Abstract
Plant growth homeostasis and defense responses are regulated by BONZAI1 (BON1), an evolutionarily conserved gene. Here, we show that growth regulation by BON1 is mediated through defense responses. BON1 is a negative regulator of a haplotype-specific Resistance (R) gene SNC1. The bon1-1 loss-of-function mutation activates SNC1, leading to constitutive defense responses and, consequently, reduced cell growth. In addition, a feedback amplification of the SNC1 gene involving salicylic acid is subject to temperature control, accounting for the regulation of growth and defense by temperature in bon1-1 and many other mutants. Thus, plant growth homeostasis involves the regulation of an R gene by BON1 and the intricate interplay between defense responses and temperature responses.
INTRODUCTION
Plant growth is controlled by internal programs and external factors. Internally, the basic cellular growth machinery is programmed in a developmental context to accumulate cell mass and increase cell number to a certain size. Positional cues are sent from neighboring cells, and hormones produced locally or distantly execute global growth regulation on individual cells. External signals, both biotic and abiotic, also greatly influence plant growth. Temperature, with its daily fluctuation and seasonal change, is one of the major environmental factors that regulate plant growth, distribution, and survival (Long and Woodward, 1988). One of the most prominent plant responses to extreme temperatures is cold acclimation, where plants tolerate freezing better if treated with low temperatures before being exposed to freezing conditions. Another response is vernalization, where a long period of cold treatment can accelerate flowering in many plants. The molecular mechanisms of these responses to extreme temperatures have been revealed recently through intensive studies (Sheldon et al., 2000; Viswanathan and Zhu, 2002). Ambient temperatures also affect various processes of growth and development. Flowering time and leaf initiation rate are modulated by thermal time (accumulative heat), with plants at higher temperature growing faster and flowering earlier (Poethig, 2003). Ambient temperature also influences the ability of plants to respond to many other hormonal, abiotic, and biotic signals. For instance, auxin-mediated hypocotyl growth, flowering time control by phytochrome, and some Resistance (R) gene–mediated disease resistances are temperature dependent (Whitham et al., 1994; Gray et al., 1998; Halliday et al., 2003).
To investigate the molecular mechanisms of growth responses to ambient temperatures, we are genetically dissecting the control of growth homeostasis at varying temperatures in Arabidopsis (Arabidopsis thaliana). Wild-type Arabidopsis plants achieve a similar size when grown at temperatures ranging from 16 to 30°C. The genetic control of this growth homeostasis was revealed by mutants (such as acaulis1, acaulis 3, acaulis4, and bonzai1 [bon1]) that cannot maintain constant size at different temperatures (Akamatsu et al., 1999; Hua et al., 2001). The loss-of-function mutant bon1-1 has a dwarf phenotype mostly because of reduced cell size at 22°C, but it resembles the wild type at 28°C, indicating an essential role of BON1 in growth homeostasis (Hua et al., 2001). BON1 encodes a member of the copine gene family that is highly conserved among protozoa, plants, nematodes, and mammals (Creutz et al., 1998). The deduced BON1 protein, like other copine proteins, has at its N terminus two calcium-dependent phospholipid binding C2 domains that are mostly found in signal transduction or membrane trafficking molecules (Rizo and Sudhof, 1998). BON1 binds to phospholipids in a calcium-dependent manner in vitro, and it is localized to the plasma membrane in plants (Hua et al., 2001). The C-terminal region of BON1 shows homology to the A domain of integrins (Williams et al., 1999) and is proposed to mediate protein–protein interaction, to possess an intrinsic protein kinase activity, or both (Caudell et al., 2000). The A domain in BON1 mediates the interaction of BON1 with its putative functional partner BAP1 that also contains a C2 domain (Hua et al., 2001). The presence of C2 domains in both BON1 and BAP1 suggests that they are involved in a biological process regulated by the membrane system, calcium state, or by both factors.
In addition to regulating growth homeostasis, BON1 is also shown to modulate defense responses. A bon1 mutant allele cpn1-1 (which we will refer to as bon1-4) exhibits precocious cell death and enhanced disease resistance under low humidity or low temperature conditions (Jambunathan et al., 2001; Jambunathan and McNellis, 2003). It is intriguing that both growth and defense are affected by bon1 mutations in a temperature-dependent manner. Emerging molecular data suggest that these two processes are intricately intertwined. Mutations or transgenes that perturb cellular metabolism and plant growth cause lesion mimic phenotypes and defense responses characteristic of systemic acquired resistance (SAR) (Mittler et al., 1995; Molina et al., 1999; Clough et al., 2000). Compromised cell growth is found in some mutants with constitutive pathogen responses, such as cpr1 and mpk4 (Bowling et al., 1994; Petersen et al., 2000). Furthermore, genes functioning in the signaling pathway of defense responses, such as TIP49a, RAR1, and SGT1, play important roles in growth and development (Austin et al., 2002; Azevedo et al., 2002; Holt et al., 2002; Liu et al., 2002). Thus, defense responses, once thought to be specific for pathogen invasions, have extensive interplay with growth regulation. Moreover, it is becoming evermore evident that defense responses are intimately connected with abiotic responses. Light, humidity, and temperature have all been demonstrated to influence certain defense responses (Dietrich et al., 1994; Yoshioka et al., 2001; Xiao et al., 2003). The molecular mechanisms of the interaction between growth, defense, and abiotic responses are yet to be revealed.
We report in this study an investigation of the molecular mechanisms of growth and defense regulated by BON1 and modulated by temperature. We found that BON1 negatively regulates a haplotype-specific R gene, SNC1. The bon1 loss-of-function mutation activates defense responses that lead to compromised cell growth in a SNC1-containing accession but not in accessions without a functional SNC1. Furthermore, we found that SNC1 is under a positive feedback regulation involving salicylic acid (SA) and that this regulatory loop is subject to temperature modulation. Thus, plant growth homeostasis controlled by BON1 involves repression of an R gene and modulation of defense responses by temperature.
RESULTS
Identification of a Natural Modifier of bon1
To better understand the molecular mechanism by which BON1 controls growth homeostasis, we took advantage of a naturally occurring bon1 modifier. The bon1 mutant alleles bon1-1 and bon1-2 confer drastically different phenotypes. At 22°C, bon1-1 mutants show greatly reduced plant size and have twisted leaves and short inflorescence stems, whereas bon1-2 mutants resemble the wild type (Figure 1). The phenotypic differences between the two mutants are unlikely caused by differences in allele strength because both appear to be loss-of-function mutations. Both bon1-1 and bon1-2 contain T-DNA insertions in exons at positions corresponding to amino acids 394 and 61, respectively, in the BON1 predicted protein. These insertions would result in truncated proteins. In addition, no wild-type BON1 transcripts were detected in either of the mutants by RNA gel blot analysis (data not shown), indicating that both are loss-of-function mutants.
Figure 1.
bon1-2 Has Wild-Type Morphology.
