Abstract
Autolysis of lactic acid bacteria (LAB) plays a vital role in dairy processing. During cheese making, autolysis of LAB affects cheese flavor development through release of intracellular enzymes and restricts the proliferation of cells in yogurt fermentation and probiotics production. In order to explore the mechanism of autolysis, the gene for the autolytic enzymes of L. bulgaricus, N-acetylmuramidase (mur), was cloned and sequenced (GenBank accession number: KF157911). Mur gene overexpression and gene knockout vectors were constructed based on pMG76e and pUC19 vectors. Recombinant plasmids were transformed into L. bulgaricus ljj-6 by electroporation, then three engineered strains with pMG76e-mur vector and fifteen engineered strains with pUC19-mur::EryBII were screened. The autolysis of the mur knockout strain was significantly lower and autolysis of the mur overexpressed strain was significantly higher compared with that of the wild type strain ljj-6. This result suggested that the mur gene played an important role in autolysis of L. bulgaricus. On the other hand, autolytic activity in a low degree was still observed in the mur knockout strain, which implied that other enzymes but autolysin encoded by mur were also involved in autolysis of L. bulgaricus.
Introduction
Lactic acid bacteria are used as starter cultures for dairy fermentation and therefore their lysis is of special interest. In cheese making, ripening is an extremely important process since it defines the flavor and texture of the cheese, which differentiates the many varieties. The process is very slow, generally between three weeks and two (or more) years. This makes cheese making an expensive business. Several technological methods have been used to accelerate ripening, such as increasing the ripening temperature, the use of modified starter cultures, and the addition of exogenous enzymes [1], [2]. Lysis of the starter strains during ripening results in the release of cytoplasmic peptidases, lipases, and other enzymes involved in amino acid catabolism, into the cheese curd [3], [4]. It is thought that these enzymes accelerate peptidolysis and remove bitter-tasting peptides. Thus, increasing starter LAB autolysis is considered essential to accelerate cheese ripening [5]. In yogurt making, autolysis of lactic acid bacteria can lead to low number of live strains in starter cultures, such that reducing starter LAB autolysis can result in more efficient yogurt starter cultures. Therefore, understanding the mechanism of LAB autolysis has important significance in dairy industry.
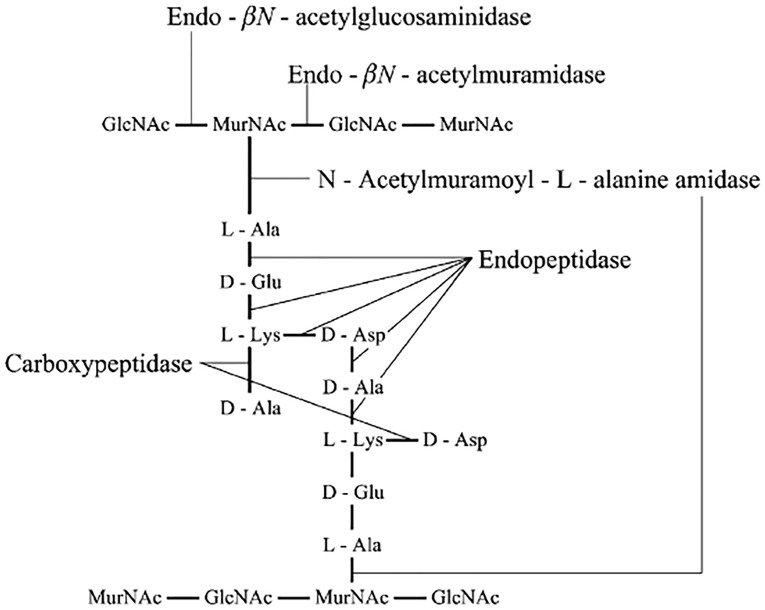
Generally, autolysis is mainly caused by lactic acid bacteria autolysins which are defined as endogenous enzymes that hydrolyze covalent bonds in the peptidoglycan of the cell wall. Five types of enzymes with lytic activity against peptidoglycan have been described for Gram-positive bacteria [6]. Figure 1 is a diagram of the peptidoglycan structure found in L. lactis with the five different types of peptidoglycan degrading activity indicated [2]. Mercier [7] cloned and sequenced the gene for N-acetylmuramidase and showed by zymogram analysis that it was the only autolysin expressed in the plasmid-free strain L. lactis subsp. cremoris MG1363. The N-acetylmuramidase open reading frame in MG1363 encodes a protein with a predicted molecular mass of 46,564 Da, which after cleavage of an N-terminal leader sequence would result in a mature N-acetylmuramidase with a predicted molecular mass of 40,264 Da.
Figure 1. A general representation of the lactococcal peptidoglycan structure showing the specificities of the different peptidoglycan hydrolases [2].
