Preface
The emerging field of computational morphodynamics aims to understand the changes that occur in space and time during development by combining three technical strategies: live imaging to observe development as it happens, image processing and analysis to extract quantitative information, and computational modelling to express and test time-dependent hypotheses. The strength of the field comes from the iterative and combined use of these techniques, which has provided important insight into plant development.
One challenge increasingly faced by developmental biologists is to understand dynamic biological processes at high spatial and temporal resolution. Time is particularly difficult to resolve because most traditional techniques achieve high spatial resolution by sample fixation, thereby preventing continued observation. Live imaging, which we define as time-lapse microscopic imaging of the same living biological sample over a defined period of time, circumvents this problem1. Live imaging has recently advanced2, 3. Today, full three-dimensional live imaging at cellular resolution is achieved by observing tissues with fluorescent proteins and stains while imaging the organism every few hours using a confocal microscope4 (Supplemental Figure 1). This method can be used to determine how cells grow and divide, to visualize patterns and changes in gene and protein expression, and to measure cellular responses to perturbations such as cell ablations and transient gene expression.
The resultant large data sets require sophisticated analysis. Computational image processing (Box 1) can automatically detect features of interest in the images, while tracking quantitative data about those features over time.
Box 1 Image processing.
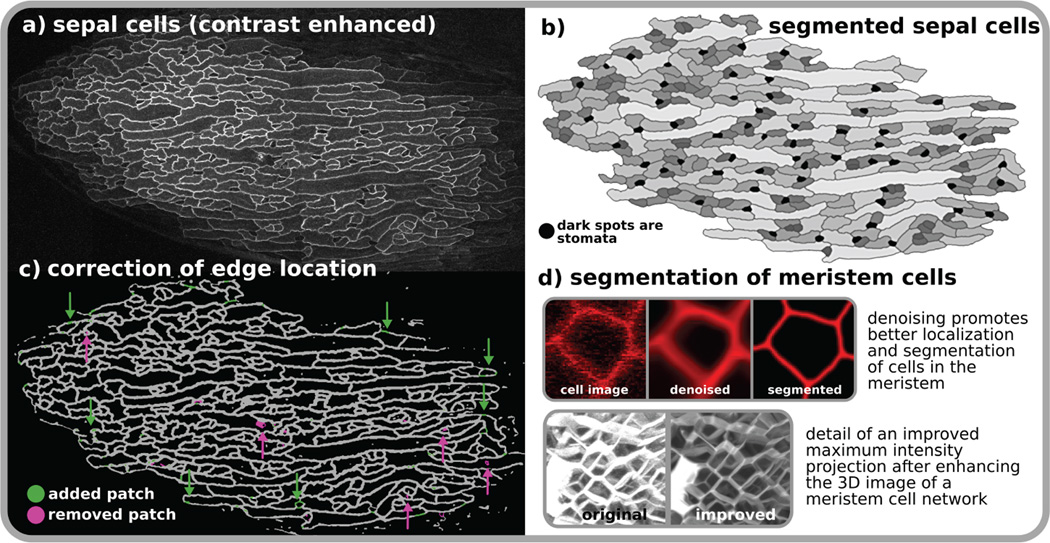
Post-acquisition image processing and analysis provide cell measurements that can assist in the formulation and validation of computational models. Two important needs in computing shape, size, connectivity and position of cells from microscope images are the generation of high quality images and the development of robust feature extraction algorithms. Noise, contrast, and spatial and temporal resolutions are some image quality attributes that largely control the development of image-processing algorithms and the effectiveness of their results. Acquired optical images are typically contaminated with shot noise (see the figure, part a); this is especially accentuated in live imaging, in which reduced light intensities are applied to avoid damaging live tissues and cells. Low image contrast, which mainly occurs in deeper parts of the tissue, hinders the separation of regions of interest from the image background. Poor spatial (few slices per 3D image) and temporal (few images over time) resolutions are detrimental to accurately resolving the true geometry and lineage of cells during development using a cell tracking software. Reducing these image aberrations leads to enhanced image quality for visualization and promotes improvements in the delineation of regions of interest (segmentation) (see the figure, parts b, d). Automatic image segmentation is a fundamental problem in image processing. Unfortunately, most methods usually produce partially good results with missing regions and edges that are sometimes difficult to automatically detect and correct. One effective approach is the semi-automatic segmentation path61, in which faulty results are eliminated by human computation55 (manual editing which can be done by crowds for a fee or free of charge) (see the figure, part c). In the future interactive computer systems may be developed where users can intervene in the segmentation process to easily and quickly repair mistakes produced by automatic programs. With the right set of interactive tools one should be able to mass distribute data for corrections and scale up semi-automatic solutions to large data sets.
Through live imaging, image processing and experimentation, biologists develop models. Computational models formalize these hypotheses by expressing them as a set of equations or rules that can be simulated using a computer5–7 (Table 1). Computer simulations allow a quantitative comparison of the model to the data. These simulations also allow visualization of nonintuitive outcomes of complex interactions and feedback loops. Thus, computational models allow narrowing down a diverse set of hypotheses to a few plausible ones that can be tested experimentally.
Table 1.
Modelling methodologies:
| Cell Circuit Dynamics |
Describe the rates of change of interacting gene and
signaling network components. Depending on the available information they are simulated in different ways: |
|---|---|
| 1 Boolean5, 18 | Individual genes are described as ON/OFF; truth tables and
state transition graphs describe gene regulatory rules. A very useful method to describe network dynamics with insufficient information (molecular mechanisms or data). However, one cannot obtain detailed dynamics of gene regulatory functions. |
| 2. Differential Equations5 |
Gene regulation described by biophysically motivated rate
laws, very often represented by Michaelis-Menten type of functions. Several analytical techniques are available for probing nonlinear network dynamics as well as for optimizing parameters. However, some parameters may have to be guessed. |
| 3. Stochastic5 | Simulations use probabilities based upon reaction rates to
decide which chemical transitions occur. Novel phenomena due to inherent stochasticity of gene regulation and signaling illuminate new principles; however, for large networks, simulations can be prohibitively computationally intensive. |
|
Mechanical Forces in Living Tissues |
The forces within and between cells which ultimately
are responsible for shaping the organ, are described by various frameworks: |
| 1. Spring Models5 |
Cell walls are described by springs, which connect to
other cells at vertices. Equilibrium is obtained by minimizing the total elastic energy. This simplified description of cells works well with cell division and growth, but it lacks resolution of finer cell wall details. |
| 2. Finite Element Methods12, 56, 57 |
Discretization of a tissue in terms of elements using
various geometries, which then implement the rules of elasticity theory. This method provides a detailed description of the elastic properties of cells, but is computationally intensive and cannot easily be modified for growing and dividing cells. |
| 3. Cellular Potts Methods42 |
Cells are described as a collection of similar spins,
which interact with each other and spins of neighboring cells. A Monte-Carlo scheme is employed to minimize the energy of the system and arrive at the equilibrium configuration. Used successfully in many biological cases; however, not adapted completely to plant systems, which require non-migrating cells. |
| 4. Subcellular Element Method65, 66 |
Coarse-grained description of cells in terms of
interacting particles, which move by sensing forces from the neighboring particles. This method has excellent spatial resolution, but could be computationally intensive for multicellular systems. |
Computational morphodynamics refers to the combined use of live imaging, image processing and computational modelling to understand morphogenesis5. In our opinion, the strength of this emerging field comes from the iterative and combined use of these techniques to understand how the dynamics of molecular signalling, cellular geometry, and mechanics dictate development.
