant J. 9, 787–797. [Google Scholar]
- Feng, Q., and Zhang, Y. (2001). The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 15, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle, W., Wang, Y., and Allis, C.D. (2003). Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15, 172–183. [DOI] [PubMed] [Google Scholar]
- Fransz, P., Armstrong, S., Alonso-Blanco, C., Fischer, T.C., Torres-Ruiz, R.A., and Jones, G. (1998). Cytogenetics for the model system Arabidopsis thaliana. Plant J. 13, 867–876. [DOI] [PubMed] [Google Scholar]
- French, S.L., Osheim, Y.N., Cioci, F., Nomura, M., and Beyer, A.L. (2003). In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 23, 1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks, F., Burgers, W.A., Brehm, A., Hughes-Davies, L., and Kouzarides, T. (2000). DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet 24, 88–91. [DOI] [PubMed] [Google Scholar]
- Furner, I.J., Sheikh, M.A., and Collett, C.E. (1998). Gene silencing and homology-dependent gene silencing in Arabidopsis: Genetic modifiers and DNA methylation. Genetics 149, 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel, A.V., Lippman, Z., Yordan, C., Colot, V., and Martienssen, R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297, 1871–1873. [DOI] [PubMed] [Google Scholar]
- Grant, P.A., Sterner, D.E., Duggan, L.J., Workman, J.L., and Berger, S.L. (1998). The SAGA unfolds: Convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 8, 193–197. [DOI] [PubMed] [Google Scholar]
- Grewal, S.I., Bonaduce, M.J., and Klar, A.J. (1998). Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150, 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt, I., and Pikaard, C.S. (2003). Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 4, 641–649. [DOI] [PubMed] [Google Scholar]
- Haizel, T., Merkle, T., Pay, A., Fejes, E., and Nagy, F. (1997). Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant J. 11, 93–103. [DOI] [PubMed] [Google Scholar]
- Jasencakova, Z., Meister, A., and Schubert, I. (2001). Chromatin organization and its relation to replication and histone acetylation during the cell cycle in barley. Chromosoma 110, 83–92. [DOI] [PubMed] [Google Scholar]
- Jasencakova, Z., Meister, A., Walter, J., Turner, B.M., and Schubert, I. (2000). Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell 12, 2087–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh, J.A., Stokes, T.L., and Richards, E.J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet 22, 94–97. [DOI] [PubMed] [Google Scholar]
- Jenuwein, T., and Allis, C.D. (2001). Translating the histone code. Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Johnson, L., Cao, X., and Jacobsen, S. (2002). Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12, 1360–1367. [DOI] [PubMed] [Google Scholar]
- Kurdistani, S.K., and Grunstein, M. (2003). Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 4, 276–284. [DOI] [PubMed] [Google Scholar]
- Kurdistani, S.K., Robyr, D., Tavazoie, S., and Grunstein, M. (2002). Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet 31, 248–254. [DOI] [PubMed] [Google Scholar]
- Lagger, G., O'Carroll, D., Rembold, M., Khier, H., Tischler, J., Weitzer, G., Schuettengruber, B., Hauser, C., Brunmeir, R., Jenuwein, T., and Seiser, C. (2002). Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 21, 2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, R.J., Earley, K., Pontes, O., Silva, M., Chen, J.Z., Neves, N., Viegas, W., and Pikaard, C.S. (2004). A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 13, 599–609. [DOI] [PubMed] [Google Scholar]
- Luger, K., Mader, A.W., Richmond, R.K., Sargent, D.F., and Richmond, T.J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Lusser, A., Brosch, G., Loidl, A., Haas, H., and Loidl, P. (1997). Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 277, 88–91. [DOI] [PubMed] [Google Scholar]
- Maluszynska, J., and Heslop-Harrison, J.S. (1991). Localization of tandemly repeated DNA sequences in Arabidopsis thaliana. Plant J. 1, 159–166. [Google Scholar]
- McKnight, S.L., and Miller, O.L., Jr. (1976). Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell 8, 305–319. [DOI] [PubMed] [Google Scholar]
- Mittelsten Scheid, O., Probst, A.V., Afsar, K., and Paszkowski, J. (2002). Two regulatory levels of transcriptional gene silencing in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 13659–13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, J.B., Mourrain, P., Beclin, C., and Vaucheret, H. (2000). DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol. 10, 1591–1594. [DOI] [PubMed] [Google Scholar]
- Morgan, G.T., Reeder, R.H., and Bakken, A.H. (1983). Transcription in cloned spacers of Xenopus laevis ribosomal DNA. Proc. Natl. Acad. Sci. USA 80, 6490–6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain, P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Murfett, J., Wang, X.J., Hagen, G., and Guilfoyle, T.J. (2001). Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell 13, 1047–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, J., Rice, J.C., Strahl, B.D., Allis, C.D., and Grewal, S.I. (2001). Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113. [DOI] [PubMed] [Google Scholar]
