Abstract
Rhadbomyosarcoma (RMS) is the most common soft-tissue sarcoma in children and is subdivided in the embryonal (ERMS) and alveolar (ARMS) subtypes, the latter being associated with the worst prognosis.
We report that sulforaphane (SFN), a broccoli-derived anticancer isothiocyanate, causes dose- and time-dependent growth inhibition and apoptosis in both ERMS and ARMS cells.
In ARMS, SFN induced the modulation of expression of crucial genes and proteins: mRNA and protein levels of PAX3-FKHR, MYCN, and MET decreased, while those of p21 and TRAIL-receptor DR5 (but not DR4) increased.
Since DR5 expression increased specifically in ARMS, we treated ARMS cells with TRAIL, SFN, or their combination. While ARMS cells (RH30 and RH4) proved to be TRAIL-resistant, SFN restored their sensitivity to TRAIL-induced cell-growth inhibition, leading to a stronger effect in combination with TRAIL.
ARMS cells transfected with siDR5 showed that SFN-induced DR5 acts as a key regulator, being directly related to the TRAIL-induced cell-growth inhibition.
The in vivo anti-tumor activity of SFN and TRAIL was evaluated in a xenograft murine model of ARMS through microPET. The results showed that the systemic treatment (3 wk) of mice with SFN or TRAIL as single agents only delayed tumor evolution, while the combined treatment of SFN and TRAIL led to tumor elimination.
These findings indicate that SFN triggers the apoptotic pathway in both alveolar and embryonal rhabdomyosarcomas and that combined treatment with SFN and TRAIL might be a promising therapy for the aggressive alveolar subtype.
Keywords: rhabdomyosarcoma, alveolar, sulforaphane, TRAIL, combination therapy
Introduction
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma in children.1 RMS is a heterogeneous tumor that expresses skeletal muscle-specific markers. Two subtypes of RMS have been identified on the basis of histopathologic features—embryonal (ERMS) and alveolar (ARMS)—each with distinct clinical and genetic characteristics, the latter being associated with the worst prognosis.1 The majority of ARMS tumors are associated with t(2;13) or t(1;13) chromosomal translocations, which generate PAX3-FKHR and PAX7-FKHR fusion products, respectively.2 These translocations modify cell growth, differentiation, and apoptotic pathways.3 Overexpression of PAX-FKHR correlated fusion genes and oncogenes is associated with poor prognosis, and the survival rate of patients with ARMS is much lower than in those with ERMS.2,4 Recent studies show that several target genes of Pax-FKHR, such as c-MET5 and MYCN,6 play important roles in ARMS tumorigenesis and are potential therapeutic targets for treating ARMS in children.5-7 Since ARMS is resistant to chemotherapy and radiotherapy, it is crucial to develop new molecules that could improve conventional anticancer treatment.
Sulforaphane (SFN), a constituent of cruciferous vegetables, is a natural isothiocyanate, with promising anti-tumor and chemopreventive activity.8 In particular, SFN anti-tumoral activity is exerted by its different pleiotropic effects that continue to increase and that actually include: suppression of cell cycle progression, induction of apoptosis, inhibition of angiogenesis,9 anti-inflammatory activity,10 specific modulation of gene expression, epigenetic effects such as the inhibition of histone deacetylases,11 activation of phase II detoxifying enzymes, and suppression of cytochrome P450 enzymes.12
In vivo studies in animals have shown that sulforaphane is rapidly absorbed, displays an absolute bioavailability of 82%, and induces a reduction in the frequency, size, and number of many tumors,13,14 while no side effects have been reported.14
The activities of SFN may suggest the identification of a rational combination of SFN with other therapeutic molecules that could trigger a potent anti-tumor effect. Tumor necrosis factor-related apoptosis inducing ligand (TRAIL) belongs to the tumor necrosis factor family of death-inducing ligands. TRAIL activates a fundamental extracellular apoptotic pathway after interaction with death receptors DR4 and DR5, and could play a critical role in anti-tumor activity.15,16
Here, for the first time, we evaluated the preclinical anti-tumor activity of SFN in vitro in RMS, and in vivo in a xenograft murine model of the aggressive ARMS subtype. Moreover, we found that SFN restores the TRAIL-mediated exogenous apoptotic pathway in TRAIL-resistant ARMS cells, and we elucidated the underlying molecular mechanisms.
Our studies provide strong preclinical evidence that sulforaphane in combination with TRAIL may be an effective new strategy for the treatment of alveolar rhabdomyosarcoma.