The field of computational morphodynamics has emerged from work with many model organisms. Insight has been gained into Dictyostelium morphogenesis8, planar cell polarity in Drosophila9, 10, assembly of the contractile ring for cytokinesis in yeast11, growth of pollen tubes12, and the pattern of leaf vasculature13, 14 among many others. In this Opinion article, we focus on the work done in plants, specifically on what has been learned through the integration of live imaging with computational modelling to better understand the role of molecular signaling, cellular geometry and biomechanics in the multicellular morphogenesis of plant meristems and lateral organs.
Regulatory networks in time and space
In our first section, we examine the conclusions reached by applying computational modelling and imaging to three molecular signaling networks underlying plant morphogenesis. These examples illustrate the different scales at which modelling and imaging can be combined: cell, tissue, and whole plant.
Root hair patterning
The Arabidopsis root epidermis is patterned in alternating cell files of specialized hair cells, trichoblasts (H), and non-hair cells, atrichoblasts (N). This specification is controlled both genetically and spatially (Figure 1a). A transcription factor complex including TRANSPARENT TESTA GLABRA (TTG)/GLABRA3 (GL3)/ENHANCER OF GLABRA3 (EGL3) interacts either with the transcription factor WEREWOLF (WER), to form an active transcriptional complex specifying N cell fate, or CAPRICE (CPC), to repress WER expression and promote H cell fate. Imaging studies have demonstrated both CPC and GL3 move from their sites of synthesis into the adjacent cell. CPC is expressed in the N cell and moves into the H cell where it is believed to repress WER, whereas GL3 is expressed in the H cell and accumulates in the nucleus of the N cell. In addition to this genetic network, N and H cell fate is determined by the position of an epidermal cell in relation to the underlying cortex layer. Epidermal cells positioned over the junction of two cortical cells adopt the H cell fate (trichoblasts) while those directly above one cortical cell develop into N cells (atrichoblasts)15. Genetic evidence indicates a transmembrane receptor protein kinase, SCRAMBLED (SCM), expressed in epidermal cells senses a signal derived from the cortical layer to repress WER in the epidermis16. It was postulated that the combined repression of WER by SCM and CPC results in specification of H cell fate while the functional TTG/GL3/EGL3/WER complex in the adjacent cell specifies N cell fate. Based on these observations two computational models have been developed to elucidate this complex interaction between spatial position and the underlying genetic network.
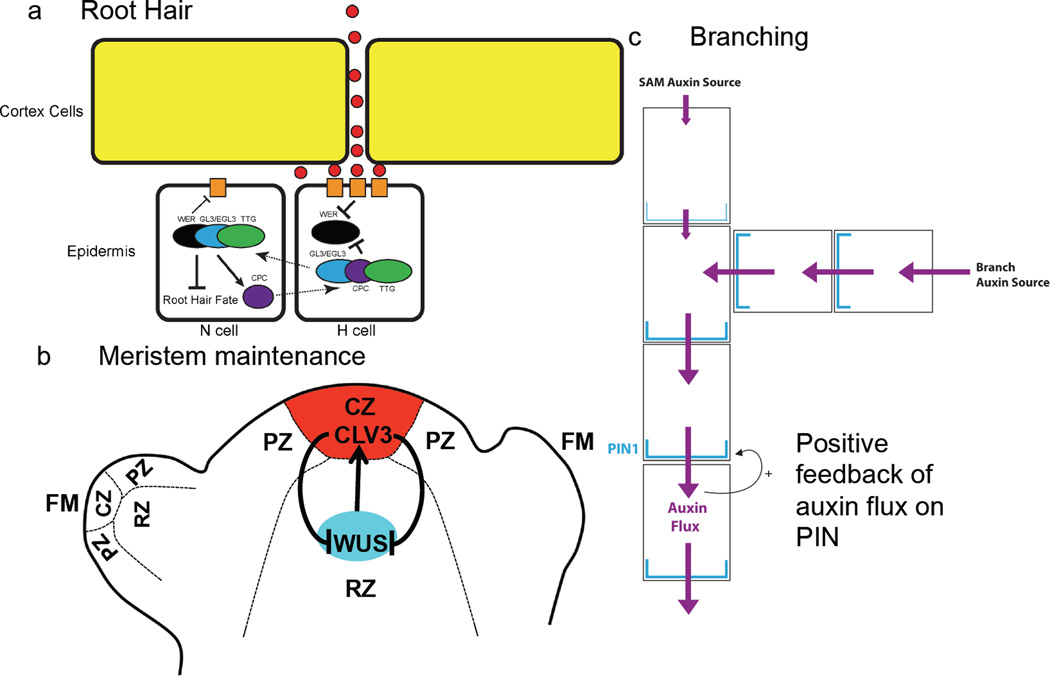
Figure 1. Gene regulatory networks?
aThe patterning of hair cell (H) and non-hair cells (N) in the root involves both spatial signaling through the SCRAMBLED receptor and activation or repression of transcriptional complexes that are influenced by the movement of proteins from one cell to another. H cell development is initiated by the combined effect of WER repression by activation of SCRAMBLED signaling by an unknown signal and CAPRICE (CPC) activity.
bThe SAM can be divided into the central zone (CZ), the peripheral zone (PZ) and the rib meristem (RM)62, 63. The CZ, comprises of a pool of pluripotent stem cells. The PZ is seated on the flanks of the SAM where new lateral organs are initiated. WUSCHEL (WUS) is expressed exclusively in the organizing center (RM) (blue) and is required to produce an unknown signal to specify the stem cell identity in the overlying cell layers. Stem cells in the CZ (red) secrete the CLAVATA3 (CLV3) small polypeptide, which activates a signalling cascade that limits the WUS transcription in the RM64. Thus, the negative feedback regulatory circuitry composed of WUS and CLV3 forms a self-correcting mechanism to maintain the stem cell homeostasis in the SAM.
cThe branching pattern of the plant can be understood by modelling the competition between auxin sources at the tip of each branch to transport auxin through regions of the plant stem (represented as large boxes). A model in which the flux of auxin (purple) positively feeds back on the amount of PIN1 auxin efflux transporter (blue) automatically establishes the competition between branches and reproduces the branching pattern of both wild type and mutant plants.