Col, bon1-1, Ws, and bon1-2 plants grown at 22°C before (A) and after (B) bolting. bon1-1 is dwarf with dark-green and twisted leaves and short inflorescence stems. bon1-2 resembles wild-type Ws.
Given that both bon1-1 and bon1-2 appear to be loss-of-function mutations, we considered the possibility that the phenotypic difference was because of differences in accession (ecotype) background. bon1-1 was isolated from the Columbia (Col) accession, whereas bon1-2 was isolated from the Wassilewskija (Ws) accession. F2 progeny of bon1-1 backcrossed to Col segregated one-quarter of dwarf plants, whereas F2 progeny of bon1-1 crossed to Ws segregated one-sixteenth of bon1-1–like dwarf plants (Table 1), suggesting the presence of a bon1 suppressor in Ws. Consistent with this view, F2 progeny of bon1-2 crossed to the Col wild type segregated approximately one-sixteenth of bon1-1–like dwarf plants, whereas F2 progeny of bon1-2 backcrossed to Ws did not segregate any dwarf plants (Table 1). These data confirm that the phenotypic differences are attributable to accession background and suggest that the modifier of the bon1 phenotype is conferred by differences in a single nuclear gene. The latter point was further supported when a quarter of the F2 progeny from a cross between bon1-1 and bon1-2 exhibited a dwarf phenotype (Table 1). Natural variations at this locus modify the bon1 phenotype, and we named this locus MOB (modifier of bon1). The Col variant of MOB is required for the dwarf phenotype of bon1-1, whereas the Ws variant of MOB dominantly suppresses this dwarf phenotype.
Table 1.
Analysis of the Genetic Basis of the Phenotypic Differences between bon1-1 and bon1-2
| F2 Progeny of | Wild Type | Mutant | Ratio Expected | P Valuea |
|---|---|---|---|---|
| bon1-1 × Col | 74 | 23 | 3:1 | 0.80 |
| bon1-1 × Ws | 100 | 7 | 15:1 | 0.90 |
| bon1-2 × Ws | >400 | 0 | NDb | ND |
| bon1-2 × Col | 117 | 8 | 15:1 | 0.94 |
| bon1-1 × bon1-2 | 314 | 115 | 3:1 | 0.39 |
P value was calculated based on χ2.
ND, not determined.
Cloning of the MOB Gene
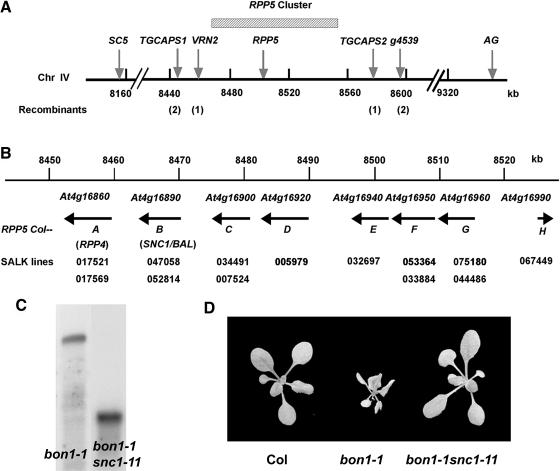
We employed a map-based cloning approach to identify the MOB gene. bon1-1 (in Col) was crossed to bon1-2 (in Ws), and the dwarf plants segregated in the F2 progeny were chosen for mapping. Codominant cleaved amplified polymorphic sequence (CAPS) markers (Konieczny and Ausubel, 1993) and simple sequence length polymorphism (SSLP) markers (Bell and Ecker, 1994) were used to locate the MOB gene on chromosome IV between markers SC5 and AG (http://www.arabidopsis.org/index.jsp) (Figure 2A). New SSLP and CAPS markers between SC5 and AG were generated between Col and Ws. Further mapping using ∼2500 plants refined the position to a 120-kb region between markers VRN2 and TGCAPS2 (Gendall et al., 2001) (Figure 2A). The recombination rate of this region is extremely low compared with the average of 200 to 250 kb/centimorgan in Arabidopsis, which is likely because of the presence of the RPP5 complex locus in this region. RPP5 is an R gene conferring resistance to the oomycete pathogen Peronospora parasitica Noco2 strain in Arabidopsis accession Landsberg erecta (Ler) (Parker et al., 1997). The locus consists of multiple copies of functional and apparently nonfunctional RPP5 homologs (Noel et al., 1999). In Col, the locus contains eight homologs of RPP5 (named Col-A to Col-H) covering a region of ∼80 kb (Figure 2C), whereas in Ler, this locus contains 10 homologs of RPP5 (named La-A to La-J) covering a region of >95 kb (Noel et al., 1999). There is a pronounced divergence and lack of synteny among the homologs in this region between Col and Ler, which may account for the significant recombination suppression noted in previous map-based cloning in this region (Gendall et al., 2001; Stokes et al., 2002). Although the RPP5 complex locus in Ws has not been sequenced, the strong recombination suppression in this region between Col and Ws suggests the presence of a divergent complex locus in Ws as well.
Figure 2.
The Col-Specific SNC1 Gene Mediates the bon1-1 Phenotype.
(A) A genetic map of the region carrying the MOB locus. The corresponding nucleotide positions (in Col) are numbered in kilobases below the line. MOB was positioned between markers VRN2 and TGCAPS2. The number of recombinants confirmed is indicated underneath the markers.
(B) A molecular map of the Col RPP5 complex locus. Predicted genes are shown by arrows indicating the direction of transcription. The locus name, the complex locus name (Noel et al., 1999), and the genetic name (if known) are given for each gene. T-DNA insertion lines from the Salk Institute are indicated below the corresponding genes.
(C) SNC1 RNA expression in bon1-1 and bon1-1 snc1-11. SNC1 transcript is ∼4.5 kb in the wild type and <1 kb in snc1-11.
(D) Suppression of bon1-1 growth phenotype by snc1-11. Col, bon1-1, and bon1-1 snc1-11 grown at 22°C are shown. bon1-1 snc1-11 resembles wild-type Col.
We reasoned that one of the RPP5 homologs differing between Col and Ws in this cluster could be the MOB gene because bon1-1 resembles the gain-of-function mutants of the second gene in the RPP5 cluster Col-B (At4g16890), also known as SNC1 and BAL (we will refer to this gene as SNC1). snc1-1 contains a missense mutation, which likely renders the mutant protein constitutively active (Zhang et al., 2003). bal is an epigenetic allele of SNC1 (we will refer to it as snc1-2), which causes overexpression of SNC1 without an alteration in the nucleotide sequences (Stokes et al., 2002). Both snc1-1 and snc1-2 mutations activate an SA-dependent defense response pathway that leads to enhanced defense responses. In addition, they both exhibit a dwarf phenotype similarly to bon1-1 (Li et al., 2001; Stokes et al., 2002). It is thus possible that MOB is one of the RPP5 homologs in the complex locus and that the activation of this homolog by bon1-1 leads to the observed growth defects. If so, the loss of function of this homolog should suppress the bon1-1 phenotype.