Results
SFN causes cell growth inhibition and apoptosis in rhabdomyosarcoma cells
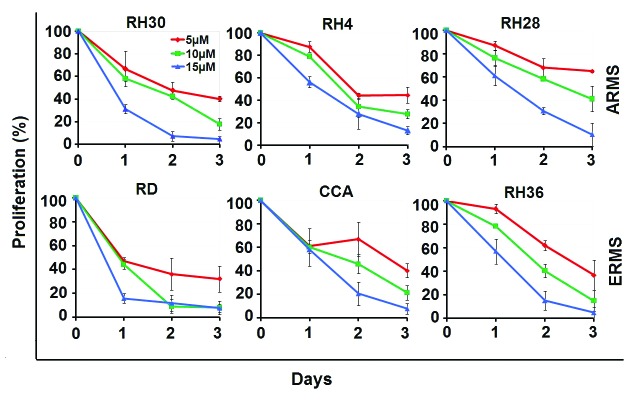
To test for the anti-tumor effect of SFN against rhabdomyosarcoma, we treated human ARMS (RH30, RH4, and RH28) and ERMS (RD, RH36, and CCA) cells with increasing concentrations of SFN (5, 10, and 15 μM), and analyzed cell viability for 3 consecutive days. SFN induced a dose-dependent inhibition of cell growth in both ARMS and ERMS cells (Fig. 1). Induction of apoptosis was evaluated in ARMS (RH30) and ERMS (RD) cells treated with SFN at 10 µM for 24 h. SFN induced a consistent percentage of apoptotic cells in both RH30 (57.7%) and RD (47.7%) cells (Fig. 2), with a significant difference from untreated control cells (*P = 0.001 for RH30 cells and *P = 0.0006 for RD cells).
Figure 1. SFN inhibits cell growth in alveolar (RH30, RH4, RH28) and embryonal (RD, RH36, CCA) RMS cells. ARMS and ERMS cells were cultured in 96-well plates. After 24 h, the cells were treated with SFN (5, 10, and 15 μM) for 3 d, and ATP was measured by the Luminescence ATP Detection Assay. Results represent the mean ± SD of 3 experiments done in triplicate.
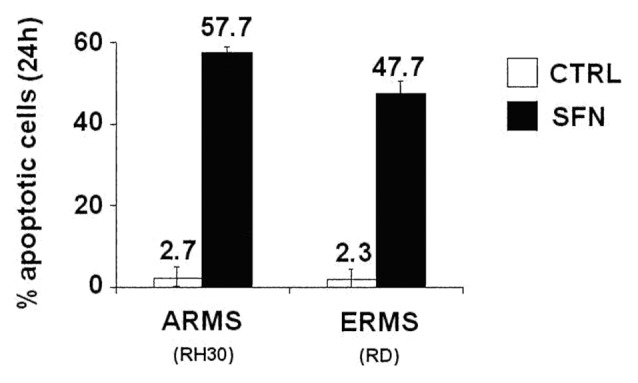
Figure 2. SFN causes apoptosis in ARMS and ERMS cells. RH30 and RD cells were cultured in slide flask. After 24 h, the cells were treated with SFN (10 μM) for 24 h, and the percentage of apoptotic cells was measured by the In Situ Cell Death Detection Kit. Results represent the mean ± SD of 3 experiments done in triplicate. Statistical analysis was performed (P = 0.001 in RH30 and P = 0.0006 in RD).
SFN modulates genes specifically in ARMS cells
We evaluated the modulation of gene expression and protein levels of highly relevant RMS-correlated oncogenes, such as PAX3-FKHR,2 PAX3,2 MYCN,6 MET,5 and oncosuppressors such as p21,19 DR5, and DR416 induced by SFN in rhabdomyosarcoma cells. Treatment with SFN in ERMS cells induced only MET mRNA decrease, and p21 mRNA increase (in two of the three cell lines) (Fig. 3A). Conversely, in all the three ARMS cell lines, SFN induced a strong dose-dependent gene expression response as follow: the levels of PAX3-FKHR, MYCN, and MET mRNA decreased, while the levels of p21 and TRAIL-receptor DR5 (but not DR4) mRNA increased (Fig. 3A).
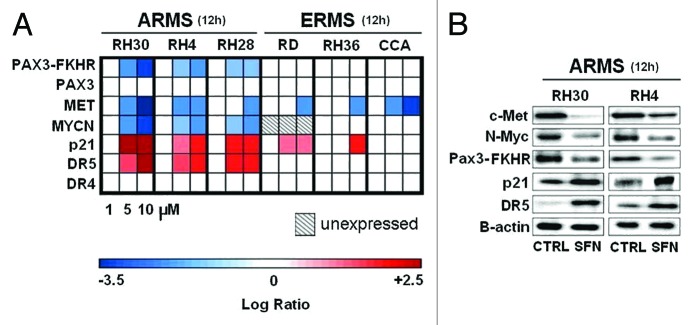
Figure 3. SFN modulates genes specifically in ARMS cells. (A) Expression of PAX3-FKHR, PAX3, MET, MYCN, MYC, p21, DR5, and DR4 mRNA in ARMS (RH30, RH4, RH28) and ERMS (RD, RH36, CCA). Log ratio was measured at 12 h after SFN treatment (1, 5, and 10 μM) in vitro by the real-time quantitative PCR assay. All expression levels were normalized to GAPDH gene in each well and each square represents the difference between the control and treated samples. Red squares indicate upregulated genes; blue squares indicate downregulated genes; white squares indicate non-significant alterations of gene expression. The squares filled with diagonal lines are non-evaluable time points. (B) Western blot analysis. ARMS cells were treated with SFN (10 μM) for 12 h. For each cell line, 30 µg of soluble proteins were analyzed by western blot and detected by anti-Met, anti-N-Myc, anti-Pax3-FKHR, anti-p21, ant-DR5 and anti-B-actin antibodies.