Benítez et al., developed a model that was based upon the assumption of WER self-activation. In their model stimulations, striped patterns of N and H cells were only obtained when the SCM signal activated the WER complex17. However, there was no evidence to support the idea of local WER self-activation. This self-activation model was recently challenged by a model developed by Savage et al. who proposed two different models that centered on the mode of regulation of WER18. The first assumed local WER self-activation with CPC repressing WER indirectly (local WER self-activation model, similar to Benítez), while the second model did not include WER self-activation but assumed uniform WER transcription that was repressed by both CPC and SCM activity (mutual support model). In model simulations run in a cpc mutant background the mutual support model closely matched the experimental observation of increased WER expression in cpc trichoblast (H) cells19. Local WER self-activation was ruled out experimentally by determining WER expression is unchanged in wild type or wer backgrounds. The mutual support model also correctly predicted wild type WER expression pattern in the gl3/egl3 double mutant background. These data provide direct experimental support for the mutual support model, ruling out WER self-activation as a mechanism for epidermal patterning in the root.
Shoot apical meristem maintenance
At the tissue level, the use of live imaging and computational modelling have recently provided some novel insights into our understanding of the mechanisms that regulate the balance between stem cell renewal and differentiation at the shoot apical meristem (SAM). The SAM gives rise to the above ground structures of flowering plants20. Stem cell maintenance in the SAM involves the function of the CLAVATA1 (CLV1) receptor kinase, its ligand CLAVATA3, and the transcription factor WUSCHEL (Figure 1b). The genes are expressed in a defined spatial pattern within the SAM: CLAVATA3 is solely expressed in the central zone, a group of pluripotent stem cells in the center of the SAM, while CLV1 and WUS are expressed directly below the CZ in the rib meristem (RM). Activation of CLV1 signaling in the RM by CLV3 results in repression of WUS, which is required for the production of a non-cell autonomous signal from the RM to maintain the pluripotent CLV3-expressing stem cells in the CZ. This feedback loop is the basis for the current model to explain the balance between stem cell renewal and differentiation. Several computational models have explored how the WUS and CLV3 gene expression domains are localized and interact within a static 2D longitudinal section of the meristem21–23. Although these models achieve spatial resolution, they do not address the dynamics of growth.
Traditional molecular genetics has been successful in identifying the key components of the signaling circuit that regulates stem cell numbers in the SAM, but does not address temporal aspects of development4. Terminal mutant phenotypes often result from the accumulation of defects over time. For example why is the shoot apical meristem enlarged in a clv3 loss of function mutant? Enlargement could occur due to faster cell divisions, slower exit of cells from the CZ or increases in the WUS-mediated CZ-inducing signal. Surprisingly, live imaging while transiently inactivating CLV3 with RNA interference, showed that none of these possibilities were correct. Instead, CZ expansion was caused by an immediate respecification of neighboring PZ cells on the boundary into CZ cells24. This observation would not have been made through static imaging of traditional genetic mutants, suggesting that live imaging of transient perturbations is an important strategy to visualize simultaneous changes in cell division and gene expression patterns. Geier et al. included these insights in a population model describing the interactions between these different cell types, in which they allowed cells to both proliferate and switch fate25. One of the challenges for the future is to integrate the genetic networks that regulate spatial patterning and respecification into a 3D growing template.
Branching
Plant branches are formed by the outgrowth of buds. Activation or repression of bud outgrowth integrates environmental inputs (such as light and nutrients), developmental signals (such as hormones and age) and genetic controls. The plasticity of bud outgrowth plays an essential role in determining plant architecture, crop yield and biomass production, all of which are important in agriculture.
To determine the timing and location of branch outgrowth in Arabidopsis, individual plants were photographed daily to measure growth26. These data have been used to build a descriptive computational model that reproduce the plant architecture26; however, the model raises the question of what molecular mechanisms controls the order in which branches grow out.
Decades ago, bud outgrowth was shown to be repressed by the downward transport of the plant hormone auxin from the shoot apex. However, the mechanism of auxin mediated inhibition of bud outgrowth is complex and indirect27, 28. For example, mutants exhibiting increased branch outgrowth can have lower (tir3), the same (axr1) or higher (max4) levels of auxin transport than wild type29. Without computational modelling, it is hard to give a plausible explanation for such contradictory observations. Prusinkiewicz et al. have developed a computational model that can simulate the branching pattern of wild type Arabidopsis plants based on the assumption of positive feedback between the flux of auxin through a region of the plant and the concentration of the auxin efflux-carrier PINFORMED1 (PIN1), which transports auxin30 (Figure 1c). Altering the parameters of this model is sufficient to produce increased branching similar to the mutant phenotypes observed. Thus, comparison of model simulations with branching pattern revealed by time-lapse imaging of whole plants and with mutant phenotypes led to a plausible mechanism for the complex patterns of branch outgrowth observed in nature.
Growth at the cellular level
While regulatory networks control developmental decisions, it is the growth and division of cells that actually leads to morphogenesis. We next illustrate how the use of computational morphodynamics has shown that the growth and division patterns of cells determine the morphology of a whole tissue.
Morphogenesis of the snapdragon petal
Analysis of the growth and cell division of plants has traditionally relied on measuring the mitotic index and analyzing the size and shape of clonal sectors, patches of marked sibling cells derived from a single progenitor cell31–33. To generate a clonal sector a random cell is marked visibly through an induced genetic change early in the development of the organ, for example, through the excision of a transposon near a gene controlling red pigmentation. The rate and direction of growth is inferred from the size and shape of the patch of progeny cells determined in the mature organ. However, sector analysis does not reveal which random cell was marked, when the progeny cells divided or how they give rise to the final patch.
In the absence of live imaging, sector analysis together with imaging of the 3D shape of organs using optical projection tomography formed the biological basis for a computational model describing how simple petal primordia grow to form the complex 3D shapes of the mouth of the snapdragon (Antirrhinum) flower34. In this model, the tissue was treated as a continuous sheet of material that can grow in 3D. A quantitative analysis of the in vivo shape of wild-type and mutant petals revealed the contributions of the dorsoventral polarity genes to the growth of each region of the petal35. These data formed the basis for the hypothesis that dorsoventral genes control local growth rates. However, a model based on this initial hypothesis was unable to replicate either the exact shape of the flower or the pattern of sectors generated in the real petals. When the model was revised such that the dorsoventral genes control both local growth rates and the activity of hypothetical organizers of tissue polarity, the model reproduced the snapdragon mouth of both wild-type and dorsoventral-mutant flowers, as assessed by comparing the result with actual petal shapes and sectors34, 35. This model enhances our understanding of how 3D shapes are generated during morphogenesis, but the precise cellular basis of these shapes remains unknown.
Live imaging of cell division and growth
To determine the cellular basis of growth, live imaging has been used to track cell lineages and record the cell division patterns in the SAMs, root meristems, floral meristems, moss buds, moss leaves, and sepals36–40 (Supplemental Box 1). These live-imaging experiments confirm the conclusions from sectoring, but they extend beyond this, with actual lineage traces showing the timing and orientation of all cell divisions from the progenitors onwards. One of the unique insights emerging is that the timing of cell division in many tissues is irregular36, 38, 39 and that this contributes in an important way to cellular patterning.