To determine whether mutations in any of the RPP5 homologs in the cluster can suppress the bon1-1 phenotype, we isolated mutants of these genes from the Salk T-DNA insertion line collections (http://signal.salk.edu/cgi-bin/tdnaexpress). Mutant lines identified for this cluster are shown in Figure 2B underneath the corresponding genes, and they are all in the Col accession background. T-DNA insertions are found in exons of SNC1, Col-C, Col-D, Col-E, Col-G, Col-F, and Col-H, presumably resulting in nonfunctional proteins even if the genes are transcribed. For Col-A (also known as RPP4), only mutants with T-DNA inserted into the introns were found, but they are likely to be loss-of-function mutants because no wild-type transcripts were detected by RNA gel blot analysis (data not shown). Each of these mutants was crossed to bon1-1, and their F2 progeny were analyzed. The bon1-1–like dwarf phenotype was observed in about one-quarter of the F2 progeny from each cross, with the exception of the crosses between bon1-1 and two snc1 mutants (data not shown). Both of the two mutants, SALK_047058 and SALK_052814, have T-DNA insertions in the first exon, and we named them snc1-11 and snc1-12, respectively. We confirmed the loss-of-function nature of snc1-11 by RNA gel blot analysis, revealing a transcript much smaller than that of the wild type in snc1-11 (Figure 2C). The F2 progeny of the crosses between bon1-1 and either snc1-11 or snc1-12 segregated the bon1-1–like phenotype in about one-sixteenth of plants, indicating that snc1-11 and snc1-12 suppress bon1-1 in a dominant manner. Furthermore, when mutants of the RPP5 cluster were crossed to bon1-2, all segregated bon1-1–like phenotypes in the F2 progeny except for crosses between bon1-2 and snc1-11 or snc1-12, confirming that SNC1 is the MOB gene that mediates the dwarf phenotype in bon1. bon1-1 snc1-11 double mutants were identified, and they are indistinguishable from wild-type Col (Figure 2D).
SNC1 Is Specific to the Col Accession
That MOB (SNC1) is functional in Col indicates that Ws does not have a functional SNC1 gene. To determine whether this is because of the absence of an SNC1 ortholog or because of a mutated SNC1 gene in Ws, we attempted to isolate SNC1-like genes from Ws. Because genes in the RPP5 cluster from Col share high sequence homology, primers specific to SNC1, but not to any of the other RPP5 homologs, were used to amplify a putative SNC1 variant from Ws using RT-PCR.
This approach produced a cDNA from a gene tightly linked to the VRN2 marker, making it a likely member of the RPP5 cluster in Ws (data not shown). Furthermore, the gene has intron splice sites consistent with those identified in other members of the RPP5 gene family (Meyers et al., 2003). We have named this gene SNW for SNC1-like in Ws. The very 3′ end of the SNW (possibly 300 bp of the coding sequence) was not obtained, as estimated by the SNW transcript size and its sequence alignment with other members. This deduced SNW protein exhibits high homology to SNC1 only at the N terminus in the Toll and interleukin-1 receptors (TIR) region (Figure 3). Its nucleotide binding (NB) and Leu-rich repeat (LRR) regions are not more similar to SNC1 than to RPP4 encoded by the first gene in the Col RPP5 cluster (Figure 3). It thus appears that there is no closely related ortholog of SNC1 in Ws. In fact, sequence comparison between Col and Ler accessions indicates that genes in the RPP5 cluster have evolved rapidly (Noel et al., 1999), and orthologous relationships may not exist for homologs in different accessions.
Figure 3.
Sequence Comparison among SNC1, SNW, and RPP4.
TIR domain and NB-Apaf-1, R proteins, and Ced4 domain are labeled. Identical amino acids are indicated by white letters with black background. High sequence identity between SNC1 and SNW is found mostly in the TIR and NB-Apaf-1, R proteins, and Ced4 domains. The C-terminal LRR region of SNW is not more similar to that of SNC1 than to that of RPP4.
To test the hypothesis that SNC1 has rapidly evolved and is likely unique to Col, we crossed bon1-1 to two other accessions: Ler and Nossen-0. F2 progeny of these two crosses segregated dwarf plants at a ratio far less than one-quarter as found in the cross to Col (data not shown), suggesting that these two accessions may not have SNC1 orthologs. The SNC1 gene is likely Col haplotype specific, and a functional SNC1 is required for the exhibition of growth defect in bon1-1.
SNC1 Mediates the Disease Resistance Phenotype of bon1
Based on our results, BON1 appears to be a negative regulator of the R gene SNC1. In this model, SNC1 would be activated in bon1-1. Activation of an R gene by the recognition of a specific pathogen avirulence (Avr) gene usually induces a local hypersensitive response, which also triggers a secondary defense response (SAR) through the signal SA (Glazebrook, 2001). Constitutive activation of SAR by the activation of SNC1 could therefore account for the enhanced disease resistance phenotype observed in bon1-4 mutants in the Col background.