The gene expression modulations induced by SFN in ARMS were also confirmed at the protein level by western blot analysis in RH30 and RH4 cells (Fig. 3B). Indeed, the treatment with SFN consistently reduced the amount of Met, Pax3-Fkhr, and N-Myc oncoproteins, while it consistently increased the amount of p21 and DR5 proteins (Fig. 3B).
SFN restores the sensitivity to TRAIL in ARMS cells
As we found that SFN enhanced levels of DR5 mRNA and protein only in the aggressive alveolar subtype, we evaluated whether SFN specifically restored the sensitivity to TRAIL in ARMS cells.
ARMS cell lines (RH30 and RH4) and ERMS cell lines (CCA and RD) were treated with TRAIL (100 ng/mL) or SFN (5 μM) (a concentration sufficient to significantly increase DR5 mRNA), or with their combination, and cell viability was evaluated for three consecutive days.
In ERMS cells, the treatment with TRAIL alone caused a consistent response that was further increased by the combination with SFN (Fig. 4).
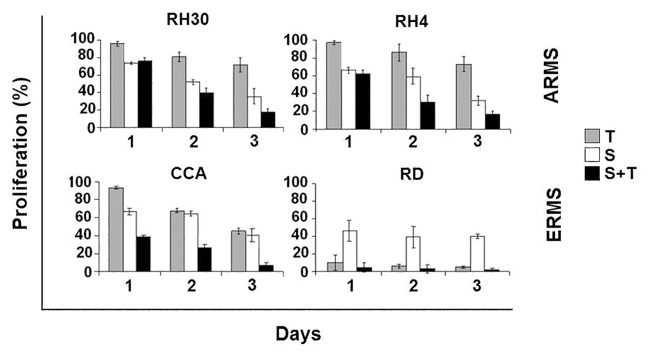
Figure 4. SFN restores the sensitivity to TRAIL in ARMS cells. ARMS and ERMS cells were cultured in 96-well plates. After 24 h, the cells were treated with SFN (5 μM) and TRAIL (100 ng/mL) for 3 d, and ATP was measured by the Luminescence ATP Detection Assay. Results represent the mean ± SD of 3 experiments done in triplicate.
Conversely, in ARMS cells, TRAIL alone only induced a minimal response, while the use of TRAIL in combination with SFN led to a strong time- and dose-dependent cell viability inhibition (higher than 80% after 72 h) (Fig. 4).
DR5 mediates SFN’s restoring activity of TRAIL-induced apoptosis in ARMS
To evaluate the direct role exerted by DR5 activation led by SFN in the TRAIL-induced apoptosis in ARMS, knockdown of DR5 was performed using a specific siRNA during SFN plus TRAIL treatment in ARMS cells.
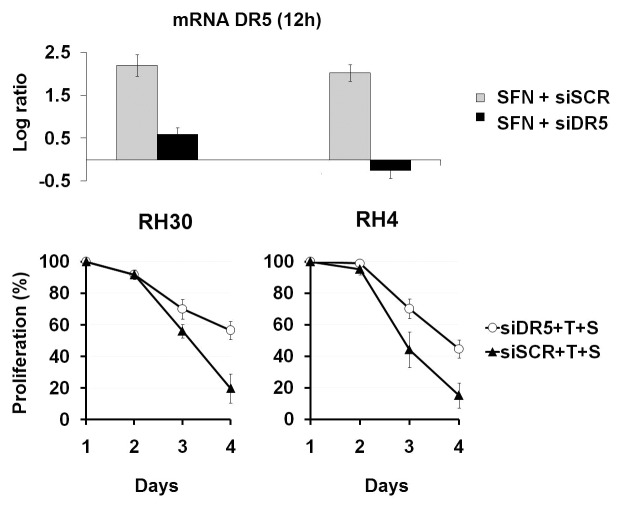
We first verified the knockdown of DR5 by a specific siRNA (siDR5) after the DR5 gene expression induction by SFN in ARMS cells (RH30 and RH4). Real-time RT-PCR of DR5 mRNA was done in ARMS cells after treatment with SFN plus siDR5, and SFN plus siSCR (200 nm). The results showed that siDR5 abrogated DR5 gene expression induced by SFN (Fig. 5A).
Figure 5. SFN’s restoring activity of TRAIL-induced apoptosis in ARMS is DR5-mediated. (A) Log ratio of DR5 mRNA was measured at 12 h after SFN treatment (10 μM) in vitro by the real-time quantitative PCR assay. ARMS cells were treated with SFN (5 µM) plus siDR5, and SFN (5 µM) plus siSCR (200 nm). All expression levels were normalized to GAPDH gene in each well and each square represents the difference between the control and treated samples. (B) ARMS cells (RH30 and RH4) were treated with SFN, TRAIL, and transfected with siDR5 (200 nM) for three days, and ATP was measured by the Luminescence ATP Detection Assay. Results represent the mean ± SD of 3 experiments done in triplicate.
ARMS cells were then treated with SFN (5 μM) plus TRAIL (100 ng/mL) and plus siDR5 or siSCR, and cell viability was evaluated over three days.