Cell division controls in patterning of sepals
For example, a computational morphodynamics approach has shown that irregular timing of cell division contributes to the cellular patterning is in sepals 39. Sepals are the leaf-like floral organs that envelop the developing bud (Figure 2a). Arabidopsis sepals are characterized by the presence of highly elongated giant cells stretching about a fifth of the length of the sepal in the outer epidermis (Figure 2b–c). The giant cells are interspersed between cells in a range of sizes, which raises the question: how is a range of cell sizes generated when cells are constrained by being tightly bound together by their cell walls?
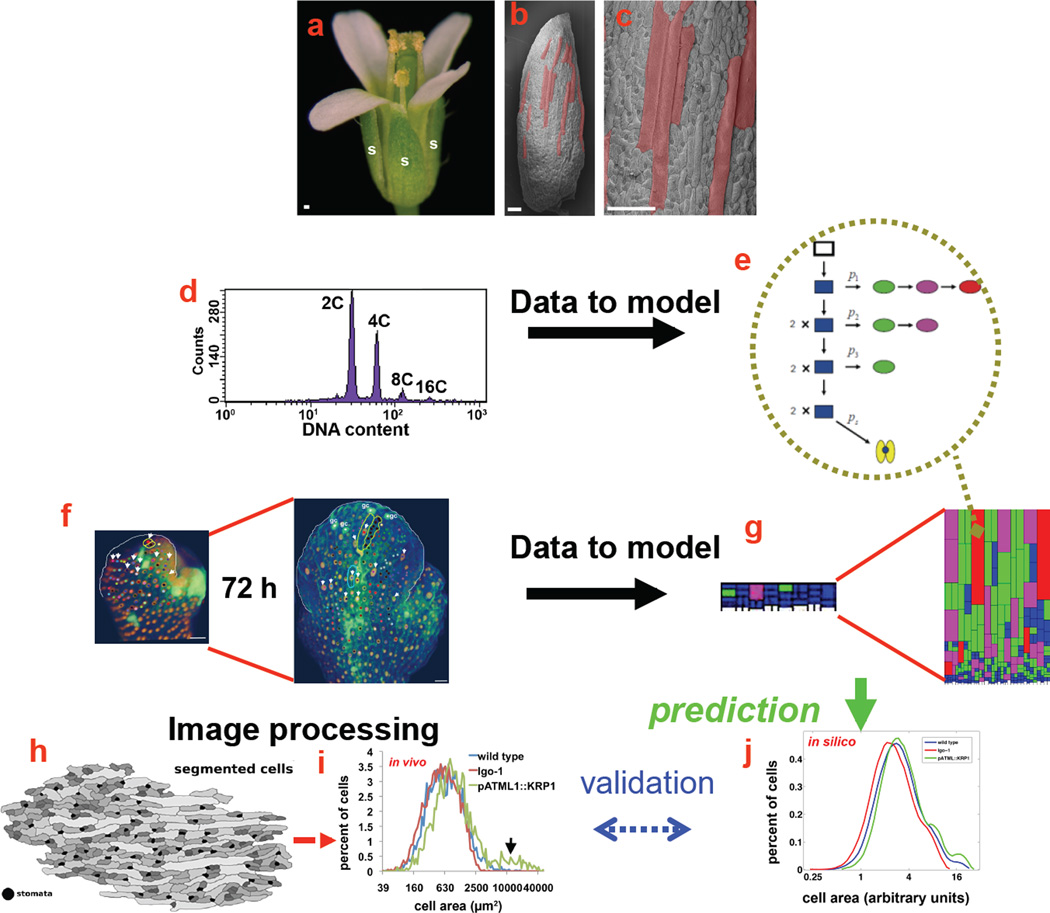
Figure 2. The iterative process of imaging, image processing and modelling in sepal patterning.
A–c The sepals (marked s) of a wild-type flower have a pattern of diverse cell sizes, ranging from giant cells (false-coloured pink) to the small cells. Scale bars: 100 µm.
d The ploidy of cells was measured using flow cytometry to determine how many rounds of endoreduplication (a cell cycle including DNA replication without division) sepal cells had undergone.
e The ploidy data formed the basis for a population model, in which cells can divide (arrow down) or endoreduplicate (arrow right). The model was used to predict the probability (p) with which each cell will enter endoreduplication at each time (subscript on p). The color represents the ploidy of the cell: blue = 2C, green = 4C, magenta = 8C, red = 16C, and yellow = stomata.
f Live imaging shows the growth of the sepal over 72 hours and the lineages of the cells (marked with the same coloured dots). Analysis of the imaging revealed that both the timing of endoreduplication and the timing of cell division are highly variable. Scale bars: 20 µm
g–A geometric growth model of sepal giant cell development based on this timing data from live imaging (f) and probability of endoreduplication as determined by the population model (e) predicts the diversity of cell areas. The color represents the ploidy of the cell: blue = 2C, green = 4C, magenta = 8C and red = 16C.
h–j A crucial step in validating the model was using image processing (h) to obtain the distribution of cell areas in vivo (i), which was compared with the model predictions (j). The model agreed well with measured areas for wild type (blue). In addition, perturbations of the model matched genetically altered plants with too many (green) or too few (red) giant cells.
Modified with permission from Roeder et al. 201039.
Cells can become enlarged through endoreduplication, in which the cells replicate their DNA but fail to divide. More than a decade ago, it was proposed that diverse cell sizes would be produced if cells enter endoreduplication at different times41. This hypothesis was impossible to test without live imaging. Recently, tracing the cell lineages in the developing sepal epidermis confirmed that large cells enter endoreduplication early and small cells enter endoreduplication later39 (Figure 2f). Based on these observations, a growing sepal was computationally modelled as an expanding template of cells, each of which could divide or enter endoreduplication with certain probabilities. These probabilities were estimated from the data (Figure2d–e). However, a simple model of this hypothesis was unable to match the actual in vivo cell size distribution (as measured using image processing (Box 2)).
A modified hypothesis arose from a detailed examination of cell divisions by live imaging, which revealed that the length of the cell cycle is highly variable and also correlated with cell size. A computational model in which cells both have a certain probability of endoreduplicating and a random cell cycle length could reproduce the observed cell areas, suggesting variability in cell division is a plausible mechanism for generating cell size diversity (Figure 2g–j). Furthermore, by changing the probability of endoreduplication in the first cell cycle, the model could predict the cell size distribution for loss-of-function mutants with too few giant cells and gain-of-function mutants with too many giant cells, validating the model (Figure 2i–j). The conclusion that the stochasticity in the timing of cell division and in the decision to endoreduplicate together produce a range of cell sizes could be reached only by combining live imaging, modelling and image processing39.
Mechanics influences organ initiation
The previous examples have discussed how gene regulatory networks and growth contribute to patterning and morphogenesis; however, these processes take place in a physical framework where mechanical forces between cells can influence the final form of the tissue. In this section, we show that a computational morphodynamics approach has shaped our understanding of how mechanical signaling impacts organ initiation.