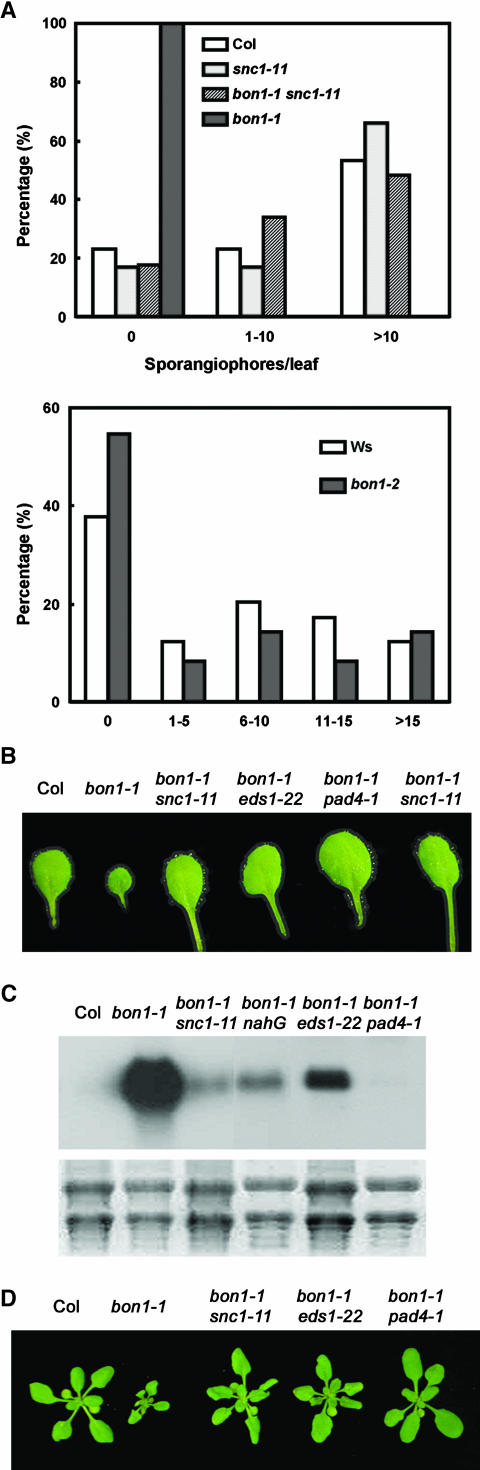
To determine whether SNC1 mediates enhanced defense responses in bon1 mutants, we analyzed and compared the disease resistance phenotypes of bon1-1, bon1-1 snc1-11, and bon1-2. Similar to previous observations with bon1-4, bon1-1 exhibited a strong resistance to the virulent pathogen P. parasitica at 22°C. Under these conditions, wild-type Col is sensitive, as shown by the formation of sporangiophores on most leaves several days after exposure to P. parasitica spores (Figures 4A and 4B). No sporangiophores were detected on any of the leaves of bon1-1, indicating a strong resistance to this pathogen (Figures 4A and 4B). Furthermore, the defense response marker gene PR1 (Pathogenesis Related1) is upregulated in bon1-1 grown at 22°C compared with that in the Col wild type (Figure 4C), suggesting that defense responses involving SAR are upregulated in bon1-1. By contrast, bon1-2, without a functional SNC1, does not exhibit an enhanced resistance to virulent pathogen P. parasitica Emwa1 strain. Sporangiophore formation is similar on bon1-2 leaves to that on the wild-type Ws leaves at 22°C (Figure 4A), and PR1 gene upregulation is usually not observed in bon1-2 (data not shown). That the heightened disease resistance in bon1-1 is mediated by SNC1 is further supported by the suppression of the resistance phenotype in the loss-of-function mutant snc1-11. bon1-1 snc1-11 is as sensitive to P. parasitica as wild-type Col, as demonstrated by the similar extent of sporangiophore formation between the double mutant and Col (Figures 4A and 4B). The upregulation of PR1 in bon1-1 is greatly suppressed by snc1-11 (Figure 4C), indicating that the enhanced disease resistance in bon1-1 is largely mediated by SNC1. These data all support a role for BON1 as a negative regulator of SNC1 in Col wild-type plants.
Figure 4.
The bon1-1 Growth Defect Is Induced by Constitutive Defense Responses.
(A) Disease resistance phenotype of bon1-1, bon1-2, and bon1-1snc1-11. No growth of P. parasitica is observed in bon1-1, whereas a similar amount of growth is found between Col and bon1-1 snc1-11 and between the Ws wild type and bon1-2. The experiment was repeated twice, and similar trends were observed.
(B) Resistance to virulent P. parasitica is suppressed by nahG, eds1-22, pad4-1, and snc1-11. Sporulations on representative leaves are shown.
(C) Suppression of PR1 gene upregulation in bon1-1 by snc1-11, eds1-22, and pad4-1 mutations and by the transgene nahG. rRNAs detected by ethidium bromide staining were used as a control.
(D) Suppression of the dwarf phenotype of bon1-1 by the transgene nahG and mutations of eds1-22 and pad4-1.
The Growth Defect of bon1-1 Is a Result of Constitutive Defense Responses
Because constitutive defense responses could lead to compromised plant growth in some mutants (Heil and Baldwin, 2002), we tested whether the dwarf phenotype of bon1-1 is a consequence of the activation of defense responses. SA is known to be a signal for local defense and for SAR (Glazebrook, 2001), we therefore introduced into bon1-1 a nahG transgene encoding an enzyme that degrades SA (Bowling et al., 1994). bon1-1 nahG plants, in great contrast with bon1-1, have close to wild-type morphology (Figure 4C). They also exhibit drastically reduced resistance to P. parasitica and reduced PR1 expression compared with bon1-1 (Figures 4B and 4C). Because the effect of nahG is more pleotropic than lowering SA level (Heck et al., 2003), we also generated a double mutant between bon1-1 and the sid2-1 mutant that is defective in SA biosynthesis (Wildermuth et al., 2001). The bon1-1 sid2-1 double mutant has a wild-type phenotype (data not shown), further indicating an involvement of SA accumulation in the growth defect of bon1-1.
SNC1 belongs to the TIR-NB-LRR type of R genes (Meyers et al., 2003), and several members of this type of R genes are shown to require the function of EDS1 and PAD4 (Aarts et al., 1998; Glazebrook, 2001). We found that both eds1 and pad4 can suppress the disease resistance phenotype as well as the growth defects of bon1-1. EDS1 exists as tandem repeats in Col, and both genes (At3g48090 and At3g48080) appear to encode functional proteins. A loss-of-function mutant of At3g48090, SALK_071051, was isolated and named eds1-22. Virulent P. parasitica sporulated on bon1-1 eds1-22, but to a less extent than on Col, indicating a partial suppression of bon1-1 resistance by eds1-22 (Figure 4B). PR1 expression is greatly reduced in bon1-1 eds1-22 compared with that in bon1-1 (Figure 4C). Furthermore, bon1-1 eds1-22 has a nearly wild-type growth phenotype (Figure 4D). The residual resistance and growth defect in bon1-1 eds1-22 is likely attributable to the presence of another functional EDS1 gene, At3g48080, a view supported by the wild-type phenotype of a double mutant generated between bon1-1 and a null eds1-1 mutant from Ws (data not shown). pad4-1 totally suppressed the bon1-1 phenotype. bon1-1 pad4-1 is as sensitive to P. parasitica as the wild-type Col (Figure 4B), and PR1 expression is absent in bon1-1 pad4-1 (Figure 4C). In addition, bon1-1 pad4-1 is indistinguishable from the wild type. By contrast, a mutation in NDR1 required for the function of several coiled coil (CC)-NB-LRR type of R genes (Aarts et al., 1998; Glazebrook, 2001) could not suppress the bon1-1 phenotype (data not shown). Thus, the activation of defense responses mediated by a TIR-NB-LRR type of R gene and SA is responsible for the growth defect in bon1-1.
SNC1 Is Subject to a Positive Feedback Regulation
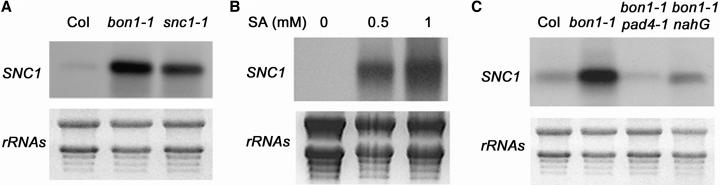
We next investigated how SNC1 is regulated by BON1 and found that the SNC1 transcript level is altered in bon1-1. RNA gel blot analysis shows that bon1-1 grown at 22°C has at least five times more SNC1 transcript compared with the wild type (Figure 5A). Overexpression of SNC1 by the strong 35S promoter of Cauliflower mosaic virus or by an epigenetic mechanism has been shown to induce constitutive defense responses (Stokes et al., 2002), making it possible to attribute the activation of SNC1 by bon1-1 to the regulation of SNC1 transcript abundance.