As expected, the treatment with SFN plus TRAIL plus siSCR induced a strong cell growth inhibition (81% in RH30 and 84% RH4 cells at 72 h); by contrast, the treatment with SFN plus TRAIL plus siDR5, in which the SFN-induced DR5 gene expression was silenced by siDR5, showed a consistent increment of viable cells (Fig. 5B).
These results demonstrate that SFN-induced DR5 acts as a key regulator, being directly related to approximately 50% (in RH30) and 30% (in RH4) of the TRAIL-induced cell growth inhibition in ARMS cells (Fig. 5B).
Combined treatment of TRAIL and SFN eliminates ARMS in mice
The in vivo anti-tumoral activity of SFN was evaluated in a xenograft murine model of ARMS by real-time molecular imaging using microPET during a 21-d treatment schedule. The incidence of the tumor was 100% (20/20 animals); all the animals had a positive PET scan two days after inoculation. The results in terms of TBR index showed that mice treated with SFN alone via systemic administration (50 mg/kg/d intraperitoneally [IP]) responded to treatment especially in the first 14 d after injection, but afterwards the tumor metabolic signal was resumed (Fig. 6). Treatment of mice (daily, IP) with 0.8 mg/kg of TRAIL alone or in combination with SFN (50 mg/kg) showed a similar trend of response (data not shown).
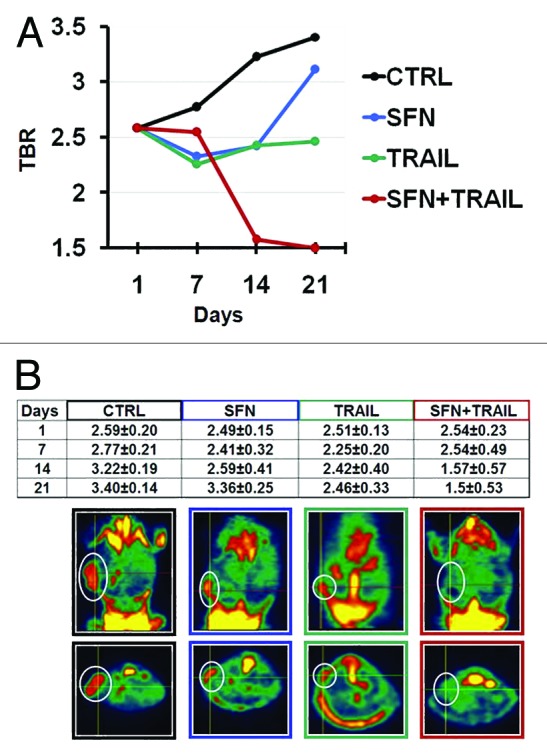
Figure 6. Combined treatment of TRAIL and SFN eliminates ARMS in mice. (A) Graphics description of TBR (Target to Background ROI) index at different time points of mice treated i.p. with SFN (50 mg/kg) or TRAIL (1.6 mg/kg) and SFN plus TRAIL. TBR means radioactive counts per gram of tissue, divided by injected dose of radioactivity per gram of animal weight. The target region of interest (ROI) was placed on the most active area of the neoplastic mass and the background ROI was placed on the lung. (B) In situ imaging of 18F-FDG uptake at the end of treatment (21 d after cell inoculation). The white circles indicate tumor mass in all representative groups of treated mice.
IP treatment with TRAIL alone at a dose of 1.6 mg/kg showed a significant reduction of tumor growth, but this was not sufficient as tumor growth was re-established soon after (Fig. 6).
However, the combined treatment (daily, IP) with TRAIL (1.6 mg/kg) plus SFN (50 mg/kg) led to a progressive reduction of the tumor, ending with tumor elimination (Fig. 6). This result was confirmed by the absence of the metabolic signal of the tumor cells (Fig. 6). The TBR was significantly different for SFN plus TRAIL compared with the placebo (P < 0.05) (Fig. 6).
Discussion
High mortality rates are still the case in the most aggressive type of rhabdomyosarcoma (ARMS), especially in patients with metastatic disease.2 Therefore, identification of new targets and new therapeutic approaches are needed for the treatment of patients with ARMS.
In the present study we evaluated the anti-tumor activity of SFN for the treatment of RMS, especially for ARMS.
Sulforaphane is an extremely interesting and promising pleiotropic natural compound, for which multiple biological and pharmacological activities have been described that could contribute to the inhibition of tumor development.8-14,20,21 Moreover, SFN from broccoli sprouts has already been evaluated in a phase I clinical trial that demonstrated its good safety profile,22 and a phase II clinical trial for treating patients with recurrent prostate cancer is currently being performed (ClinicalTrials.gov, NCT01228084).
Here, for the first time, we evaluated the anti-tumor activity of SFN in RMS. We found that SFN exerted time- and dose-dependent inhibition of cellular proliferation and apoptosis in both ARMS and ERMS subtypes, with a remarkably stronger effect in the aggressive ARMS subtype.
At the molecular level, SFN induced gene expression modifications in a subtype specific manner, with downregulation of key oncogenes (such as MYCN, c-MET, and PAX3-FKHR), and upregulation of key oncosuppressors such as p21 and DR5 (but not DR4), only in ARMS.