Phyllotaxis and rhizotaxis by polarized auxin transport
In both the shoot and root, local maxima in the concentration of the plant hormone auxin specify the location of organ outgrowth. Modelling has shown that polarized auxin transport through the auxin efflux carrier PINFORMED (PIN) proteins can generate these localized auxin maxima 42–44.
In the root, lateral root primordia are initiated from the twin files of pericycle cells (specialized cells, located outside the vascular tissues) at irregular intervals. Lateral root formation commonly correlates with higher than average levels of auxin perception, especially the outer convex side of the root45, 46: reduction in auxin perception or transport in the root decreases the density of lateral roots, and activation of auxin synthesis in pericycle cells initiates the formation of lateral root primordia 47–49.
Similarly, early biochemical and genetic experiments showed that transport of the hormone auxin is necessary for the specification of organs in the SAM (Okada et al, 1991). Experiments with the tomato SAM further confirmed that local auxin accumulation was both necessary and sufficient for the induction of organ growth around the SAM50. But by the end of the 20th century it was still not known how auxin acted in the SAM to form organs. Subsequent live-imaging experiments showed that the polarization of the auxin efflux carrier PIN1 in the SAM epidermis resulted in local auxin maxima where organs would later arise 51–54. These live-imaging data were used to construct two separate computer models of auxin-based patterning in the shoot. In both models the observed auxin maxima were generated when PIN1 was polarized toward the neighbouring cell with the highest concentration of auxin. However, these models required hypothetical mechanisms whereby cells needed to sense the concentration of auxin in their neighbours 44, 55 The mechanism by which this polarization could occur remained a mystery until more recent examination of tissue mechanics using computational morphodynamics.
Mechanics orients cell polarity
Biologists have long understood that the final form of a tissue is connected with specific genetic programmes. But how the mechanical properties of those tissues affect their final form was hard to study due to inherent technical limitations, such as measuring cellular stress within a tissue. In plants this is particularly relevant because cells adhere to their neighbours; therefore, any local mechanical change is propagated throughout the tissue. Recently, live imaging and computational modelling were used to test the hypothesis that mechanical properties of the SAM epidermis determine the positions of new organs56, 57. It has long been observed that the orientations of cell wall microfibrils align with the orientation of cellular microtubules, as microtubules serve as tracks for the enzymes involved in cellulose synthesis. Using a live imaging data set, a realistic SAM template was extracted from a confocal image stack for modelling simulations.
Computational analysis of stress patterns revealed that the direction of stress of each cell accurately predicted the microtubule orientations observed by live imaging56 (Figure 3a). Furthermore, the model could predict the circumferential reorientation of microtubules around a wound site following cell ablation (a mechanical perturbation), as observed by live imaging (Figure 3c). Likewise, live imaging showed that PIN1 polarized towards anticlinal cell walls parallel to the microtubule arrays, which suggested that stress patterns have a role in PIN polarization, and therefore in auxin transport directions57 (Figure 3a–c). Simulations of an updated model in which PIN1 localizes towards the most stressed walls were able to produce a phyllotactic pattern. Thus, live imaging and modelling were instrumental in reaching the conclusion that in plants mechanics is a plausible mechanism for coordinating the extent of growth (via auxin) with the direction of growth (via microtubules).
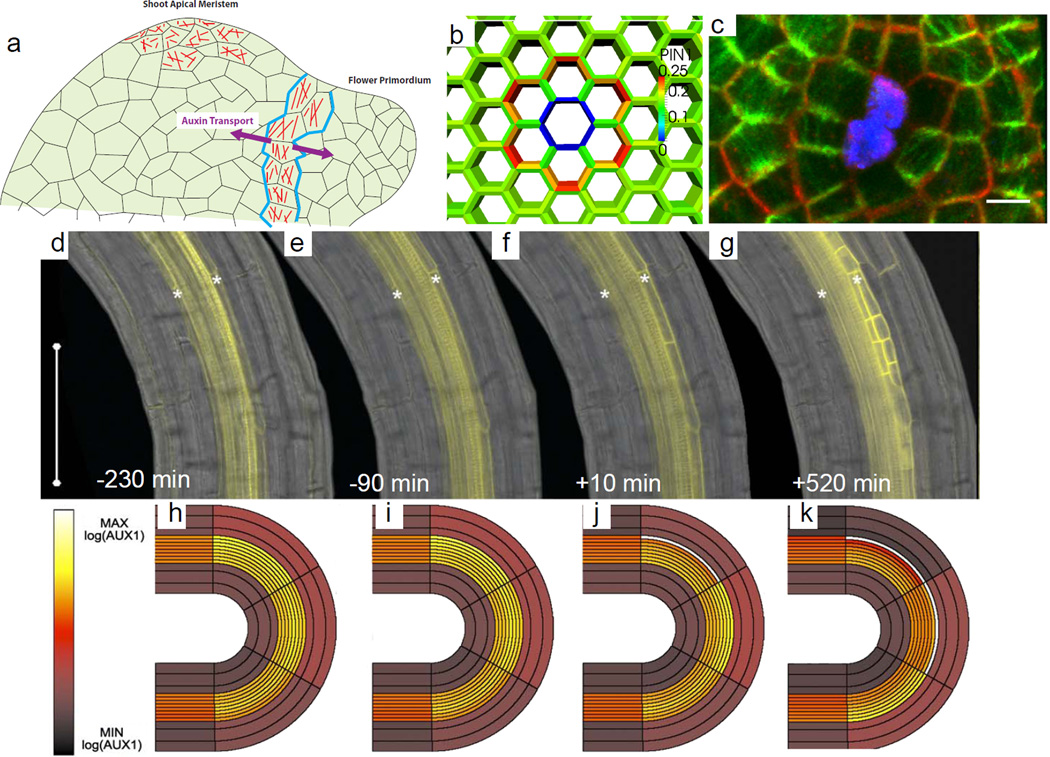
Figure 3. Mechanics.
a At the boundaries between the shoot apical meristem and the flower primordia, the anisotropic mechanical stress of the cells orients the microtubules (red) circumferential to the primordia. The stress also polarizes PIN1 (blue) such that auxin is transported both towards the new organ as well as towards the site of the next organ formation. By contrast, in the center of the meristem, patterns of both PIN and microtubules are more irregular, reflecting isotropic stress patterns.
b–C A model of the change in the mechanical stress pattern after the ablation of a cell (blue) predicts that PIN1 (red) will relocalize to point away form from the ablated cell. Live imaging (c) shows that both PIN1 (red) and microtubules (green) reorient to point away from the ablation site (blue). Scale bar: 5 µm. Reproduced with permission from Heisler et al., 201057.
d–g Live imaging of AUX1 (yellow) accumulation in the outer pericycle cell layer (asterisks) at sites of curvature before and after the first cell division (time 0) marking the formation of a lateral root primordia. Reproduced with permission from Laskowski et al., 200858.
h–k Modelling results based on live imaging show cell elongation can cause an increase in auxin concentration when positive feedback of AUX1 is added to the model parameters. Reproduced with permission from Laskowski et al., 200858.
