Abstract
The high morbidity and mortality of colorectal cancer pose a significant public health problem worldwide. Here we assessed the pro-cancer efficacy and mechanism of action of CCNB1 in different colorectal cancer cells. We provided evidence that CCNB1 mRNA and protein level were upregulated in a subset of human colorectal tumors, and positively correlated with Chk1 expression. Repression of Chk1 caused a significant decrease in cell proliferation and CCNB1 protein expression in colorectal cancer cells. Furthermore, downregulation of CCNB1 impaired colorectal cancer proliferation in vitro and tumor growth in vivo. Specifically, suppression of CCNB1 caused a strong G2/M phase arrest in both HCT116 and SW480 cells, interfering with the expression of cdc25c and CDK1. Additionally, CCNB1 inhibition induced apoptotic death in certain colorectal cancer cells. Together, these results suggest that CCNB1 is activated by Chk1, exerts its oncogenic role in colorectal cancer cells, and may play a key role in the development of a novel therapeutic approach against colorectal cancer.
Keywords: CCNB1, colorectal cancer, Chk1, cell growth, cell cycle, apoptosis
Introduction
Colorectal cancer is one of the major causes of cancer-related morbidity and mortality, ranking as the third most common malignancy in males and the second in females in the worldwide, with an estimated incidence of 1 233 700 new cancer cases and 608 700 deaths occurring in 2008.1 The development of colorectal cancer is a multistep process characterized by accumulation of epigenetic and genetic events, and influenced by lifestyle.2 Despite a number of molecular events having been discovered, the process of tumor initiation and progression is still unclear and molecules that play a crucial role in this process remain to be identified.
CCNB1, an important member of cyclin family, is a key initiator and rigorous quality control step of mitosis. It has a pivotal role in regulating cyclin-dependent kinase 1 (CDK1) and forming complex with it, which phosphorylates their substrates to promote the transition of cell cycle from G2 phase to mitosis.3,4 Increasing evidence demonstrates that CCNB1 is involved in checkpoint control, whose dysfunction is an early event in tumorigenesis, and that its deregulated expression is observed in a number of different human cancers including breast cancer, cervical cancer, lung cancer, esophageal squamous cell carcinoma, and melanoma.5-9 In parallel, evidence has showed that inhibition of CCNB1 expression renders breast cancer cells more sensitive to chemotherapy drug taxol,10 and CCNB1 is an independent predictor of HBV-related hepatocellular carcinoma recurrence.11 Furthermore, it has been revealed that wild-type p53 represses the transcription of CCNB1 through Sp1 transcription factor in tumor cells,12 and the protein level of CCNB1 is undetectable in HCT116 p53+/+ cells, while overexpressed in HCT116 p53−/− cells.13 These data highlight the potential pivotal roles of CCNB1 in the tumor development. However, detailed functional characteristics of CCNB1 are still unknown in colorectal cancer to date.
Here, we validated Chk1 was a right regulator of CCNB1 and observe the influence of modulating CCNB1 expression on biobehaviors of colorectal cancer cells and growth of transplanted tumor through different modern molecular biology techniques, accordingly elucidate the key role and mechanism of CCNB1 overexpression in promoting colorectal tumorigenesis.
Results
CCNB1 is overexpressed in human colorectal cancer tissues
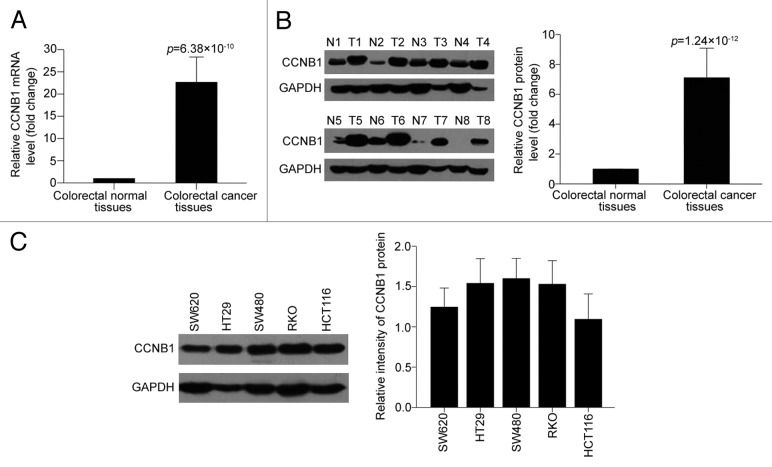
To study the expression pattern of CCNB1, we compared CCNB1 mRNA expression between colorectal cancer tissues and their pair-matched adjacent normal colorectal tissues from 30 individual patients using qRT-PCR analysis. As shown in Figure 1A, CCNB1 was frequently overexpressed in 93.3% (28/30) in colorectal cancer tissues compared with the matched adjacent nontumorous tissues from the same patient (Tumor/Normal > 2-fold), with up to 100-fold increases in cancer tissues. We simultaneously confirmed the protein level of CCNB1 expression by western blot in the same cases for CCNB1 mRNA dectection. Consistent with the mRNA data, CCNB1 protein level was also visibly elevated in colorectal cancer tissues with an average value of ~7 compared with the normal tissue as 1.0 (Fig. 1B).