The p53 pathway is frequently inactivated in RMS through somatic mutations or functional inactivation.23 Remarkably, SFN restored p53-independent transcriptional and translational regulation of major p53 downstream target genes in ARMS, in terms of activation of p21 and DR5.
As the anti-tumor in vivo effect of systemic repeated treatment of SFN as a single agent against ARMS xenograft in mice showed only temporally limited activity but did not eliminate tumors, we further evaluated SFN in a rational combined treatment, taking into account its molecular pleiotropic effects.
In particular, as we found that SFN induced the upregulation of the DR5 (TRAIL receptor) mRNA and protein specifically in ARMS cells, we evaluated the administration of the recombinant human TRAIL protein in a rational combination with SFN in ARMS. The TRAIL protein is physiologically expressed in the human system and plays a critical role in anti-tumor immunity. TRAIL interacts with death receptors, DR4 and DR5, and activates an intracellular apoptotic pathway in cancer cells.15 A phase I trial has established a good profile of safety and tolerability of the recombinant human TRAIL (rhTRAIL, also named dulanermin) in patients with advanced cancer,24 while phase II trials are currently evaluating the anti-tumor efficacy of TRAIL as a single agent or in combination with established cancer therapeutics25 in a wide range of both solid and nonsolid malignant neoplasms. However, signaling through TRAIL death receptors may also induce anti-apoptotic responses and proliferation pathways. These pro-survival responses may, therefore, limit the feasibility of TRAIL in mono-therapies. Thus, many clinical trials are evaluating TRAIL in combination therapy.25
We found that ARMS cells were resistant to TRAIL compared with ERMS cells.
However, we showed that SFN restored the sensitivity to TRAIL in ARMS cells, as documented by a remarkable anti-tumor response to the combined treatment of SFN plus TRAIL. The restoration of the TRAIL sensitivity induced by SFN in ARMS cells is mainly mediated by the SFN-induced upregulation of DR5 expression.
Based on these promising in vitro results, we evaluated the anti-tumor efficacy of the in vivo combined treatment of SFN plus TRAIL in a xenograft mice model of ARMS by FDG microPET molecular imaging. We showed that the systemic IP treatment with SFN or TRAIL as single agents only delayed ARMS tumor growth, but were not sufficient to eliminate the tumor metabolic signals. On the contrary, the combined use of SFN plus TRAIL led to a remarkable result, achieving a progressive reduction of tumor metabolic signals up to a complete elimination.
Overall, our results show for the first time that SFN is effective against RMS and specifically restores the responsiveness to TRAIL in ARMS, through the induction of DR5 expression. Moreover, the potent in vivo responsiveness of SFN combined with TRAIL in ARMS shows clinically relevant promise for the treatment of this aggressive and poor responsive RMS subtype.
Materials and Methods
Cell culture and reagents
RH30, RH4, and RH28 cell lines derive from ARMS and express PAX3-FKHR fusion gene, while CCA, RH36, and RD cell lines derive from ERMS. RH30 and RD cells were acquired from DSMZ and ATCC respectively, CCA were kindly provided by Prof PL Lollini (University of Bologna) while RH4, RH28, and RH36 were kindly provided by Prof Peter Houghton (St Jude Children’s Hospital) and Dr Angelo Rosolen (University of Padova). Sulforaphane (D,L-SFN 99% pure) was purchased from LKT Laboratories and it was added to all cell lines at concentrations of 5, 10, and 15 μM. Cells were harvested and counted at 24, 48, and 72 h.
The soluble human recombinant TRAIL (Biomol) was added to cell lines at a concentration of 100 ng/mL.
Viability assay
Cell viability was determined using the Luminescence ATP Detection Assay System (ATPlite) (PerkinElmer). Cells were plated in triplicate using 96-well cluster plates: 20 × 103 CCA cells were plated with 100 μL of OPTI-MEM (GIBCO BRL) containing 4% FCS, 2 mM l-glutamine, and 1% penicillin/streptomycin. The remaining cell lines were seeded at 5000 cells/well at the same conditions. Cells were incubated at 37 °C for various periods and viability was measured according to the manufacturer’s protocol.
Apoptosis analysis
ARMS (RH30) and ERMS (RD) cells (2 × 105) were plated in slide flasks with 2 mL of OPTI-MEM (GIBCO BRL) containing 10% FBS, 2 mM l-glutamine, and 1% penicillin/streptomycin. SFN was added at 10 μM to the cells and apoptosis analysis was performed after 24 h using the In Situ Cell Death Detection TUNEL Kit (Roche) according to the supplier’s instructions.
Trasfection by siRNA
Small interfering RNA (siRNA) specific for DR5 mRNA (siDR5) and scrambled siRNA (siSCR) (from MWG) had the following sequences: siDR5: 5′-AUGAGAUAAA GGUGGCUAAT TdTdT-3′ and siSCR: 5′-UAGCGACUAA ACACAUCAAd TdT-3′.
Cells were seeded in Opti-MEM serum-free media (GIBCO/BRL). Serum containing medium was added 4 h after transfection. An analysis of cell viability was performed after cells were treated with siDR5 or siSCR alone (200 nM) or in combination with 5 μM of SFN and 100 ng/mL of TRAIL every 24 h for 3 consecutive days.