The aim of the present study was to clone and sequence the N-acetylmuramidase gene from the L. bulgaricus Ljj-6, exploring the relationship between N-acetylmuramidase gene and L. bulgaricus autolysis, and laying the foundation for the directional control of lactic acid bacteria autolysis in future studies.
Materials and Methods
Bacterial cultures, media and growth conditions
The parental strains and plasmids that were used in this study are listed in Table 1. Escherichia coli cells were grown in Luria-Bertani broth (LB) medium with aeration at 37°C. L. bulgaricus were routinely grown at 37°C in Man-Rogosa-Sharpe (MRS) medium (Beijing Land Bridge Technology Co., Ltd. CM187) under static conditions. MRS was autoclaved for 15 min at 121°C. The strain morphology was observed by light microscopic Olympus CX41.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant genotype or description | Reference and/or source |
| Strains | ||
| E. coli DH5α | F−, φ80d lacZ △M15, △(lacZYA-argF) U169, deoR, recA1, endA1, hsdR17 (rk−, mk+ ), phoA, supE44, λ−, thi-1, gyrA96, relA1 | TaKaRa |
| L. bulgaricus ljj-6 | Wild-type L. bulgaricus isolated from Yogurt samples; low rate of autolysis | this study |
| L. bulgaricus 11842 | Wild-type L. bulgaricus with normalrate of autolysis | ATCC 11842 |
| L. bulgaricus k12 | mur knockout mutant of L. bulgaricusljj-6; mur::EryBII | this study |
| L. bulgaricus O3 | mur overexpression mutant of L. bulgaricus ljj-6; ljj-6 introductionof a functional mur gene on expression vector pMG76e; | this study |
| Plasmids | ||
| pMD18T | clone vector; Ampr | TaKaRa |
| pMG76e | pMG36e derived integration vector; Emr | College of food science and Nutritional Engineering, China Agricultural University |
| pMD18T-mur | pMD18T derived integration vector containing the murljj-6 gene with salI, sphI restriction enzyme sites; Ampr | this study |
| pMG76e-mur | pMG76e derived expression vector containing the murljj-6 gene; Emr | this study |
| pUC19 | clone vector; Ampr | TaKaRa |
| pUC19-mur | pUC18 derived integration vector containing the murljj-6 gene with BstEII, HinCII restriction enzyme sites; Ampr | this study |
| pUC19-mur::EryBII | pUC18-mur; Emr; Ampr | this study |
DNA manipulations
Routine molecular biology techniques were performed according to standard procedures. Restriction and modifying enzymes (New England Biolabs) were used as recommended by the manufacturer. Plasmid DNA was prepared from E. coli and L. bulgaricus cells by use of BIOMIGA Plasmid Miniprep kits (PD1211-01). Chromosomal DNA was isolated from L. bulgaricus cells by use of BIOMIGA Bacteriall gDNA kits (GD2411-01).
Amplification of the mur gene
DNA was extracted from L. bulgaricus ljj-6 using the sodium dodecyl sulfate/proteinase K/cetyltrimethylammonium bromide (CTAB) method [8] with minor modifications. The mur gene was amplified from the DNA template by PCR with ExTaq DNA polymerase (Takara) and primers containing an salI site and an sphI site (Primer 3,4 in Table 2). The PCR reaction mixtures contained 250 ng of genomic DNA as template, 100 pmol of each primer, 5 U of ExTaq DNA polymerase, 12.5 mM MgCl2, and 200 mM each dNTP in deionized water to a final volume of 50 mL. The reaction was performed using the PCR thermocycler (Hangzhou Bioer Technology Company, China) with a program consisting of 1 cycle at 95°C for 4 min, then 35 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 2 min, and finally 1 cycle of 72°C for 8 min. Products were quantified by comparing the intensity of the product DNA band to that of a 2000 bp marker standard in 1% agarose. Each DNA fragment was ligated into the pGEM-T Easy vector (Promega) according to the manufacturer’s instructions at 16°C overnight. The recombinant plasmid was then transformed into Escherichia coli DH5α competent cells. Positive clones of transformed cells were selected and sequenced at Shanghai Invitrogen Company, China.
Table 2. Primers used in this study.