The observation that lateral root primordia are initiated on the outer surfaces of root curvatures implies a role for mechanical stresses in lateral root initiation as well. A recent study coupled live imaging with computational modelling and showed that alterations in cell length at the sites of curvature can induce auxin maxima that are required for lateral root initiation58. Imaging of the expression levels and patterns of PIN proteins in the different developmental zones of the root was used to parameterize a model of root auxin flux. This showed that the organization of PIN in the root leads to the formation of local auxin reflux loops that are further reinforced by the flux of auxin through the root tissue at sites of curvature. Live imaging of auxin transporter 1 (AUX1; which imports auxin into cells) showed that it accumulates on the outer facing membrane face at sites of root curvature in the pericycle layer (Figure 3d–g). Updating the model to include a positive feedback loop for AUX1 in auxin accumulation indicated that auxin peaks become localized to a few outer pericycle cells that correspond to where in silico roots are curved in model simulations (Figure 3h–k). These studies reveal that mechanics is involved in producing local auxin maxima and therefore in generating developmental patterns in both the root and the shoot.
Perspectives
The question of how cellular signals interact with tissue mechanics has been difficult to address with traditional methods; therefore, morphogenesis is poorly understood. Computational morphodynamics is a way forward in integrating aspects of physics, chemistry and computer science with biology to understand how genes regulate the behaviour of cells, how cells interact to give rise to tissues, and how tissues are organized into organs within the final form of an organism.
Although the computational morphodynamic approach is still in its infancy, its use in both plants and animals is expanding6. Many studies have used either live imaging or computational modelling5–7 effectively, and the integration of these approaches has led to novel conclusions that could not have been reached otherwise. Computational modelling is helpful for examining systems with many variables or parameters that are difficult to directly measure. Live imaging reveals dynamic processes that cannot be fully understood by looking at fixed samples. Additionally, image processing is required to quantitatively understand the collected data in a robust and repeatable way. Through the course of this Opinion we have highlighted the insights gained from a few studies in plants where imaging, image processing, and computational modelling are combined.
The limitations of the computational morphodynamics approach are both technical and computational, but these limitations are being addressed by new methods. For example, new fluorescent sensors, probes and markers allow new systems such as small molecules, hormones gradients and protein-protein interactions to be imaged. New microscopes may allow access to cells deeper in tissues, and higher resolution. Novel image processing strategies allow automatic or semi-automatic identification and tracking of cells such that greater quantities of increasingly complex data can be used40, 59, 60. New modeling methodologies may allow more accurate models of the properties of growing and dividing cells. While technology will no doubt provide us with better tools, we must understand that technology alone will not deliver a complete and holistic understanding of development. Arriving at the ultimate goal of constructing “computable” plants and animals will also require cogent experiments and creative hypotheses; progress in computational morphodynamics needs both new technical approaches and new ideas.
Supplementary Material
Acknowledgements
We apologize to the numerous members of these fields whose work we could not include owing to space limitations. We thank E. Mjolsness, W. Li, Y. Zhou, and L. Ben-Ghaly for insightful comments. We acknowledge support from the Gordon and Betty Moore Cell Center at Caltech, National Institutes of Health grants F32GM090543 to P.T.T. and R01 GM086639 to E.M.M., and U.S. Department of Energy grant DE-FG02-88ER13873 and U.S. National Science Foundation grant IOS-0846192 to E.M.M.
Biographies
Adrienne Roeder is a postdoctoral scholar in biology at Caltech in the laboratory of Elliot Meyerowitz. Previously she was a graduate student with Martin Yanofsky at UC San Diego. Her interests are in using computational morphodynamics to determine the role of cell division in the simultaneous patterning and morphogenesis of plant organs.
Paul Tarr received his Ph.D. from the University of California at Los Angeles in 2008. He is currently a postdoctoral scholar in Elliot Meyerowitz's laboratory at the California Institute of Technology. His research focuses on function of plant hormones in the patterning and maintenance of the Arabidopsis shoot apical meristem.
Cory Tobin is a graduate student in the Biology Division at Caltech. He got his B.S. in Biochemistry from California Lutheran University. His thesis is on the mechanisms controlling phyllotaxis in Arabidopsis.
Xiaolan Zhang has been a postdoctoral scholar with Elliot Meyerowitz at the California Institute of Technology, Pasadena, California, USA, since September 2007. She carried out her doctoral work with Michael Scanlon and Kelly Dawe at the University of Georgia, Athens, GA, USA. She is interested in studying the interplay mechanisms among plant boundary formation, meristem maintenance and lateral organ initiation.
Vijay Chickarmane is senior research fellow, biology division, Caltech, in Elliot Meyerowitzs laboratory. Previously he held postdoctoral positions at Caltech, and at KGI, Claremont. His doctoral work is on theoretical physics, at IUCAA, Pune, India. His interests are in biophysics of cell growth and mathematical modeling of inter-cellular signaling and genetic networks.
Alexandre Cunha is a Computational Scientist with the Center for Advanced Computing Research at the California Institute of Technology. He is a member of Caltech's Center for Integrative Study of Cell Regulation where he works developing algorithms and tools for bioimaging processing and for computational plant simulation. He has a doctoral degree in Computational Science & Engineering from Carnegie Mellon University, Pennsylvania, USA, and has done postdoctoral work at the Center for Computational Biology at the University of California, Los Angeles, USA. His research interests are in fast and effective algorithms for image processing, geometry extraction from images, and variational modeling.
Elliot Meyerowitz has been a faculty member in the Division of Biology at the California Institute of Technology since 1980. His laboratory works on the developmental biology of plants, with a particular emphasis on the development of computational models of multicellular tissue growth and pattern formation from stem cell populations.
Contributor Information
Adrienne H. K. Roeder, Division of Biology 156-29, California Institute of Technology, 1200 E. California Blvd., Pasadena, CA 91125, aroeder@caltech.edu, (626) 395-6895, FAX (626) 449-0756
Paul T. Tarr, Division of Biology 156-29, California Institute of Technology, 1200 E. California Blvd., Pasadena, CA 91125, paultarr@caltech.edu, (626) 395-6895, FAX (626) 449-0756
Cory Tobin, Division of Biology 156-29, California Institute of Technology, 1200 E. California Blvd., Pasadena, CA 91125, ctobin@caltech.edu, (626) 395-4936, FAX (626) 449-0756.
Xiaolan Zhang, Division of Biology 156-29, California Institute of Technology, 1200 E. California Blvd., Pasadena, CA 91125, zhangxl@caltech.edu, (626) 395-8438, FAX (626) 449-0756.
Vijay Chickarmane, Division of Biology 156-29, California Institute of Technology, 1200 E. California Blvd., Pasadena, CA 91125, vchickar@caltech.edu, (626) 395-6895, FAX (626) 449-0756.
Alexandre Cunha, Center for Advanced Computing Research MC 158-79, California Institute of Technology, 1200 E. California Blvd., Pasadena, CA 91125, cunha@cacr.caltech.edu, (626) 395-8031.