Figure 5.
SNC1 Is under a Positive Feedback Regulation.
(A) Upregulation of the SNC1 RNA transcripts in bon1-1 and snc1-1 compared with Col.
(B) Induction of the SNC1 transcript by SA application. RNAs were extracted 2 d after plants were sprayed with 0, 0.5, and 1 mM of SA.
(C) Suppression of SNC1 upregulation in bon1-1 by pad4-1 and nahG. Wild-type level of the SNC1 transcripts is observed in bon1-1 pad4-1 and bon1-1 nahG.
Further study shows that this regulation may be indirect because of the existence of positive feedback regulation of SNC1. We found that SNC1 is transcriptionally upregulated by SA. Wild-type plants treated with SA have drastically increased amounts of SNC1 transcript compared with control plants treated with water (Figure 5B). It is thus possible that the bon1 mutation causes activation of the SNC1 protein, which leads to SA accumulation and a subsequent elevation of the SNC1 transcript. Consistent with this idea, we observed an increase in SNC1 transcript abundance in snc1-1 (Figure 5A), although snc1-1 is caused by a missense mutation likely affecting the SNC1 protein activity (Zhang et al., 2003). To test the hypothesis that the upregulation of the SNC1 transcript is a secondary effect, we used pad4 and nahG to block this feedback regulation. In bon1-1 pad4 and bon1-1 nahG mutants, the induction of SNC1 by bon1-1 is eliminated (Figure 5C), indicating that BON1 likely acts on the SNC1 transcript abundance through feedback regulation.
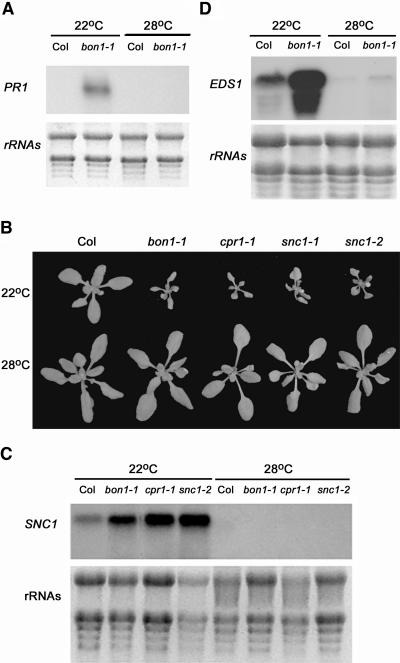
The SNC1 Pathway Is Temperature Regulated
BON1 regulates plant growth in a temperature-dependent manner as indicated by the fact that at a higher temperature (28°C) the growth defects that bon1-1 exhibits at 22°C are not observed. The enhanced disease resistance phenotype of bon1-4 regulation on defense responses is also temperature dependent (Jambunathan and McNellis, 2003). PR1 expression is upregulated at 22°C but is not detected at 28°C in bon1-1 (Figure 6A). We asked whether this temperature regulation could act upon the SNC1 pathway by analyzing the temperature responses of the snc1 gain-of-function mutants (snc1-1 and snc1-2) and an SNC1-related mutant, cpr1. The cpr1 mutation has not been molecularly identified. However, the CPR1 gene was mapped close to the SNC1 locus and is proposed to be an epigenetic allele of SNC1 (Stokes and Richards, 2002). All these mutants have constitutive expression of SAR and exhibit dwarf phenotypes resembling bon1-1 at 22°C (Figure 6B). When these mutants were grown at 28°C, they all had a wild-type morphology (Figure 6B), indicating that temperature could regulate SNC1 or its downstream components independently of BON1. Thus, temperature regulation of the bon1-1 phenotype could be attributed to its modulation of the SNC1 pathway.
Figure 6.
Temperature Regulation of the bon1-1 Phenotype.
(A) PR1 expression in bon1-1 at 22°C and 28°C. The PR1 gene is upregulated in bon1-1 at 22°C but not at 28°C.
(B) Phenotypes of Col, bon1-1, cpr1-1, snc1-1, and snc1-2 grown at 22°C and 28°C. Higher temperature (28°C) suppresses the dwarf phenotype of these mutants exhibited at 22°C.
(C) SNC1 RNA expression in the wild type, bon1-1, cpr1-1, and snc1-2 grown at 22°C and 28°C. Induction of the SNC1 transcript in these mutants at 22°C is abolished at 28°C.
(D) EDS1 RNA expression in the wild type and bon1-1 at 22°C and 28°C. The EDS1 transcript level is higher at 22°C than at 28°C in both the wild type and bon1-1.
We found that temperature regulates the level of components in the feedback regulation loop consisting of SNC1, EDS1/PAD4, and SA. As in bon1-1, the SNC1 RNA transcript at 22°C is more abundant in snc1-1, snc1-2, and cpr1 than in the wild type (Figure 6C). The SNC1 RNA transcript is not detectable at 28°C by RNA gel blot analysis in any of these mutants (Figure 6C), indicating that temperature regulates the RNA transcript abundance of SNC1 in the mutants. Transcripts of EDS1 genes are also upregulated in bon1-1, and this upregulation is abolished by high temperature (Figure 6D). Similar regulation is found for PAD4 transcript level and SA amount in bon1-1 (data not shown). Thus, in bon1-1, all components in the regulatory loop tested have a higher level at 22°C than at 28°C. Interestingly, EDS1 transcripts appear to be regulated by temperature in wild-type Col as well. We consistently see more EDS1 transcripts at 22°C than at 28°C (Figure 6D), suggesting that some components in the SNC1 pathway might be modulated by temperature even when the autoregulatory loop is not activated.
DISCUSSION
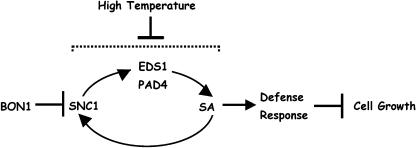
In this study, we further investigated the molecular mechanisms of growth homeostasis and defense responses regulated by BON1. We identified a natural modifier of bon1, SNC1, which is a haplotype-specific R gene. We propose that BON1 is a negative regulator of the haplotype-specific R gene SNC1 (Figure 7). Activation of SNC1 by bon1-1 leads to constitutive defense responses, and through this effect, plant growth is compromised (Figure 7). Furthermore, SNC1 is under positive feedback regulation involving EDS1, PAD4, and SA. Temperature primarily modulates one or several of the components in this feedback regulation, conferring a downregulation of all components in the pathway by higher temperatures (Figure 7). This provides a mechanism for the temperature regulation of the bon1 phenotype.
Figure 7.
Model for Growth Homeostasis Controlled by BON1.