| No. | Primer | Sequence | Reference |
| 1 | Lb-murF1 | 5′-ATGGCTGGACACAGAAA-3′ | this study |
| 2 | Lb-murR1 | 5′-TTAATTATCGTACTTATTCAGGT-3′ | this study |
| 3 | Mur-SalI-F1 | 5′-ACGCGCATGCATGGCTGGACACAGAAA-3′ | this study |
| 4 | Mur-sphI-R1 | 5′-ACGCGCATGCTTAATTATCGTACTTATTCAGGT-3′ | this study |
| 5 | Mur-sphI-F1 | 5′-ACGCGCATGCATGGCTGGACACAGAAA-3′ | this study |
| 6 | Mur-EcorI-R2 | 5′-ACATGAATTCTTAATTATCGTACTTATTCAGGT-3′ | this study |
| 7 | EryB-HincII | 5′-CGGTCAACATGACCACCGACGCCGCGACG-3′ | this study |
| 8 | EryB-BstEII | 5′-CGGGTAACCTCACTGCAACCAGGCTTCCGG-3′ | this study |
Construction of recombinant plasmids
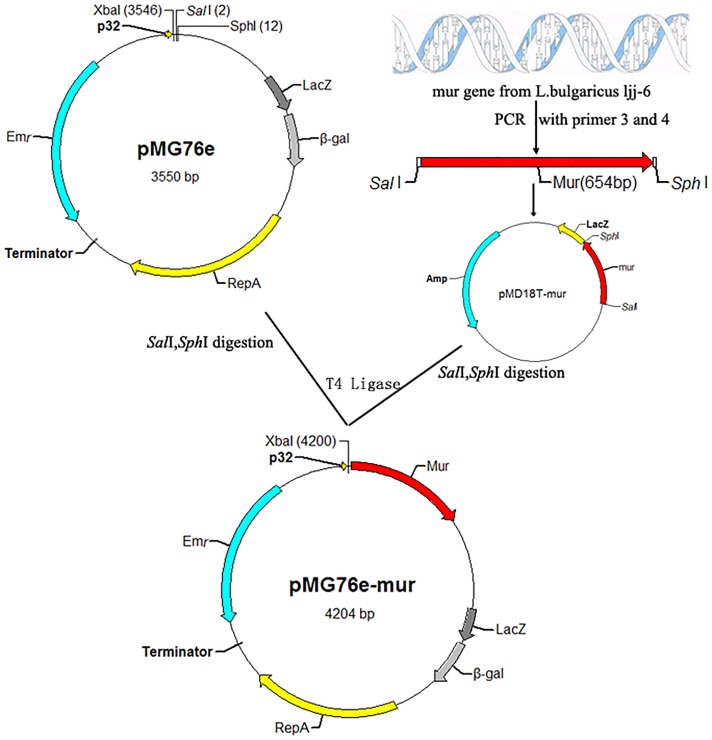
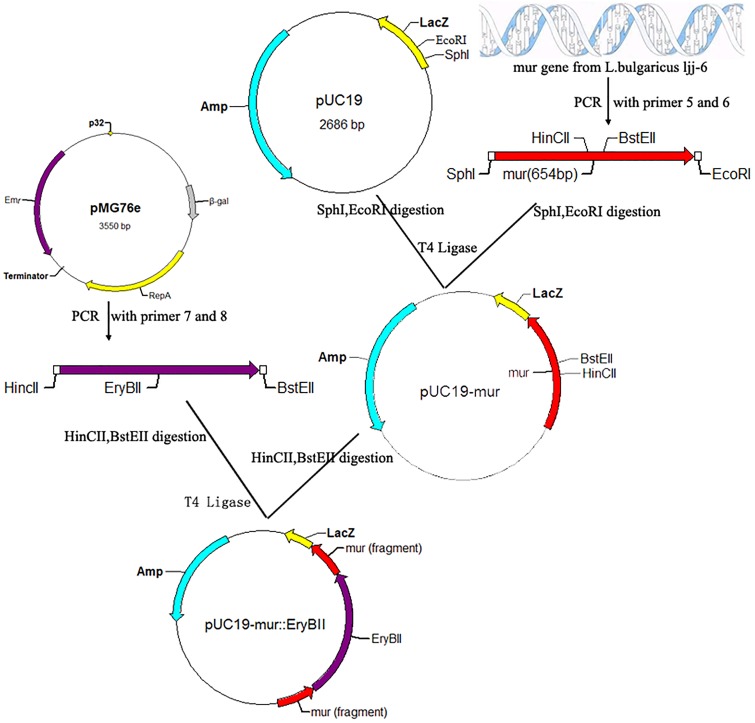
The mur gene was subcloned into pMD18-T in E. coli DH5α to generate the recombinant plasmids pMD18T-mur. Plasmid pMD18T-mur and pMG76e were extracted from E. coli DH5α, respectively. The recombinant plasmid pMD18T-mur and the vector pMG76e were digested with salI and sphI, and purified with the QIAprep Spin Miniprep kit. Ligase was used to construct the recombinant of pMG76e-mur (Figure 2). The recombinant shuttle vector was transformed into E. coli DH5α by the heat-shock method and purified with the same kits. Erythromycin was maintained in the growth media at 200 ug/mL to select for positive transformants. Construction of recombinant plasmids pUC19-mur::EryBII was similar to the above method (Figure 3) [9].
Figure 2. Construction of recombinant plasmids pMG76e-mur.
Figure 3. Construction of recombinant plasmids pUC19-mur::EryBII.