Elliot M. Meyerowitz, Division of Biology 156-29, California Institute of Technology, 1200 E. California Blvd., Pasadena, CA 91125, meyerow@caltech.edu, (626) 395-6889, FAX (626) 449-0756
References
- 1.Reddy GV, Roy-Chowdhury A. Live-Imaging and Image Processing of Shoot Apical Meristems of Arabidopsis thaliana. Methods Mol Biol. 2009;553:305–316. doi: 10.1007/978-1-60327-563-7_15. [DOI] [PubMed] [Google Scholar]
- 2.Ball E. Cell division patterns in living shoot apices. Phytomorphology. 1960;10:377–396. [Google Scholar]
- 3.Dumais J, Kwiatkowska D. Analysis of surface growth in shoot apices. Plant Journal. 2002;31:229–241. doi: 10.1046/j.1365-313x.2001.01350.x. [DOI] [PubMed] [Google Scholar]
- 4.Reddy GV, Gordon SP, Meyerowitz EM. Unravelling developmental dynamics: transient intervention and live imaging in plants. Nat Rev Mol Cell Biol. 2007;8:491–501. doi: 10.1038/nrm2188. [DOI] [PubMed] [Google Scholar]
- 5.Chickarmane V, et al. Computational Morphodynamics: A Modeling Framework to Understand Plant Growth. Annu Rev Plant Biol. 2010;61:65–87. doi: 10.1146/annurev-arplant-042809-112213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oates AC, Gorfinkiel N, Gonzalez-Gaitan M, Heisenberg CP. Quantitative approaches in developmental biology. Nat Rev Genet. 2009;10:517–530. doi: 10.1038/nrg2548. [DOI] [PubMed] [Google Scholar]
- 7.Grieneisen VA, Scheres B. Back to the future: evolution of computational models in plant morphogenesis. CurrOpin Plant Biol. 2009;12:606–614. doi: 10.1016/j.pbi.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Sawai S, Thomason PA, Cox EC. An autoregulatory circuit for long-range self-organization in Dictyostelium cell populations. Nature. 2005;433:323–326. doi: 10.1038/nature03228. [DOI] [PubMed] [Google Scholar]
- 9.Aigouy B, et al. Cell flow reorients the axis of planar polarity in the wing epithelium of Drosophila. Cell. 142:773–786. doi: 10.1016/j.cell.2010.07.042. [DOI] [PubMed] [Google Scholar]
- 10.Amonlirdviman K, et al. Mathematical modeling of planar cell polarity to understand domineering nonautonomy. Science. 2005;307:423–426. doi: 10.1126/science.1105471. [DOI] [PubMed] [Google Scholar]
- 11.Vavylonis D, Wu JQ, Hao S, O'Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:397–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 12.Fayant P, et al. Finite element model of polar growth in pollen tubes. Plant Cell. 2010;22:2579–2593. doi: 10.1105/tpc.110.075754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scarpella E, Marcos D, Friml J, Berleth T. Control of leaf vascular patterning by polar auxin transport. Genes Dev. 2006;20:1015–1027. doi: 10.1101/gad.1402406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bayer EM, et al. Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev. 2009;23:373–384. doi: 10.1101/gad.497009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galway ME, et al. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol. 1994;166:740–754. doi: 10.1006/dbio.1994.1352. [DOI] [PubMed] [Google Scholar]
- 16.Kwak SH, Shen R, Schiefelbein J. Positional signaling mediated by a receptorlike kinase in Arabidopsis. Science. 2005;307:1111–1113. doi: 10.1126/science.1105373. [DOI] [PubMed] [Google Scholar]
- 17.Benitez M, Espinosa-Soto C, Padilla-Longoria P, Diaz J, Alvarez-Buylla ER. Equivalent genetic regulatory networks in different contexts recover contrasting spatial cell patterns that resemble those in Arabidopsis root and leaf epidermis: a dynamic model. IntJ Dev Biol. 2007;51:139–155. doi: 10.1387/ijdb.062183mb. [DOI] [PubMed] [Google Scholar]
- 18.Savage NS, et al. A mutual support mechanism through intercellular movement of CAPRICE and GLABRA3 can pattern the Arabidopsis root epidermis. PLoS Biol. 2008;6:e235. doi: 10.1371/journal.pbio.0060235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee MM, Schiefelbein J. Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell. 2002;14:611–618. doi: 10.1105/tpc.010434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steeves TA, Sussex IM. Patterns in Plant Development. Cambridge, England: Cambridge University Press; 1989. [Google Scholar]
- 21.Hohm T, Zitzler E, Simon R. A dynamic model for stem cell homeostasis and patterning in Arabidopsis meristems. PLoS One. 2010;5:e9189. doi: 10.1371/journal.pone.0009189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonsson H, et al. Modeling the organization of the WUSCHEL expression domain in the shoot apical meristem. Bioinformatics. 2005;21:I232–I240. doi: 10.1093/bioinformatics/bti1036. [DOI] [PubMed] [Google Scholar]
- 23.Nikolaev SV, Penenko AV, Lavreha VV, Mjolsness ED, Kolchanov NA. A Model Study of the Role of Proteins CLV1, CLV2, CLV3, and WUS in Regulation of the Structure of the Shoot Apical Meristem. Russian Journal of Developmental Biology. 2007;38:383–388. [PubMed] [Google Scholar]
- 24.Reddy GV, Meyerowitz EM. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science. 2005;310:663–667. doi: 10.1126/science.1116261. [DOI] [PubMed] [Google Scholar]
- 25.Geier F, et al. A quantitative and dynamic model for plant stem cell regulation. PLoS One. 2008;3:e3553. doi: 10.1371/journal.pone.0003553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mundermann L, Erasmus Y, Lane B, Coen E, Prusinkiewicz P. Quantitative modeling of Arabidopsis development. Plant Physiol. 2005;139:960–968. doi: 10.1104/pp.105.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall SM, Hillman JR. Correlative Inhibition of Lateral Bud Growth in Phaseolus-Vulgaris L Timing of Bud Growth Following Decapitation. Planta. 1975;123:137–143. doi: 10.1007/BF00383862. [DOI] [PubMed] [Google Scholar]
- 28.Thimann KV, Skoog F, Kerckhoff WG. On the inhibition of bud development and other functions of growth substance in Vicia Faba. Proceedings of the Royal Society of London Series B-Containing Papers of a Biological Character. 1934;114:317–339. [Google Scholar]
- 29.Stirnberg P, Ward S, Leyser O. Auxin and strigolactones in shoot branching: intimately connected? Biochem Soc Trans. 2010;38:717–722. doi: 10.1042/BST0380717. [DOI] [PubMed] [Google Scholar]
- 30.Prusinkiewicz P, et al. Control of bud activation by an auxin transport switch. Proc Natl Acad Sci USA. 2009;106:17431–17436. doi: 10.1073/pnas.0906696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bossinger G, Smyth DR. Initiation patterns of flower and floral organ development in Arabidopsis thaliana. Development. 1996;122:1093–1102. doi: 10.1242/dev.122.4.1093. [DOI] [PubMed] [Google Scholar]