BON1 is a negative regulator of the R gene SNC1. SNC1, when activated, can induce SA accumulation and defense responses that in turn inhibit cell growth. A positive feedback regulation exists between SNC1 and SA, and high temperature suppresses this feedback amplification or components in the feedback regulation.
Interaction of Growth and Defense
Our dissection of growth homeostasis regulated by BON1 underlines the importance of regulating defense responses in plant growth control. BON1 normally represses the function of the R gene SNC1. The loss of BON1 function activates SNC1, leading to constitutive defense responses and subsequently compromising cell growth. It has long been recognized that disease resistance has a fitness cost. Compromised cell growth is sometimes found in mutants with constitutive pathogen responses, possibly because of resource reallocation from growth to defense, the metabolic burden of defense responses on plants, or both (Heil and Baldwin, 2002). There can be a fitness cost even with the acquisition of a resistance trait such as an R gene RPM1 (Tian et al., 2003). Thus, defense responses need to be repressed under most conditions and only need to be induced in the presence of pathogen invasion.
Creation of new R genes is beneficial for plants because each new gene can enable the plant to recognize new varieties of pathogens. Plants appear to drive R gene duplication through complex R gene loci like that of RPP5, in which duplicated genes have evolved to confer resistance to distinct pathogens (Parker et al., 1997; Noel et al., 1999; van der Biezen et al., 2002). The advantage of multiple R genes, however, is counterbalanced by the fitness cost of R gene misexpression, making it critical that each gene be expressed only when needed. A negative regulation of the R gene activity by endogenous plant genes like BON1 can confer tight control over the activation of defense responses and may be prevalent among the R genes.
Regulation of SNC1 by BON1
The activation of SNC1 by the bon1-1 mutation demonstrates that BON1 is a negative regulator of SNC1. This is one of the few recently uncovered examples where an R gene is regulated by a plant gene. The plant R gene was proposed to specifically recognize the pathogen Avr gene in the gene-for-gene model (Flor, 1971) whose simplest biochemical interpretation is areceptor-ligand model where the Avr protein directly interacts and activates the R protein (Van der Biezen and Jones, 1998). The limited repertoire of R proteins compared with the varieties of pathogens led to the guard hypothesis. Under the guard hypothesis, non-R plant proteins are guarded by R proteins. The non-R plant proteins sense the pathogen Avr proteins and mediate the activation of the guard R proteins (Dangl and Jones, 2001; Martin et al., 2003). One of the examples is the Arabidopsis RIN4 protein, a bacterial virulence target guarded by two R proteins, RPM1 and RPS2. Phosphorylation or elimination of the RIN4 protein by the Avr proteins activates RPM1 and RPS2, respectively, and triggers defense responses (Mackey et al., 2002, 2003; Axtell and Staskawicz, 2003).
The interaction between SNC1 and BON1 supports the guard hypothesis that R genes are regulated by plant genes and that there is an indirect interaction between Avr and R proteins. The R protein SNC1 appears to be normally repressed by a negative regulator as suggested by the gain-of-function snc1-1 mutation (Zhang et al., 2003). Whether BON1 is the negative regulator of SNC1 protein and guarded by SNC1 in analogy to RIN4 by RPS2 is yet to be determined. The pathogen effector Avr for SNC1, if any exists, is not known. It is therefore currently not possible to test whether an Avr protein targets BON1 for modification or elimination, an event that would then be recognized by SNC1. BON1 is a protein evolutionarily conserved from paramecium to human, and it likely has a cellular function in an essential process such as membrane trafficking or signal transduction. This cellular function also could contribute to defense responses (by facilitating PR protein secretion or by transducing calcium signals) and thus may be targeted by pathogen Avr proteins. Plants may counteract this targeting by using SNC1 to monitor the BON1 protein or other molecules in the BON1 pathway to recognize pathogen invasion.
The molecular mechanism of how BON1 negatively regulates SNC1 is not known. SNC1 transcript levels are regulated by BON1, but this regulation could be indirect and through feedback regulation of SNC1. We have shown that SA upregulates SNC1 transcripts and that the elevated level of SNC1 could be because of an accumulation of SA by an activated SNC1 protein. Similar positive feedback regulation between an R gene and SA has been observed in plants carrying a mutant form of the R gene SSI4. Though SSI4 transcript is upregulated in the mutant, the gain-of-function missense mutation, but not the overexpression of SSI4, results in an enhanced disease resistance phenotype (Shirano et al., 2002). In addition, SA is demonstrated to induce some R genes of the TIR-NB-LRR type but not the CC-NB-LRR type (Shirano et al., 2002). Thus, defense responses in bon1-1 could be amplified through a positive autoregulation of SNC1 and the upregulation of other R genes by SA. This view would suggest that the increase in SNC1 transcription in bon1-1 mutants is indirect and may not be indicative of the primary mechanism by which BON1 acts in pathogen response.
It is possible that BON1 regulates the protein activity of SNC1 more directly and BON1 might be guarded by SNC1. BON1 and SNC1 proteins could potentially have direct physical interaction. BON1 is localized to the plasma membrane (Hua et al., 2001). The subcellular localization of SNC1 is not known, but it could be plasma membrane associated as shown for the CC-NB-LRR type of R protein, RPM1 (Boyes et al., 1998). The association of BON1 with SNC1 could inhibit the activity of SNC1, and the elimination of BON1 could therefore activate the SNC1 activity. Alternatively, there is no physical interaction between BON1 and SNC1, and the direct regulation of SNC1 is performed by downstream components of BON1. The regulation of SNC1 by BON1 could be through other mechanisms, including transcriptional control. For instance, a slight upregulation of SNC1 transcription in bon1-1 (beyond our detection sensitivity) could be amplified by the positive feedback regulation to induce full activation of SNC1.
It is intriguing that the loss-of-function snc1 mutation is dominant over wild-type SNC1 in suppressing bon1-1 phenotype. This phenomenon could be caused by haploid insufficiency. A threshold of SNC1 activity might be required for it to activate defense responses, and one copy of SNC1 is below the threshold. A feedback regulation could amplify the activity above the threshold, conferring a strong activation of defense responses with two copies of wild-type SNC1 but no apparent activation with one copy of SNC1.
Though defense responses regulated by BON1 are mostly mediated by SNC1, an SNC1-independent defense-related pathway also appears to be regulated by BON1. In bon1-1 snc1-11 double mutants, we consistently observed residual PR1 expression, although at a much reduced level than in bon1-1. We also occasionally observed PR1 upregulation in bon1-2 but not in the Ws wild type. Thus, there seems to be a weak upregulation of defense responses by bon1 mutations in the absence of SNC1. This could be because of a weak activation of other SNC1-related R genes. Alternatively, it could be because of some basal activation of defense responses by the bon1 mutation, and SNC1 acts as a specific amplifier of the basal resistance.