Transformation of L. bulgaricus
L. bulgaricus was transformed with pMG76e-mur and pUC19-mur::EryBII by electroporation according to the protocol described previously by Holo and Nes [10] and Kim et al. [11] with minor modifications. Briefly, L. bulgaricus was cultured in MRS medium until reaching an OD600 of 0.6–0.1. The cell pellet was then washed in deionized water and resuspended in 1/200 volume of 0.5 M sucrose containing 10% glycerol. Competent cells were added to the ligation mixture, and the mixture was treated using the Gene Pulser Apparatus (BioRad, Richmond, CA, USA) according to the manufacturer’s instructions. The electroporated mixture was immediately diluted with 1 mL of MRS broth and incubated for 2 h at 36°C and then plated on solid MRS media containing erythromycin (200 ug/mL) and incubated for 48 h at 36°C.
Autolysis detection of lactic acid bacteria
Detection of cell autolysis rate was done using the PI-FCM method: the cultured cells in liquid were collected after centrifugation (5000×g, 5 min), followed by washing two times with PBS buffer and then resuspended. The bacterial suspension absorption value (OD650) was adjusted to about 1 to control the number of cells in the bacterial suspension to about 1×107 cells/mL. Then 1 mL bacteria liquid was centrifuged at 4°C (3000×g, 5 min) after which the supernatant was discarded. The cells were resuspended in 1 mL PI-PBS solution (20 mmol/L). Cell suspensions were dyed by PI-PBS in a dark environment for 30 min at 4°C. A 630 nm long pass filter was used to collect the red fluorescence (FL3488 nm) and 1×105 cells were collected from each sample. Test time ranged from 40 to 100 s, depending on the concentrations of the cell suspensions. Data were analyzed by CellQuest online analysis system. FCM enumerations were accurate to 104 cells/mL. The rate of autolysis was defined as the ratio of the positive cell number with PI dying to the total cell number. Comparisons between groups were tested by One - Way ANOVA analysis and LSD test.
Results
Amplification of the full-length sequence of mur gene
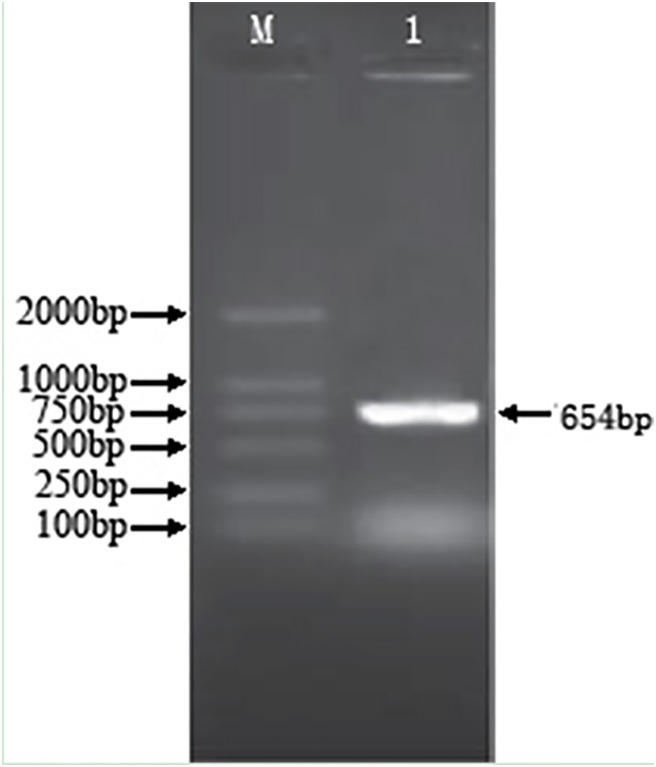
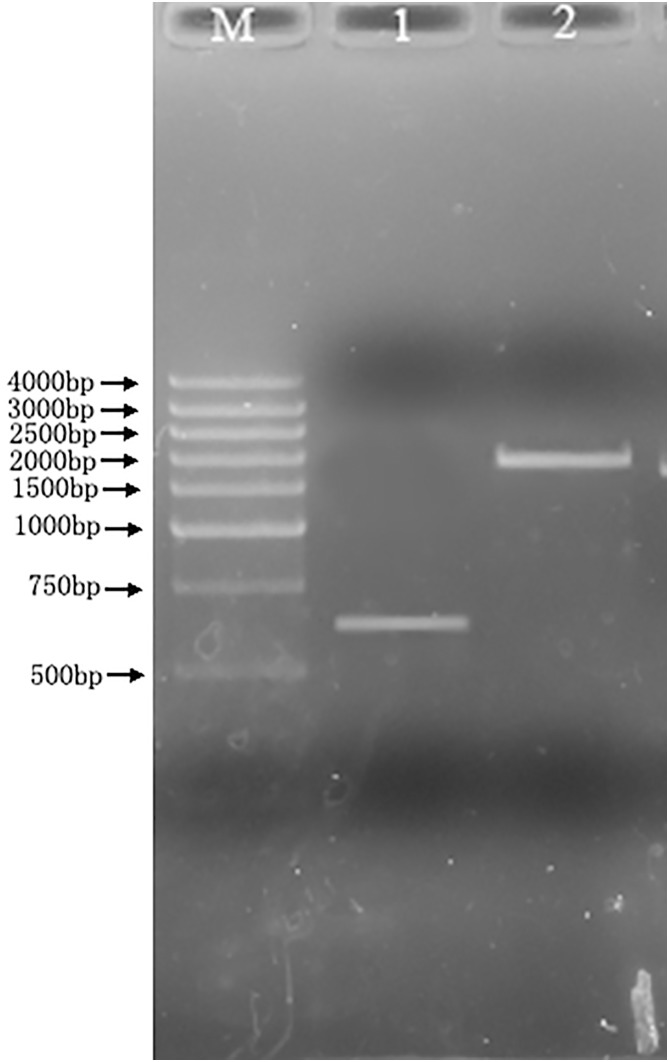