- 32.Irish VF, Sussex IM. A Fate Map of the Arabidopsis Embryonic Shoot Apical Meristem. Development. 1992;115:745. [Google Scholar]
- 33.Poethig RS, Sussex IM. The Cellular-Parameters of Leaf Development in Tobacco - a Clonal Analysis. Planta. 1985;165:170–184. doi: 10.1007/BF00395039. [DOI] [PubMed] [Google Scholar]
- 34.Green AA, Kennaway JR, Hanna AI, Bangham JA, Coen E. Genetic Control of Organ Shape and Tissue Polarity. PLoSBiol. 2010;8:e1000537. doi: 10.1371/journal.pbio.1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui ML, Copsey L, Green AA, Bangham JA, Coen E. Quantitative Control of Organ Shape by Combinatorial Gene Activity. PLoS Biol. 2010;8:e1000538. doi: 10.1371/journal.pbio.1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campilho A, Garcia B, Toorn HV, Wijk HV, Scheres B. Time-lapse analysis of stem-cell divisions in the Arabidopsis thaliana root meristem. Plant J. 2006;48:619–627. doi: 10.1111/j.1365-313X.2006.02892.x. [DOI] [PubMed] [Google Scholar]
- 37.Harrison CJ, Roeder AH, Meyerowitz EM, Langdale JA. Local cues and asymmetric cell divisions underpin body plan transitions in the moss Physcomitrella patens. CurrBiol. 2009;19:461–471. doi: 10.1016/j.cub.2009.02.050. [DOI] [PubMed] [Google Scholar]
- 38.Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM. Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development. 2004;131:4225–4237. doi: 10.1242/dev.01261. [DOI] [PubMed] [Google Scholar]
- 39.Roeder AHK, et al. Variability in the Control of Cell Division Underlies Sepal Epidermal Patterning in Arabidopsis thaliana. PLoS Biol. 2010;8:e1000367. doi: 10.1371/journal.pbio.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernandez R, et al. Imaging plant growth in 4D: robust tissue reconstruction and lineaging at cell resolution. Nat Methods. 2010;7:547–553. doi: 10.1038/nmeth.1472. [DOI] [PubMed] [Google Scholar]
- 41.Traas J, Hulskamp M, Gendreau E, Hofte H. Endoreduplication and development: rule without dividing? Curr Opin Plant Biol. 1998;1:498–503. doi: 10.1016/s1369-5266(98)80042-3. [DOI] [PubMed] [Google Scholar]
- 42.Grieneisen VA, Xu J, Maree AF, Hogeweg P, Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature. 2007;449:1008–1013. doi: 10.1038/nature06215. [DOI] [PubMed] [Google Scholar]
- 43.Jonsson H, Heisler MG, Shapiro BE, Meyerowitz EM, Mjolsness E. An auxin-driven polarized transport model for phyllotaxis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1633–1638. doi: 10.1073/pnas.0509839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith RS, et al. A plausible model of phyllotaxis. Proc NatlAcadSciU SA. 2006;103:1301–1306. doi: 10.1073/pnas.0510457103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fortin MC, Pierce FJ, Poff KL. The pattern of secondary root formation in curving roots of Arabidopsis thaliana (L.) Heynh. Plant Cell Environ. 1989;12:337–339. doi: 10.1111/j.1365-3040.1989.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 46.De Smet I, et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development. 2007;134:681–690. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- 47.Boerjan W, et al. Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell. 1995;7:1405–1419. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dubrovsky JG, et al. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci USA. 2008;105:8790–8794. doi: 10.1073/pnas.0712307105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- 50.Reinhardt D, Mandel T, Kuhlemeier C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell. 2000;12:507–518. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benkova E, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 52.Heisler MG, et al. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Current Biology. 2005;15:1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 53.Reinhardt D, et al. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;426:225–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- 54.Vernoux T, Kronenberger J, Grandjean O, Laufs P, Traas J. PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development. 2000;127:5157–5165. doi: 10.1242/dev.127.23.5157. [DOI] [PubMed] [Google Scholar]
- 55.Jonsson H, Heisler MG, Shapiro BE, Meyerowitz EM, Mjolsness E. An auxin-driven polarized transport model for phyllotaxis. Proc Natl Acad Sci USA. 2003;103:1633–1638. doi: 10.1073/pnas.0509839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamant O, et al. Developmental patterning by mechanical signals in Arabidopsis. Science. 2008;322:1650–1655. doi: 10.1126/science.1165594. [DOI] [PubMed] [Google Scholar]
- 57.Heisler MG, et al. Alignment between PIN1 Polarity and Microtubule Orientation in the Shoot Apical Meristem Reveals a Tight Coupling between Morphogenesis and Auxin Transport. PLoS Biol. 2010;8:e1000516. doi: 10.1371/journal.pbio.1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laskowski M, et al. Root system architecture from coupling cell shape to auxin transport. PLoS Biol. 2008;6:e307. doi: 10.1371/journal.pbio.0060307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gor V, Elowitz M, Bacarian T, Mjolsness E. Proceedings of the 2005 IEEE Computer Society conference on Computer Vision and Pattern Recognition. 2005;42:142. [Google Scholar]
- 60.Liu M, Roy-Chowdhury AK, Reddy GV. IEEE Computer Society Workshop on Mathematical Methods in Biomedical Image Analysis. 2009 [Google Scholar]
- 61.Cunha A, Roeder A, Meyerowitz EM. 32nd Annual International Conference of the IEEE EMBS 5338–5342 (Argentina, 2010). [Google Scholar]
- 62.Clark SE. Cell signalling at the shoot meristem. Nat Rev Mol Cell Biol. 2001;2:276–284. doi: 10.1038/35067079. [DOI] [PubMed] [Google Scholar]
- 63.Nishimura A, Tamaoki M, Sato Y, Matsuoka M. The expression of tobacco knotted1-type class 1 homeobox genes correspond to regions predicted by the cytohistological zonation model. Plant J. 1999;18:337–347. doi: 10.1046/j.1365-313x.1999.00457.x. [DOI] [PubMed] [Google Scholar]
- 64.Xie M, Tataw M, Venugopala Reddy G. Towards a functional understanding of cell growth dynamics in shoot meristem stem-cell niche. Semin Cell Dev Biol. 2009;20:1126–1133. doi: 10.1016/j.semcdb.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 65.Sandersius SA, Newman TJ. Modeling cell rheology with the Subcellular Element Model. PhysBiol. 2008;5:015002. doi: 10.1088/1478-3975/5/1/015002. [DOI] [PubMed] [Google Scholar]
- 66.Van Liedekerke P, et al. A particle-based model to simulate the micromechanics of single-plant parenchyma cells and aggregates. Phys Biol. 7:026006. doi: 10.1088/1478-3975/7/2/026006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.