RNA extraction and real time PCR
Total RNA was extracted from RH30, RH4, CCA, and RD cells using the RNeasy Mini Kit (QIAGEN) from cells untreated and treated with 5 μM and 10 μM SFN for 12 h, or with 200 nM siDR5 or siSCR for 24 h. Each RNA sample was quantified twice with the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). First-strand cDNA was synthesized using 500 ng of total RNA and the cDNA Synthesis Kit for RT-PCR (Roche) according to the manufacturer’s standard procedures. A Light Cycler 480 (Roche) was used to perform real-time RT-PCR in triplicate starting with 10 ng of cDNA in a final volume of 20 μL, using the SYBR Green Master Mix 2× (Applied Biosystems). RT-PCR reaction conditions were 2 min at 50 °C, 10 min at 95 °C, 15 s at 95 °C, and 60 s at 60 °C for 40 cycles.
Levels of mRNA were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or to human β-actin (hACTB); primer sequences are reported in Table S1.
Western blot analysis and reagents
Western blot analysis was performed on ARMS cells (RH30 and RH4) untreated and treated with 10 μM SFN for 12 h. Total proteins (N-Myc and Met) and nuclear proteins (Pax3-FKHR, DR4, DR5, and p21) were extracted with a hypotonic buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 1 mM DTT, and protease inhibitors) and a high salt buffer (20 mM HEPES, pH 7.9, 0.4 M NaCl, 1 mM EDTA, 10% glycerol, 1 mM DTT, and protease inhibitors). For each western blot analysis of three identical experiments, 30 µg of proteins were resolved in a 10% SDS-polyacrylamide gel and transferred onto a Hybond-P filter (Amersham Biosciences). The following primary antibodies were used: monoclonal mouse IgG N-Myc (Oncogene), rabbit polyclonal IgG p21 (Santa Cruz Biotechnology), polyclonal rabbit IgG Met (Santa Cruz Biotechnology), polyclonal goat IgG FKHR (Santa Cruz Biotechnology), and monoclonal mouse IgG DR5 (Calbiochem). The following secondary antibodies were used: sheep anti-mouse IgG-HRP (horseradish peroxidase) (Amersham Biosciences), anti-rabbit IgG-HRP (Amersham Biosciences), and donkey anti-goat IgG-HRP (Santa Cruz Biotechnology), and they were diluted 1:2000 (for Met) and 1:10 000 (for all the others) in PBS, Tween 0.2%, 3.5% BSA and incubated for 1 h at room temperature in constant agitation. The survey was done at ChemiDoc (Biorad) with the program QuantityOne after covering the membrane for 1–2 min in the dark with ECL Advance (Amersham).
Experimental tumor models and treatments
Female athymic mice (CD-1 nude 6-wk-old) were purchased from Charles River Laboratories. Mice were given food and water ad libitum and allowed to acclimatize one week before beginning the experiments. All the experiments were approved by the Scientific Ethics Committee of Bologna University (13825-X/10) and conducted at the Centre of Applied Biomedical Research (CRBA, Policlinico S. Orsola-Malpighi).
A total of 10 × 106 ARMS cells (RH30) were injected subcutaneously in the dorsal tissue with an insulin syringe under anesthesia with Advertin (400 µL, intraperitoneally [IP]). Mice were randomized in the following 4 groups (5 animals each group) and treated IP for 21 consecutive days respectively with: saline (controls); SFN (50 mg/kg); TRAIL (1.6 mg/kg); SFN plus TRAIL at the same concentrations.
Mice were treated when tumors reached a mass of approximately 100 mm3 in size. Micro-PET scans were performed at the following time points: 1, 7, 14, and 21 d of treatment.
Micro-PET analysis
The animals underwent four small animal PET (microPET) scans with 18F-Fluorodeoxyglucose d(18F-FDG)17 at 1, 7, 14, and 21 d of treatment. The entire diagnostic procedure was performed under a warm light to maintain body temperature, especially during anesthesia. The procedure was performed as previously described18 and consisted of the following steps: animals were anesthetized with gas anesthesia (Sevofluorane 3–5% and oxygen 1 L/min) and were injected with 20 MBq of 18F-FDG, in a volume of 0.1 mL, via the tail vein with an insulin syringe. The animals were subsequently allowed to wake up for the uptake period (60 min), during which they were allowed to move freely. The residual dose was measured to verify the effective dose injected. Finally, anesthesia was induced a second time in the same way to allow the performance of the scan. Each anesthetized animal was placed on the scanner bed in the prone position. Images were acquired with a microPET tomograph (GE, eXplore Vista DR) for a total acquisition time of 20 min. As the axial field of view was 4 cm, one bed position was sufficient to cover the whole body. Once the scan had been completed, the gas anesthesia was interrupted and the animal was placed in a warm recovery box until complete recovery. FDG-PET images were reconstructed iteratively (OSEM 2D) and read in three planes (axial, sagittal, and coronal). Scans were considered positive if areas of increased FDG uptake were present at locations consistent with inoculation site. Necrosis was diagnosed when a cold central area appeared within the neoplastic mass. Semi-quantitative analysis was performed for each identified tumor using the target to background ratio (TBR). The target region of interest (ROI) was placed on the most active area of the neoplastic mass and the background ROI was placed on the lung. TBR was finally calculated with the following formula: TBR = Max Count in the Target ROI / Mean Count in the Background ROI.