The PCR amplification was performed with the genome DNA plate from L. bulgaricus ljj-6, and the amplified product was detected by 1% agarose gel electrophoresis. The results showed that a 650 bp gene fragment was obtained (Figure 4), which was consistent with other lactic acid bacteria. The sequencing results revealed four mutations in mur gene between ljj-6 and ATCC11842 (Figure 5). However, the amino acid sequences of these two kinds of strain were consistent, which indicated that the mur gene of ljj-6 was cloned. The sequencing results were submitted to GenBank (accession number KF157911).
Figure 4. Amplification of the full-length sequence of mur gene.
1: The cloned 654 bp fragment from ljj-6 genome with primer 1,2; M: DNA marker DL2000.
Figure 5. Mur gene fragment cloned from L. bulgaricus ljj-6 BLAST with L. bulgaricus ATCC 11842 complete genome.
(Identities: 650/654).
Construction of recombinant plasmids
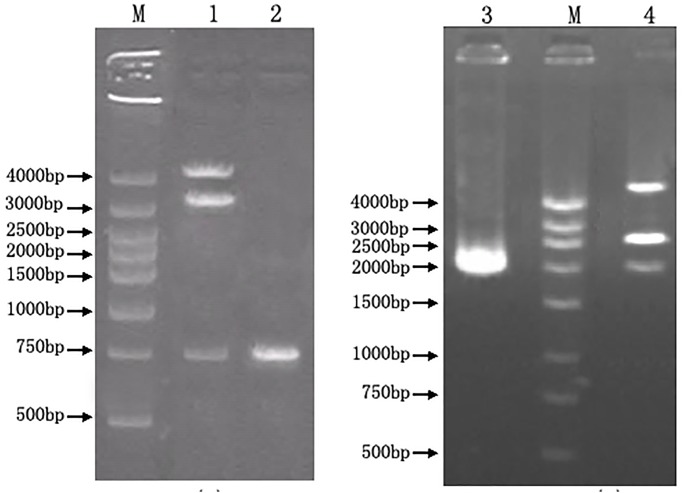
Sequence of mur gene approximately 650 bp in size was amplified. SalI and SphI restriction sites were introduced to the mur gene to facilitate its insertion into the multiple cloning site of pMG76e. The recombinant plasmid was amplified and isolated from E. coli DH5α transformants and digested with SalI and SphI. Analysis of the digestion mixtures by 1.0% agarose gel electrophoresis revealed that the cells contained both the 4.2 kb pMG76e-mur fragment and the 654 kb mur fragment, indicating that the mur gene had been successfully incorporated into pMG76e, in addition PCR reaction (Primer 1, 2 in Table 2) also verified this conclusion (Figure 6A).
Figure 6. Identification of recombinant plasmids by digestion and PCR.
A: pMG76e-mur; B:pUC19-mur::EryBII. 1: pMG76e-mur was digested by SalI and sphI to 4.2 kb, 3.5 kb, 0.65 kb fragments; 2: Identification of mur gene from pMG76e-mur by PCR with primer 1,2; M: DNA marker DGL4000; 3: Identification of mur gene from pUC19-mur::EryBII by PCR with primer 1,2; 4: pUC19-mur::EryBII was digested by EcorI and sphI to 4.7 kb, 2.7 kb, 2.0 kb fragments.
The identification of recombinant plasmid pUC19-mur::EryB was performed using the same method as in pMG76e-mur. Analysis of the digestion revealed that the cells contained both the 4.7 kb pUC19-mur::EryBII and the 2.0 kb mur::EryBII fragment, indicating that the mur::EryBII gene had been successfully incorporated into pUC19. Additionally, PCR reaction with primers 1 and 2 also verified this conclusion (Figure 6B).