Interaction of Disease Resistance and Temperature Responses
Disease resistance is known to be modulated by abiotic factors, such as light, temperature, and humidity, but the molecular basis for this modulation is largely unexplored except for a few cases. In one case, temperature has been shown to modulate RNA silencing–mediated defense (Szittya et al., 2003). At a low temperature, both virus and transgene-triggered RNA silencing are inhibited, and the amount of small interfering RNAs are gradually reduced from higher to lower temperatures (27 to 15°C). In another case, temperature is shown to regulate the synthesis of SA (Malamy et al., 1992). High temperature prevents induction of PR genes and disease resistance mediated by the R gene N, and this is correlated with an inhibition of SA accumulation, although SA does not appear to be the sole signal regulated by temperature.
In this study, we found that the amount of components in the feedback regulatory loop (including SNC1, EDS1, PAD4, and SA) is greatly reduced at high temperatures in bon1-1 mutants. This likely accounts for the modulation of bon1 phenotype by temperature. It is conceivable that common signaling molecules and similar feedback regulation exist in defense responses induced by some other R genes. Temperature regulation of the level of individual components provides a mechanism for the modulation of defense responses by environmental signals in many other cases.
Because of the feedback regulation among SNC1, PAD4/EDS1, and SA, the regulation of these components by temperature may not be independent. Temperature may primarily regulate one component in the pathway and subsequently regulate all components in the pathway through the feedback regulation. The primary target of temperature regulation is yet to be determined. It is interesting to note that the EDS1 genes have a higher expression at 22°C than at 28°C even in the wild type, suggesting that temperature regulation could exist independently of feedback amplification. Further study of the growth homeostasis controlled by BON1 and temperature will shed more light on the interplay between growth, defense, and temperature response as well as the evolution and function of R genes.
METHODS
Plant Growth Conditions and Treatments
Plants were grown at 22°C or 28°C under constant light with 30 to 70% relative humidity unless stated otherwise. For SA treatment, 3-week-old Col plants were sprayed with SA in 0.005% Silwet L-77 or only 0.005% Silwet L-77 (for control), and tissues were collected 2 d later for RNA extraction. Pathogen resistance test on P. parasitica was as described with modifications (Kim and Delaney, 2002). Briefly, 2-week-old plants were sprayed with strains Noco2 (for plants in Col background) or Emwa1 (for plants in Ws background) and kept at 22°C under high humidity (covered with plastic dome). The number of sporangiophores on the leaves was counted a week later. Approximately 70 leaves were counted for each genotype.
Plant Materials
bon1-2 was isolated from the Wisconsin Knockout Facility by PCR-based screening following the procedures instructed by the facility (http://www.biotech.wisc.edu/Arabidopsis/). The T-DNA insertion site in bon1-2 was identified by sequencing the PCR product amplified with a T-DNA border primer and a BON1-specific primer. T-DNA insertion lines for genes in the RPP5 cluster and for EDS1 were isolated from the SalkT-DNA line collection (http://signal.salk.edu/cgi-bin/tdnaexpress) available through the ABRC. PCR-based genotyping was used to identify homozygous single mutants and double mutants.
Map-Based Cloning of MOB
The mapping population was created by crossing bon1-1 to bon1-2, and dwarf plants from the F2 progeny were chosen for mapping. Bulk segregation analysis was performed on pools of 50 plants with SSLP and CAPS markers available for Col and Ws (Lukowitz et al., 2000), and the MOB gene was located on the lower arm of chromosome IV. Subsequently, the MOB gene was placed between markers SC5 and AG. A larger population of mutant plants was surveyed with SC5 and AG, and recombinants between MOB and SC5 or MOB and AG were selected. Information on the existing markers can be found at The Arabidopsis Information Resource (http://www.arabidopsis.org/index.jsp). New markers we identified as polymorphic between Col and Ws are as follows. SC5, AccI digest, Col (0.55 kb) and Ws (0.3 kb and 0.25 kb); VRN2, Col (1.5 kb) and Ws (0.8 kb); and TGCAP2, DdeI digest, Col (0.5 kb and smaller fragments) and Ws (0.35 kb and smaller fragments).
Isolation of the SNW Gene
SNC1-specific primers ColB-1 (5′-ATATGGAGATAGCTTCTTCTTCTG-3′) and ColB-8 (5′-CTTGGAGAGAGGTTCAACCAC-3′) were used to amplify close to full length of the SNW gene from the Ws genomic DNA. Oligo(dT) (18-mer) was used for cDNA synthesis by reverse transcription reaction on RNA extracted from Ws. Primers ColB-1 and WsColB2(5′-CCTTGATGACACCTCTACAGTC-3′) were subsequently used to amplify the cDNA of SNW from the cDNA pools.
DNA Analysis and Genotyping
Standard molecular techniques were used (Sambrook et al., 1989). Genomic DNAs were prepared from a small piece of leaves for PCR genotyping as described (Konieczny and Ausubel, 1993). Thirty-five cycles were used to amplify the genomic DNA, and each cycle consists of 94°C for 30 s, 52°C for 30 s, and 72°C for 2 min, and the extension time varied according to the length of the PCR products.
RNA Analysis
Total RNAs were extracted using Tri Reagent (Molecular Research, Cincinnati, OH) from 3-week-old plants unless stated otherwise. Ten or twenty micrograms of total RNAs per sample were used for RNA gel blot analysis. PR1, EDS1, and PAD4 genes were amplified from the genomic DNA using PCR. SNC1-specific gene fragment was amplified from exon 1 or exons 1 and 2 of SNC1. The PCR products were then used for single-stranded DNA probe synthesis by 30 cycles of Taq polymerase reaction with only the antisense primers. Standard hybridization procedure was followed (Sambrook et al., 1989).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY510018 (partial SNW genomic sequence).
Acknowledgments
We thank G. Fink for discussions; P. Grisafi, H. Garnsey, and M. Callahan for technical assistance; K. Roberg-Perez for critical reading of the manuscript; and X. Li, X. Dong, E. Richards, B. Staskawicz, J. Dewdney, and the ABRC for mutant seeds.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jian Hua (jh299@cornell.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.020479.