Statistical analysis
Comparison of the effects of various treatments was performed using one way analysis of variance and a two tailed Student t test. Differences with a P value < 0.05 were considered statistically significant. Statistical examination of in vivo animal survival data utilized log rank statistical analyses between the different treatment groups.
As regards the micro-PET data analysis, the first scan TBR was compared with the TBR at the second, third, and fourth scan (t test). The mean TBR was calculated for each scan and normalized to subcutaneous tissues or lung tissues. Statistical significance between experimental values was determined by t testing. P < 0.05 was considered significant for all statistical tests.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank the Centre of Applied Biomedical Research (CRBA, Policlinico S. Orsola-Malpighi). The authors thank Dr Susan West for the english language revision. The research was supported by grants from MIUR, FIRB piattaforme—reti CHEM-PROFARMA-NET, Fondazione del Monte di Bologna e Ravenna.
Author Contributions
R.T. and P.H. designed the study and jointly wrote the manuscript. E.B. performed cellular and molecular biology studies, and the in vivo studies in mice. C.Q. performed the in vivo microPET studies, C.N. designed and evaluated the data of the microPET analysis, S.F. supervised the microPET studies. A.P. and G.C.-F. contributed to the design and supervision of all the experiments, and critically revised the manuscript.
Glossary
Abbreviations:
- SFN
sulforaphane
- ARMS
alveolar rhabdomyosarcoma
- ERMS
embryonal rhabdomyosarcoma
- DR5
death receptor 5
- TRAIL
tumor necrosis factor-related apoptosis inducing ligand
- siRNA
small interference RNA
- TBR
target to background ROI
- ROI
region of interest
- IP
intraperitoneal
- 18F-FDG
18F-fluorodeoxyglucose
References
- 1.Wexler LH, Meyer WH, Helman LJ. Rhabdomyosarcoma and the undifferentiated sarcomas Principles and practice of pediatric oncology. Eds: Pizzo PA, Poplack DG, 5th ed, Lippincott. 2006; 971-1002. [Google Scholar]
- 2.Sorensen PH, Lynch JC, Qualman SJ, Tirabosco R, Lim JF, Maurer HM, Bridge JA, Crist WM, Triche TJ, Barr FG. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children’s oncology group. J Clin Oncol. 2002;20:2672–9. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- 3.Davicioni E, Finckenstein FG, Shahbazian V, Buckley JD, Triche TJ, Anderson MJ. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66:6936–46. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- 4.Fredericks WJ, Galili N, Mukhopadhyay S, Rovera G, Bennicelli J, Barr FG, Rauscher FJ., 3rd The PAX3-FKHR fusion protein created by the t(2;13) translocation in alveolar rhabdomyosarcomas is a more potent transcriptional activator than PAX3. Mol Cell Biol. 1995;15:1522–35. doi: 10.1128/mcb.15.3.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taulli R, Scuoppo C, Bersani F, Accornero P, Forni PE, Miretti S, Grinza A, Allegra P, Schmitt-Ney M, Crepaldi T, et al. Validation of met as a therapeutic target in alveolar and embryonal rhabdomyosarcoma. Cancer Res. 2006;66:4742–9. doi: 10.1158/0008-5472.CAN-05-4292. [DOI] [PubMed] [Google Scholar]
- 6.Tonelli R, McIntyre A, Camerin C, Walters ZS, Di Leo K, Selfe J, Purgato S, Missiaglia E, Tortori A, Renshaw J, et al. Antitumor activity of sustained N-myc reduction in rhabdomyosarcomas and transcriptional block by antigene therapy. Clin Cancer Res. 2012;18:796–807. doi: 10.1158/1078-0432.CCR-11-1981. [DOI] [PubMed] [Google Scholar]
- 7.Toffolatti L, Frascella E, Ninfo V, Gambini C, Forni M, Carli M, Rosolen A. MYCN expression in human rhabdomyosarcoma cell lines and tumour samples. J Pathol. 2002;196:450–8. doi: 10.1002/path.1068. [DOI] [PubMed] [Google Scholar]
- 8.Fimognari C, Hrelia P. Sulforaphane as a promising molecule for fighting cancer. Mutat Res. 2007;635:90–104. doi: 10.1016/j.mrrev.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Bertl E, Bartsch H, Gerhäuser C. Inhibition of angiogenesis and endothelial cell functions are novel sulforaphane-mediated mechanisms in chemoprevention. Mol Cancer Ther. 2006;5:575–85. doi: 10.1158/1535-7163.MCT-05-0324. [DOI] [PubMed] [Google Scholar]