- Nishioka, K., Chuikov, S., Sarma, K., Erdjument-Bromage, H., Allis, C.D., Tempst, P., and Reinberg, D. (2002). Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 16, 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, R., Muller, A., Napoli, C.A., Selinger, D.A., Pikaard, C.S., Richards, E.J., Bender, J., Mount, D.W., and Jorgensen, R.A. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30, 5036–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, A.H., Mermoud, J.E., O'Carroll, D., Pagani, M., Schweizer, D., Brockdorff, N., and Jenuwein, T. (2002). Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 30, 77–80. [DOI] [PubMed] [Google Scholar]
- Pikaard, C.S. (2002. a). Nucleolar dominance: Uniparental gene silencing on a multi-megabase scale in genetic hybrids. Plant Mol. Biol. 43, 163–177. [DOI] [PubMed] [Google Scholar]
- Pikaard, C.S. (2002. b). The epigenetics of nucleolar dominance. Trends Genet. 16, 495–500. [DOI] [PubMed] [Google Scholar]
- Pontes, O., Lawrence, R.J., Neves, N., Silva, M., Lee, J.H., Chen, Z.J., Viegas, W., and Pikaard, C.S. (2003). Natural variation in nucleolar dominance reveals the relationship between nucleolus organizer chromatin topology and rRNA gene transcription in Arabidopsis. Proc. Natl. Acad. Sci. USA 100, 11418–11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst, A.V., Fransz, P.F., Paszkowski, J., and Mittelsten Scheid, O. (2003). Two means of transcriptional reactivation within heterochromatin. Plant J. 33, 743–749. [DOI] [PubMed] [Google Scholar]
- Robyr, D., Suka, Y., Xenarios, I., Kurdistani, S.K., Wang, A., Suka, N., and Grunstein, M. (2002). Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109, 437–446. [DOI] [PubMed] [Google Scholar]
- Rundlett, S.E., Carmen, A.A., Kobayashi, R., Bavykin, S., Turner, B.M., and Grunstein, M. (1996). HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93, 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett, S.E., Carmen, A.A., Suka, N., Turner, B.M., and Grunstein, M. (1998). Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392, 831–835. [DOI] [PubMed] [Google Scholar]
- Sandmeier, J.J., French, S., Osheim, Y., Cheung, W.L., Gallo, C.M., Beyer, A.L., and Smith, J.S. (2002). RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 21, 4959–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro, R., Li, J., and Grummt, I. (2002). The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 32, 393–396. [DOI] [PubMed] [Google Scholar]
- Saze, H., Mittelsten Scheid, O., and Paszkowski, J. (2003). Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 34, 65–69. [DOI] [PubMed] [Google Scholar]
- Selker, E.U. (1998). Trichostatin A causes selective loss of DNA methylation in Neurospora. Proc. Natl. Acad. Sci. USA 95, 9430–9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe, W.J., Jasencakova, Z., Houben, A., Kakutani, T., Meister, A., Huang, M.S., Jacobsen, S.E., Schubert, I., and Fransz, P.F. (2002). DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21, 6549–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimer, A., Amedeo, P., Afsar, K., Fransz, P., Scheid, O.M., and Paszkowski, J. (2000). Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell 12, 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner, D.E., and Berger, S.L. (2000). Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl, B.D., and Allis, C.D. (2000). The language of covalent histone modifications. Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Strohner, R., Nemeth, A., Jansa, P., Hofmann-Rohrer, U., Santoro, R., Langst, G., and Grummt, I. (2001). NoRC—A novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 20, 4892–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq, M., Saze, H., Probst, A.V., Lichota, J., Habu, Y., and Paszkowski, J. (2003). Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. USA 100, 8823–8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton, J., Hassig, C.A., and Schreiber, S.L. (1996). A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408–411. [DOI] [PubMed] [Google Scholar]
- Tian, L., and Chen, Z.J. (2001). Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA 98, 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer, M., Wu, J., Suka, N., and Grunstein, M. (2000). Global histone acetylation and deacetylation in yeast. Nature 408, 495–498. [DOI] [PubMed] [Google Scholar]
- Vongs, A., Kakutani, T., Martienssen, R.A., and Richards, E.J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260, 1926–1928. [DOI] [PubMed] [Google Scholar]
- Wu, K., Malik, K., Tian, L., Brown, D., and Miki, B. (2000). Functional analysis of a RPD3 histone deacetylase homologue in Arabidopsis thaliana. Plant Mol. Biol. 44, 167–176. [DOI] [PubMed] [Google Scholar]
- Yang, X.J., and Seto, E. (2003). Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr. Opin. Genet. Dev. 13, 143–153. [DOI] [PubMed] [Google Scholar]
- You, A., Tong, J.K., Grozinger, C.M., and Schreiber, S.L. (2001). CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc. Natl. Acad. Sci. USA 98, 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., and Reinberg, D. (2001). Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes Dev. 15, 2343–2360. [DOI] [PubMed] [Google Scholar]