Transformation of L. bulgaricus
The recombinant plasmids pMG76e-mur and pUC19-mur::EryBII were transformed into L. bulgaricus ljj-6 by electroporation. The electroporated cells were incubated for 2 h at 37°C and then plated on solid MRS containing 0.5 M sucrose and erythromycin (200 µg/mL). Colonies of positive transformants were visible after 48 h of incubation at 37°C. Three engineered strains (L. bulgaricusO1-O3) with pMG76e-mur vector and fifteen candidate engineered strains (L. bulgaricus k1-k15) with pUC19-mur::EryBII were screened. The fifteen candidate colonies (k1-k15) were then transferred onto MRS agar plate containing ampicillin (50 µg/ml). Possible double-crossover mutants had lost their capability to grow in the presence of ampicillin but were still resistant to erythromycin. Finally six double-crossover mutants (k7, k8, k10, k11, k12, k13) were chosen by the second round screening.
Identification of mur gene knockout L. bulgaricus
The genome DNA was extracted from the the mur knockout strain k12 which has been cultured in MRS broth for 24 hours. The PCR amplification for the mutant genome DNA and wildtype genome DNA was performed using the Primer 3 and 4 (Figure 7). The results indicated that 650 bp and 2000 bp amplified products were obtained from wildtype and mutant genome DNA respectively. The nearly 1.4 kb difference existed between two products, which was consistent with the length of inserted erythromycin resistance gene fragment (1378 bp). This implied that the mur gene knockout strain was constructed successfully through insertion of erythromycin resistance gene.
Figure 7. Identification of mur gene knockout Lactobacillus delbrueckii subsp. bulgaricus.
M: DNA marker DGL4000; Lane 1, Lactobacillus delbrueckii subsp. bulgaricus; Lane 2, mur gene knockout Lactobacillus delbrueckii subsp. bulgaricus.
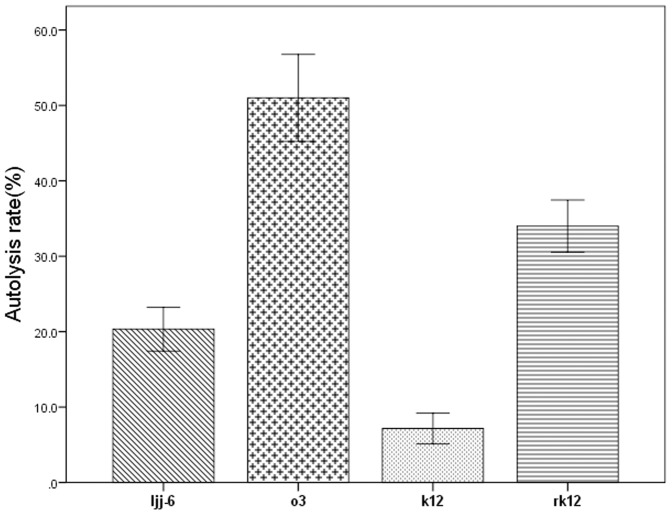
The complementation experiment was carried out to exclude downstream effects of the introduced knock-out gene on other genes. To generate the mur reconstituted strain, pMG76e-mur was introduced into the mur deletion mutant k12 by electroporation. The mur-reconstituted strain was designated rk12. The autolysis of rk12 was detected and the result (Figure 8) showed that autolysis of strain rk12 was significantly higher compared with that of the k12 and ljj-6, this indicated that mur reconstituted strain restored its autolytic ability.
Figure 8. The result of autolysis detection on four strains.
Morphology observation of different autolysis lactic acid bacteria
Autolysis differences of lactic acid bacteria can result in significantly different variation on cell morphology. As shown in Figure 9, mycelial morphology showed a uniform rod and the boundary was clear between the cells after the wild type L. bulgaricus was cultured for 16 h in MRS broth at 37°C. However, for the mur knockout strain K12, cells were connected and became filamentous, and the boundary was blurred between the cells. For the mur overexpressed strain O3, the growth speed of cells was significantly slower, and the size and density of cells were smaller than wild type cells.
Figure 9. Light microscopic observation of different cellular morphology of Ljj-6 and engineering bacteria.
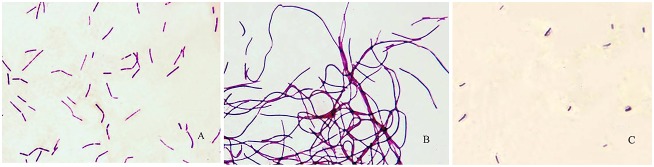
A: L. bulgaricus Ljj-6 (wild type); B: L. bulgaricus K12(pUC19-mur::EryBII); C: L. bulgaricus O3(pMG76e-mur). Magnification: ×1000.