Figure 1. CCNB1 is overexpressed in colorectal cancer. (A) qRT-PCR analysis of CCNB1 mRNA expression in 30 paired human colorectal cancer tissues and the matched adjacent normal tissues. GAPDH was used as internal control and for normalization of the data. The data shown were average fold changes of CCNB1 expression (2−ΔCT) in colorectal cancer tissues relative to the matched adjacent normal tissues. (B) Western blot validation of CCNB1 protein in the 30 paired human colorectal cancer tissues and the matched adjacent normal tissues. CCNB1 protein expression was normalized to GAPDH and expressed relative to the matched adjacent normal tissues. (C) Expression of CCNB1 protein by western blot in five colon cancer cell lines. Data were presented as mean ± SD.
We then examined the expression of CCNB1 in a panel of five human colorectal cancer cell lines (named HCT116, RKO, HT29, SW480, and SW620) by western blot (Fig. 1C), and chose HCT116 and SW480 cell lines, where CCNB1 was expressed respectively lower and higher among the five cell lines, for further investigation.
Inhibition of CCNB1 suppresses the proliferation of colorectal cancer cells in vitro and tumorigenicity in vivo
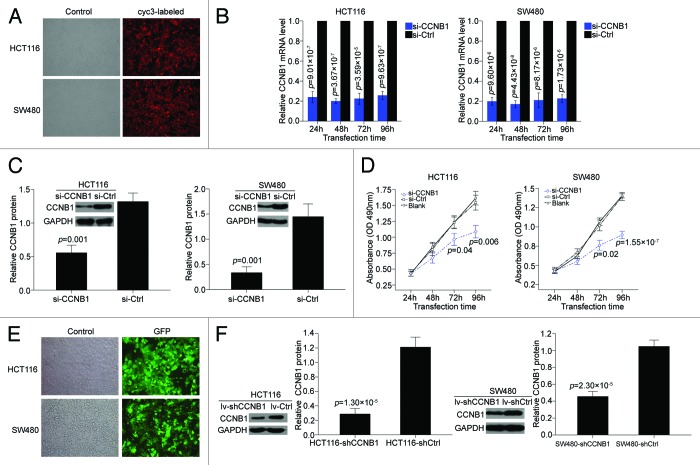
To examine whether repression of CCNB1 has the effects on cellular proliferation, the two colorectal cancer cell lines HCT116 and SW480 were transfected with specific CCNB1 siRNA. The transfection efficiency of siRNA in both cells was viewed by microscopy at 24 h after transfection of a cyc3-labled siRNA negative control (Fig. 2A), the expression of CCNB1 mRNA was determined at 24, 48, 72, and 96 h postransfection (Fig. 2B), and the expression of CCNB1 protein was determined at 72 h postransfection (Fig. 2C). MTT assay revealed that the inhibition of CCNB1 significantly decreased the growth rate of both colorectal cancer cell lines, compared with the cells tranfected with siRNA negative control (Fig. 2D).
Figure 2. Transfection efficiency and expression level of CCNB1 inhibition both under transient and steady-state conditions. Inhibition of CCNB1 suppresses the proliferation of colon cancer cells, HCT116 and SW480. (A) HCT116 and SW480 cells were transfected with cyc3-labled siRNA negative control at 24 h after plating, and then viewed at 24 h posttransfection by light microscopy (control) and fluorescence microscopy (cyc3-labled). (B and C) HCT116 and SW480 cells were transfected with CCNB1 siRNA or siRNA negative control (50 nM), and then the relative expression of CCNB1 mRNA was determered by qPCR at 24, 48, 72, and 96 h posttransfection. The relative expression of CCNB1 protein was determered at 72 h posttransfection. Data were presented as mean ± SD (n = 3). (D) At 24, 48, 72, and 96 h after transfection, cell proliferation was determined by the MTT assay. Data were presented as mean ± SD from three independent experiments. (E and F) Infection efficiency of HCT116 and SW480 cells stably expressing shRNA for CCNB1 by lentiviral transduction were viewed by a green fluorescent protein gene, GFP, and further determined by western blot. Data shown were representative of three individual western blot analyses.