- 10.Heiss E, Herhaus C, Klimo K, Bartsch H, Gerhäuser C. Nuclear factor kappa B is a molecular target for sulforaphane-mediated anti-inflammatory mechanisms. J Biol Chem. 2001;276:32008–15. doi: 10.1074/jbc.M104794200. [DOI] [PubMed] [Google Scholar]
- 11.Myzak MC, Dashwood WM, Orner GA, Ho E, Dashwood RH. Sulforaphane inhibits histone deacetylase in vivo and suppresses tumorigenesis in Apc-minus mice. FASEB J. 2006;20:506–8. doi: 10.1096/fj.05-4785fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angeloni C, Leoncini E, Malaguti M, Angelini S, Hrelia P, Hrelia S. Modulation of phase II enzymes by sulforaphane: implications for its cardioprotective potential. J Agric Food Chem. 2009;57:5615–22. doi: 10.1021/jf900549c. [DOI] [PubMed] [Google Scholar]
- 13.Conaway CC, Yang YM, Chung FL. Isothiocyanates as cancer chemopreventive agents: their biological activities and metabolism in rodents and humans. Curr Drug Metab. 2002;3:233–55. doi: 10.2174/1389200023337496. [DOI] [PubMed] [Google Scholar]
- 14.Kallifatidis G, Labsch S, Rausch V, Mattern J, Gladkich J, Moldenhauer G, Büchler MW, Salnikov AV, Herr I. Sulforaphane increases drug-mediated cytotoxicity toward cancer stem-like cells of pancreas and prostate. Mol Ther. 2011;19:188–95. doi: 10.1038/mt.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren YG, Wagner KW, Knee DA, Aza-Blanc P, Nasoff M, Deveraux QL. Differential regulation of the TRAIL death receptors DR4 and DR5 by the signal recognition particle. Mol Biol Cell. 2004;15:5064–74. doi: 10.1091/mbc.E04-03-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim H, Kim EH, Eom YW, Kim WH, Kwon TK, Lee SJ, Choi KS. Sulforaphane sensitizes tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-resistant hepatoma cells to TRAIL-induced apoptosis through reactive oxygen species-mediated up-regulation of DR5. Cancer Res. 2006;66:1740–50. doi: 10.1158/0008-5472.CAN-05-1568. [DOI] [PubMed] [Google Scholar]
- 17.Nanni C, Di Leo K, Tonelli R, Pettinato C, Rubello D, Spinelli A, Trespidi S, Ambrosini V, Castellucci P, Farsad M, et al. FDG small animal PET permits early detection of malignant cells in a xenograft murine model. Eur J Nucl Med Mol Imaging. 2007;34:755–62. doi: 10.1007/s00259-006-0288-y. [DOI] [PubMed] [Google Scholar]
- 18.Dandekar M, Tseng JR, Gambhir SS. Reproducibility of 18F-FDG microPET studies in mouse tumor xenografts. J Nucl Med. 2007;48:602–7. doi: 10.2967/jnumed.106.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hecker RM, Amstutz RA, Wachtel M, Walter D, Niggli FK, Schäfer BW. p21 Downregulation is an important component of PAX3/FKHR oncogenicity and its reactivation by HDAC inhibitors enhances combination treatment. Oncogene. 2010;29:3942–52. doi: 10.1038/onc.2010.145. [DOI] [PubMed] [Google Scholar]
- 20.Pham NA, Jacobberger JW, Schimmer AD, Cao P, Gronda M, Hedley DW. The dietary isothiocyanate sulforaphane targets pathways of apoptosis, cell cycle arrest, and oxidative stress in human pancreatic cancer cells and inhibits tumor growth in severe combined immunodeficient mice. Mol Cancer Ther. 2004;3:1239–48. [PubMed] [Google Scholar]
- 21.Singh AV, Xiao D, Lew KL, Dhir R, Singh SV. Sulforaphane induces caspase-mediated apoptosis in cultured PC-3 human prostate cancer cells and retards growth of PC-3 xenografts in vivo. Carcinogenesis. 2004;25:83–90. doi: 10.1093/carcin/bgg178. [DOI] [PubMed] [Google Scholar]
- 22.Shapiro TA, Fahey JW, Dinkova-Kostova AT, Holtzclaw WD, Stephenson KK, Wade KL, Ye L, Talalay P. Safety, tolerance, and metabolism of broccoli sprout glucosinolates and isothiocyanates: a clinical phase I study. Nutr Cancer. 2006;55:53–62. doi: 10.1207/s15327914nc5501_7. [DOI] [PubMed] [Google Scholar]
- 23.Xia SJ, Pressey JG, Barr FG. Molecular pathogenesis of rhabdomyosarcoma. Cancer Biol Ther. 2002;1:97–104. doi: 10.4161/cbt.51. [DOI] [PubMed] [Google Scholar]
- 24.Herbst RS, Eckhardt SG, Kurzrock R, Ebbinghaus S, O’Dwyer PJ, Gordon MS, Novotny W, Goldwasser MA, Tohnya TM, Lum BL, et al. Phase I dose-escalation study of recombinant human Apo2L/TRAIL, a dual proapoptotic receptor agonist, in patients with advanced cancer. J Clin Oncol. 2010;28:2839–46. doi: 10.1200/JCO.2009.25.1991. [DOI] [PubMed] [Google Scholar]
- 25.Hellwig CT, Rehm M. TRAIL signaling and synergy mechanisms used in TRAIL-based combination therapies. Mol Cancer Ther. 2012;11:3–13. doi: 10.1158/1535-7163.MCT-11-0434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.