Autolysis detection
The result of autolysis detection on ljj-6, o3, k12 and rk12 by flow cytometry indicated that the autolysis of the mur knockout strain k12 was significantly lower and autolysis of the mur overexpressed strain o3 was significantly higher compared with that of the wile type strain ljj-6 (Figure 8). This result suggested that the mur gene played an important role in autolysis of L. bulgaricus. On the other hand, autolytic activity in a low degree was still observed in the mur knockout strain, which implied that other enzymes but autolysin encoded by mur were also involved in autolysis of L. bulgaricus.
Discussion
The rate of starter autolysis is an important factor controlling cheese ripening and flavor development. This is because many starter enzymes that affect cheese ripening, such as peptidases, lipases and enzymes that catalyze amino acid conversions, are located intracellularly (not all of the latter remain functional after being released into cheese, as they may require intracellular co-factors). After the initial breakdown of milk caseins by the lactococcal cell envelope proteinase (lactocepin; EC 3.4.21.96), autolysis of starters causing release of these enzymes usually has beneficial consequences [12], [13]. For example, it has been reported that Cheddar cheese was bitter in flavour when made using the non-autolytic starter strain L. lactis subsp. cremoris HP which contains a type-I lactocepin [14], [15]. However, Cheddar cheese made using the autolytic strain L. lactis subsp. cremoris LW1484, which contains the same specificity-type lactocepin as HP [16], [17], was not bitter [18]. On the other hand, semi–hard Saint Paulin cheese made with the autolytic starter L. lactis subsp. cremoris RD251 remained bitter due to a low level of total intracellular peptidase activity [19]. Although starter autolysis is usually beneficial, undesirable consequences such as insufficient acid production and removal of residual lactose can result if autolysis is too rapid. Autolysis of lactococci used as starter cultures in the manufacture of fermented milk results in the leakage of lipases, proteases and peptidases and other intracellular components, which play an important role in flavor development during ripening. Hence, autolysis properties of lactic acid bacteria are crucial for their applications as dairy starters.
Autolysis of lactic acid bacteria in media and buffer systems has previously been studied by Sugahara et al. [4], Østlie et al. [20], Zhang et al. [21] and Østlie et al. [22]. These studies showed that autolysis varied both among species and between strains and took place at pH 5.2 to 7.0, 30 to 40°C as optimal autolysis temperature but also moderate autolysis at 20°C and ionic strength of 0.3, all conditions comparable with the conditions in Swiss-type cheese. The pH and specificity studies Østlie et al. [20] suggest that at least two different autolytic enzymes are involved in autolysis of propionibacteria. Later, Douillard et al. [23] investigated autolysis of Lactobacillus helveticus and Propionibacterium freudenreichii in Swiss cheeses by using species-specific lysis markers and demonstrated that autolysis of this P. freudenreichii strain occurred late during ripening and was tardy. However, the importance of autolysis of propioni bacteria in cheese ripening needs further investigation since only one strain was studied for a relatively short ripening period [24].
Differences in mur sequences is difficult to explain the huge variation in the extent of autolysis found in different L. bulgaricus strains. In addition, the completed Lactobacillus bulgaricus ATCC11842 genome sequence shows the presence of several open reading frames that putatively encode cell wall hydrolases having up to 47% predicted amino acid identity to mur gene. These enzymes may have roles in the autolysis of L. bulgaricus. Further studies are needed to clarify the number and types of autolytic enzymes within different strains and species of L. bulgaricus and also how they work.
Funding Statement
The authors acknowledge the financial support provided by the China Postdoctoral Science Foundation (2013M541091). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pillidge CJ, Rallabhandi PSVS, Tong X-Z, Gopal PK, Farley PC, et al. (2002) Autolysis of Lactococcus lactis. International Dairy Journal 12: 133–140. [Google Scholar]
- 2. Crow VL, Coolbear T, Gopal PK, Martley FG, McKay LL, et al. (1995) The role of autolysis of lactic acid bacteria in the ripening of cheese. International Dairy Journal 5: 855–875. [Google Scholar]
- 3. Chung W, Hancock REW (2000) Action of lysozyme and nisin mixtures against lactic acid bacteria. International Journal of Food Microbiology 60: 25–32. [DOI] [PubMed] [Google Scholar]