To confirm the in vitro phenotype of CCNB1 expression, we further tested the effect of CCNB1 suppression on the growth of colorectal cancer tumors in vivo. First, we engineered HCT116 and SW480-based cells, which were stably expressing short hairpin RNA (shRNA) for CCNB1 by lentiviral transduction. The shRNA plasmid pL/shRNA/F contained a green fluorescent protein gene GFP which indicated the successful lentivirus transfection. The highest infection efficiency was obtained at multiplicities of infection of 10 for HCT116 and 100 for SW480 cells (Fig. 2E). To know the knockdown efficiency, endogenous expression of CCNB1 protein was examined by western blot. The HCT116 cells transduced with shCCNB1 vector (HCT116-shCCNB1) showed an at least 75% reduction in the expression of CCNB1 protein compared with the cells transduced with shControl vector (HCT116-shCtrl), and the SW480-shCCNB1 cell showed an 60% reduction compared with the SW480-shCtrl cells (Fig. 2F). These results indicated that the two stable cell lines were efficient and specific in downregulating CCNB1 expression, thus confirming the validity of following assay.
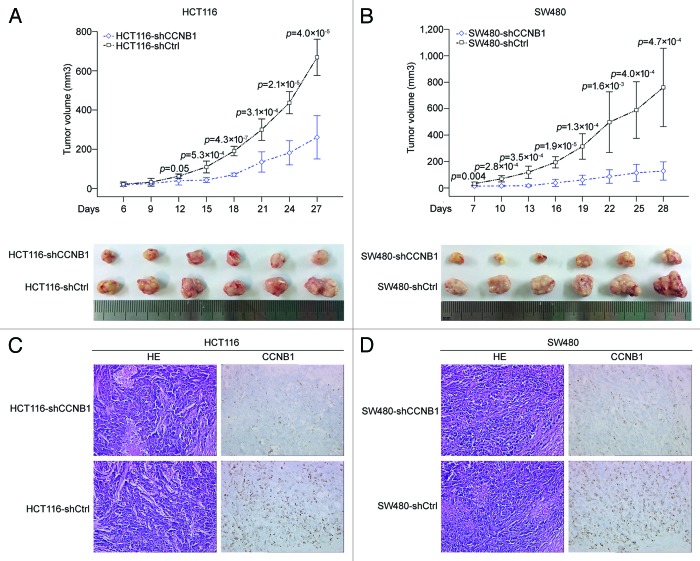
The HCT116-shCCNB1, HCT116-shCtrl, SW480-shCCNB1, and SW480-shCtrl cells were subcutaneously inoculated into BALB/c nude mice. As shown in Figure 3A, the tumors with HCT116-shCCNB1 grew more slowly than the tumors with HCT116-shCtrl (P < 0.001). Similar results were obtained using SW480 cells (P < 0.001; Fig. 3B). IHC staining confirmed that the tumors with CCNB1-repressing displayed much lower CCNB1 level than that of controls (Fig. 3C and D). Collectively, our data suggest that CCNB1 exerts a growth-promoting function in human colorectal cancer.
Figure 3. Downregulation of CCNB1 suppresses tumorigenicity of colorectal cancer cells in vivo. (A and B) Tumor xenograft model. The HCT116-shCCNB1, HCT116-shCtrl, SW480-shCCNB1 and SW480-shCtrl cells were injected into the left-side axilla of nude mice (n = 6/each group). Data points are presented as the mean ± SD tumor volume. (C and D) Histopathology of xenograft tumors. The tumor sections were under H&E staining and IHC staining using antibody against CCNB1. Data were presented as mean ± SD (n = 6).