References
- Aarts, N., Metz, M., Holub, E., Staskawicz, B.J., Daniels, M.J., and Parker, J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu, T., Hanzawa, Y., Ohtake, Y., Takahashi, T., Nishitani, K., and Komeda, Y. (1999). Expression of endoxyloglucan transferase genes in acaulis mutants of Arabidopsis. Plant Physiol. 121, 715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin, M.J., Muskett, P., Kahn, K., Feys, B.J., Jones, J.D., and Parker, J.E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J., and Staskawicz, B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Bowling, S.A., Guo, A., Cao, H., Gordon, A.S., Klessig, D.F., and Dong, X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6, 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes, D.C., Nam, J., and Dangl, J.L. (1998). The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. USA 95, 15849–15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudell, E.G., Caudell, J.J., Tang, C.H., Yu, T.K., Frederick, M.J., and Grimm, E.A. (2000). Characterization of human copine III as a phosphoprotein with associated kinase activity. Biochemistry 39, 13034–13043. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., Fengler, K.A., Yu, I.C., Lippok, B., Smith, R.K., Jr., and Bent, A.F. (2000). The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA 97, 9323–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creutz, C.E., Tomsig, J.L., Snyder, S.L., Gautier, M.C., Skouri, F., Beisson, J., and Cohen, J. (1998). The copines, a novel class of C2 domain-containing, calcium-dependent, phospholipid-binding proteins conserved from Paramecium to humans. J. Biol. Chem. 273, 1393–1402. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Dietrich, R.A., Delaney, T.P., Uknes, S.J., Ward, E.R., Ryals, J.A., and Dangl, J.L. (1994). Arabidopsis mutants simulating disease resistance response. Cell 77, 565–577. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Gendall, A.R., Levy, Y.Y., Wilson, A., and Dean, C. (2001). The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107, 525–535. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2001). Genes controlling expression of defense responses in Arabidopsis—2001 status. Curr. Opin. Plant Biol. 4, 301–308. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., Ostin, A., Sandberg, G., Romano, C.P., and Estelle, M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 7197–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday, K.J., Salter, M.G., Thingnaes, E., and Whitelam, G.C. (2003). Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J. 33, 875–885. [DOI] [PubMed] [Google Scholar]
- Heck, S., Grau, T., Buchala, A., Metraux, J.P., and Nawrath, C. (2003). Genetic evidence that expression of NahG modifies defence pathways independent of salicylic acid biosynthesis in the Arabidopsis-Pseudomonas syringae pv. tomato interaction. Plant J. 36, 342–352. [DOI] [PubMed] [Google Scholar]
- Heil, M., and Baldwin, I.T. (2002). Fitness costs of induced resistance: Emerging experimental support for a slippery concept. Trends Plant Sci. 7, 61–67. [DOI] [PubMed] [Google Scholar]
- Holt, B.F., III, Boyes, D.C., Ellerstrom, M., Siefers, N., Wiig, A., Kauffman, S., Grant, M.R., and Dangl, J.L. (2002). An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Dev. Cell 2, 807–817. [DOI] [PubMed] [Google Scholar]
- Hua, J., Grisafi, P., Cheng, S.H., and Fink, G.R. (2001). Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev. 15, 2263–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan, N., and McNellis, T.W. (2003). Regulation of Arabidopsis COPINE 1 gene expression in response to pathogens and abiotic stimuli. Plant Physiol. 132, 1370–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan, N., Siani, J.M., and McNellis, T.W. (2001). A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell 13, 2225–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.S., and Delaney, T.P. (2002). Arabidopsis SON1 is an F-box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell 14, 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Li, X., Clarke, J.D., Zhang, Y., and Dong, X. (2001). Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol. Plant Microbe Interact. 14, 1131–1139. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Serino, G., Deng, X.W., and Dinesh-Kumar, S.P. (2002). Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell 14, 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, S.P., and Woodward, F.I. (1988). Plants and Temperature. (Cambridge, UK: Society for Experimental Biology).
- Lukowitz, W., Gillmor, C.S., and Scheible, W.R. (2000). Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123, 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Holt, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754. [DOI] [PubMed] [Google Scholar]
- Malamy, J., Hennig, J., and Klessig, D.F. (1992). Temperature-dependent induction of salicylic acid and its conjugates during the resistance response to Tobacco mosaic virus infection. Plant Cell 4, 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G.B., Bogdanove, A.J., and Sessa, G. (2003). Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54, 23–61. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R.W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15, 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R., Shulaev, V., and Lam, E. (1995). Coordinated activation of programmed cell death and defense mechanisms in transgenic tobacco plants expressing a bacterial proton pump. Plant Cell 7, 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina, A., Volrath, S., Guyer, D., Maleck, K., Ryals, J., and Ward, E. (1999). Inhibition of protoporphyrinogen oxidase expression in Arabidopsis causes a lesion-mimic phenotype that induces systemic acquired resistance. Plant J. 17, 667–678. [DOI] [PubMed] [Google Scholar]
- Noel, L., Moores, T.L., van Der Biezen, E.A., Parniske, M., Daniels, M.J., Parker, J.E., and Jones, J.D. (1999). Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11, 2099–2112. [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Coleman, M.J., Szabo, V., Frost, L.N., Schmidt, R., van der Biezen, E.A., Moores, T., Dean, C., Daniels, M.J., and Jones, J.D. (1997). The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell 9, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Poethig, R.S. (2003). Phase change and the regulation of developmental timing in plants. Science 301, 334–336. [DOI] [PubMed] [Google Scholar]
- Rizo, J., and Sudhof, T.C. (1998). C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 273, 15879–15882. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sheldon, C.C., Finnegan, E.J., Rouse, D.T., Tadege, M., Bagnall, D.J., Helliwell, C.A., Peacock, W.J., and Dennis, E.S. (2000). The control of flowering by vernalization. Curr. Opin. Plant Biol. 3, 418–422. [DOI] [PubMed] [Google Scholar]
- Shirano, Y., Kachroo, P., Shah, J., and Klessig, D.F. (2002). A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14, 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes, T.L., Kunkel, B.N., and Richards, E.J. (2002). Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 16, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes, T.L., and Richards, E.J. (2002). Induced instability of two Arabidopsis constitutive pathogen-response alleles. Proc. Natl. Acad. Sci. USA 99, 7792–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya, G., Silhavy, D., Molnar, A., Havelda, Z., Lovas, A., Lakatos, L., Banfalvi, Z., and Burgyan, J. (2003). Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 22, 633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D., Traw, M.B., Chen, J.Q., Kreitman, M., and Bergelson, J. (2003). Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423, 74–77. [DOI] [PubMed] [Google Scholar]
- van der Biezen, E.A., Freddie, C.T., Kahn, K., Parker, J.E., and Jones, J.D. (2002). Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29, 439–451. [DOI] [PubMed] [Google Scholar]
- Van der Biezen, E.A., and Jones, J.D. (1998). Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23, 454–456. [DOI] [PubMed] [Google Scholar]
- Viswanathan, C., and Zhu, J.K. (2002). Molecular genetic analysis of cold-regulated gene transcription. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 877–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham, S., Dinesh-Kumar, S.P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: Similarity to toll and the interleukin-1 receptor. Cell 78, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C., Dewdney, J., Wu, G., and Ausubel, F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Williams, S.C., Hinshelwood, J., Perkins, S.J., and Sim, R.B. (1999). Production and functional activity of a recombinant von Willebrand factor-A domain from human complement factor B. Biochem. J. 342, 625–632. [PMC free article] [PubMed] [Google Scholar]
- Xiao, S., Brown, S., Patrick, E., Brearley, C., and Turner, J.G. (2003). Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15, 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka, K., Kachroo, P., Tsui, F., Sharma, S.B., Shah, J., and Klessig, D.F. (2001). Environmentally sensitive, SA-dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J. 26, 447–459. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Goritschnig, S., Dong, X., and Li, X. (2003). A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15, 2636–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]