- 4. Sugahara K, Yokoi K-j, Nakamura Y, Nishino T, Yamakawa A, et al. (2007) Mutational and biochemical analyses of the endolysin LysgaY encoded by the Lactobacillus gasseri JCM 1131T phage φgaY. Gene 404: 41–52. [DOI] [PubMed] [Google Scholar]
- 5. Ledala N, Wilkinson BJ, Jayaswal RK (2006) Effects of oxacillin and tetracycline on autolysis, autolysin processing and atl transcription in Staphylococcus aureus. International Journal of Antimicrobial Agents 27: 518–524. [DOI] [PubMed] [Google Scholar]
- 6. Croux C, García J (1991) Sequence of the lye gene encoding the autolytic lysozyme of Clostridium acetobutylicum ATCC824: comparison with other lytic enzymes. Gene 104: 25–31. [DOI] [PubMed] [Google Scholar]
- 7. Mercier C, Domakova E, Tremblay J, Kulakauskas S (2000) Effects of a muramidase on a mixed bacterial community. FEMS Microbiology Letters 187: 47–52. [DOI] [PubMed] [Google Scholar]
- 8. Mullaney EJ, Ullah AH (2003) The term phytase comprises several different classes of enzymes. Biochem Biophys Res Commun 312: 179–184. [DOI] [PubMed] [Google Scholar]
- 9. Zhang XM, Shang N, Zhang X, Gui M, Li PL (2013) Role of plnB gene in the regulation of bacteriocin production in Lactobacillus paraplantarum L-XM1. Microbiological Research 168: 305–310. [DOI] [PubMed] [Google Scholar]
- 10. Holo H, Nes IF (1989) High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. cremoris Grown with Glycine in Osmotically Stabilized Media. Appl Environ Microbiol 55: 3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim SJ, Lee JY, Jun DY, Song JY, Lee WK, et al. (2009) Oral administration of Lactococcus lactis expressing Helicobacter pylori Cag7-ct383 protein induces systemic anti-Cag7 immune response in mice. FEMS Immunol Med Microbiol 57: 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crow VL, Gopal PK, Wicken AJ (1995) Cell surface differences of lactococcal strains. International Dairy Journal 5: 45–68. [Google Scholar]
- 13. Fox PF, Wallace JM, Morgan S, Lynch CM, Niland EJ, et al. (1996) Acceleration of cheese ripening. Antonie Van Leeuwenhoek 70: 271–297. [DOI] [PubMed] [Google Scholar]
- 14. Hickey DK, Kilcawley KN, Beresford TP, Sheehan EM, Wilkinson MG (2007) Starter strain related effects on the biochemical and sensory properties of Cheddar cheese. J Dairy Res 74: 9–17. [DOI] [PubMed] [Google Scholar]
- 15. Kaleta P, Callanan MJ, O’Callaghan J, Fitzgerald GF, Beresford TP, et al. (2009) Exploitation of the diverse insertion sequence element content of dairy Lactobacillus helveticus starters as a rapid method to identify different strains. J Microbiol Methods 79: 32–36. [DOI] [PubMed] [Google Scholar]
- 16. Sheehan A, Cuinn GO, Fitzgerald RJ, Wilkinson MG (2006) Proteolytic enzyme activities in Cheddar cheese juice made using lactococcal starters of differing autolytic properties. J Appl Microbiol 100: 893–901. [DOI] [PubMed] [Google Scholar]
- 17. Sheehan A, O’Loughlin C, O’Cuinn G, Fitzgerald RJ, Wilkinson MG (2005) Cheddar cheese cooking temperature induces differential lactococcal cell permeabilization and autolytic responses as detected by flow cytometry: implications for intracellular enzyme accessibility. J Appl Microbiol 99: 1007–1018. [DOI] [PubMed] [Google Scholar]
- 18. Sallami L, Kheadr EE, Fliss I, Vuillemard JC (2004) Impact of autolytic, proteolytic, and nisin-producing adjunct cultures on biochemical and textural properties of cheddar cheese. J Dairy Sci 87: 1585–1594. [DOI] [PubMed] [Google Scholar]
- 19. Collins YF, McSweeney PL, Wilkinson MG (2003) Evidence of a relationship between autolysis of starter bacteria and lipolysis in cheddar cheese during ripening. J Dairy Res 70: 105–113. [DOI] [PubMed] [Google Scholar]
- 20. Østlie H, Vegarud G, Langsrud T (1995) Autolysis of Dairy Propionibacteria in Buffer Systems. Journal of Dairy Science 78: 2315–2325. [Google Scholar]
- 21. Zhang X (2012) Diversity analysis of peptidoglycan hydrolases in lactic acid bacteria and their phages. International Dairy Journal 25: 60–65. [Google Scholar]
- 22. Østlie HM, Vegarud G, Langsrud T (2007) Autolysis of propionibacteria: Detection of autolytic enzymes by renaturing SDS-PAGE and additional buffer studies. International Journal of Food Microbiology 117: 167–174. [DOI] [PubMed] [Google Scholar]
- 23. Douillard FP, Mahony J, Campanacci V, Cambillau C, van Sinderen D (2011) Construction of two Lactococcus lactis expression vectors combining the Gateway and the NIsin Controlled Expression systems. Plasmid 66: 129–135. [DOI] [PubMed] [Google Scholar]
- 24. Jebava I, Plockova M, Lortal S, Valence F (2011) The nine peptidoglycan hydrolases genes in Lactobacillus helveticus are ubiquitous and early transcribed. Int J Food Microbiol 148: 1–7. [DOI] [PubMed] [Google Scholar]