Elevated CCNB1 expression in colorectal cancer is dependent on Chk1 expression
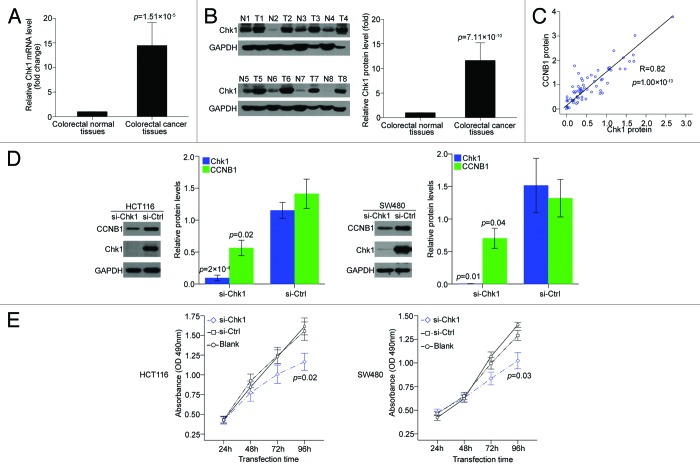
In mammalian cells, a large proportion of human cancers are thought to be highly reliant on the checkpoint kinase 1 (Chk1) expression,14,15 and CCNB1 was previously suggested to serve as a biomarker predictive of the efficacy of Chk1 inhibitors.16 To verify whether Chk1 could functionally regulate CCNB1 expression in colorectal cancer, we detected Chk1 expression in the 30 paired clinical colorectal cancer samples and matched adjacent normal tissues and revealed that Chk1 was overexpressed in both mRNA and protein levels in the cancer tissues (Fig. 4A and B) and correlated positively with CCNB1 protein expression (R = 0.82, P = 1.9 × 10−18; Fig. 4C). To further confirm the potential effect of Chk1 on CCNB1 expression, we transfected specific siRNA targeting Chk1 to HCT116 and SW480 cells. The result showed that downregulation of Chk1 suppressed CCNB1 protein expression in both two cell lines, compared with siRNA negative control (Fig. 4D). Moreover, MTT assay revealed that inhibition of Chk1 significantly decreased the growth rate of both colorectal cancer cell lines, compared with the negative control-transfected cells (Fig. 4E), implying Chk1 might be the right regulator of CCNB1 in colorectal cancer.
Figure 4. CCNB1 expression is dependent on Chk1 expression in colorectal cancer. (A and B) mRNA and protein levels of Chk1 in 30 paired human colorectal cancer tissues and the matched adjacent normal tissues. The expression of GAPDH was applied for normalization. Bars represented the average fold changes of Chk1 expression in colorectal cancer tissues relative to the matched adjacent normal tissues. Mean ± SD. (C) Expressin of CCNB1 protein positively correlated with expression of Chk1 protein in 30 paired human colorectal cancer tissues and the matched adjacent normal tissues. (D) Seventy-two hours after transfection with 50 nM of Chk1 siRNA or siRNA control, the endogenous protein levels of Chk1 and CCNB1 in HCT116 and SW480 cells were detected by western blot. Bars represented the relative protein levels that were normalized to GAPDH. Data were presented as mean ± SD (n = 3). (E) At 24, 48, 72, and 96 h after transfection, cell proliferation was determined by the MTT assay. Data were presented as mean ± SD from three independent experiments.
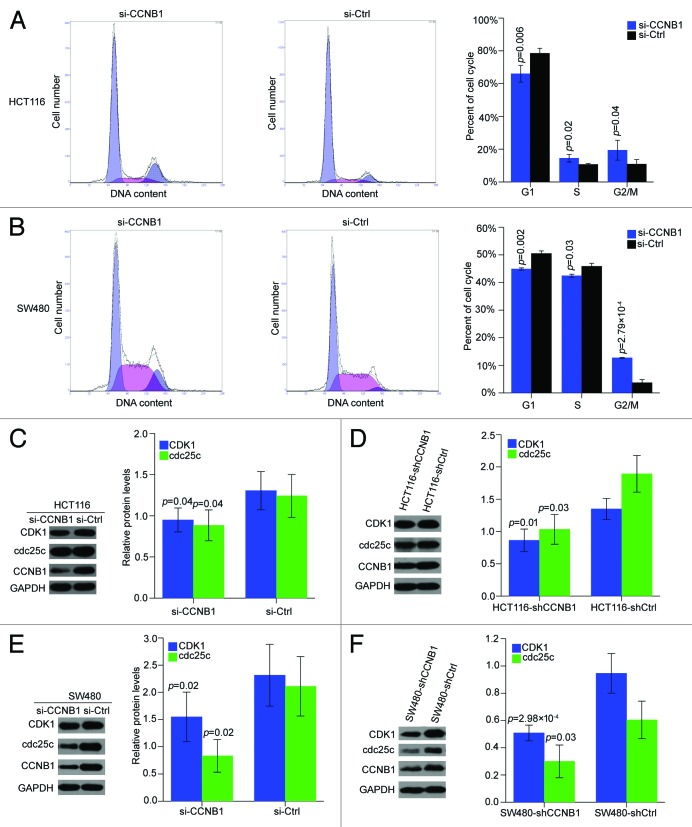
Knockdown of CCNB1 blocks cell cycle progression and regulates cell cycle factors cdc25c/CDK1 in both two colorectal cancer cells
We then determined the cell cycle distribution of HCT116 and SW480 cells by PI staining, to further explore the possible mechanisms by which CCNB1 regulates colorectal cancer cell growth. Suppression of CCNB1 increased in the percentage of cells in the G2/M phase from 13% to 24% and decreased in the percentage of cells in the G1 phase from 80% to 65% in HCT116 cells (Fig. 5A). Notably, repression of CCNB1 could also triggered an accumulation of cells at G2/M phase, and decreased the number of cells at G1 phase in SW480 cells (Fig. 5B). Furthermore, we detected the expression levels of critical mitosis regulators, cdc25c and CDK1. As expected, cdc25c and CDK1 protein leves were downregulated in both HCT116 and SW480 cells transfected with CCNB1 siRNA (Fig. 5C and E). In addition, the levels of cdc25c and CDK1 were also strongly repressed in the tumor xenografts generated from HCT116-shCCNB1 and SW480-shCCNB1 cells, compared with the respective vector control tumor xenografts (Fig. 5D and F). Thus, we hypothesized that the deregulation of CCNB1 might activate cell cycle progression through cdc25c and CDK1 in colorectal cancer cells.
Figure 5. Inhibition of CCNB1 inhibits cell-cycle progression in HCT116 and SW480 cells in vitro and in vivo. (A and B) 72h after transfection, cells were collected, stained with PI, and cell cycle was analyzed with FACS. Results were average from three separate experiments. Data were presented as mean ± SD of three independent experiments. (C and D) Western blot analysis of cdc25c and CDK1 proteins in CCNB1-repressed HCT116 cells and tumor xenografts generated from HCT116-shCCNB1 cells. The data shown were mean ± SD of three individual experiments. (E and F) Western blot analysis of cdc25c and CDK1 proteins in CCNB1-repressed SW480 cells and tumor xenografts generated from SW480-shCCNB1 cells. The data shown were mean ± SD of three individual experiments.
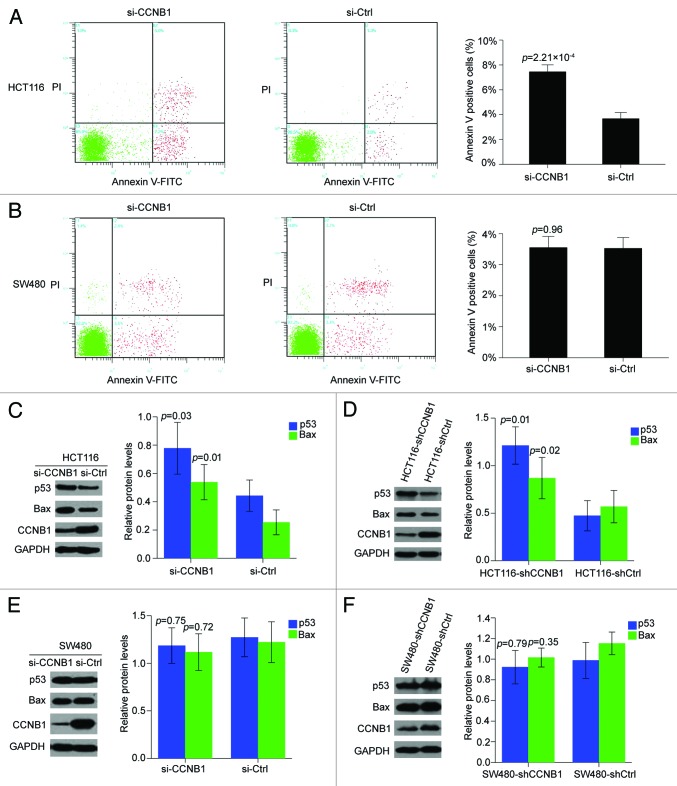
CCNB1 silencing induces apoptosis and regulates apoptosis-related factors p53/Bax in p53-wild HCT116 cells, but not in p53-mutant SW480 cells
To determine whether apoptosis also contributed to the growth inhibition of CCNB1 siRNA, we performed flow-cytometry-based analysis of HCT116 and SW480 cells after transfection of CCNB1 siRNA or control siRNA. As shown in Figure 6A, an increase in the early apoptosis population upon the repression of CCNB1 was found in HCT116 cells. However, the Annexin V-FITC data showed that knockdown of CCNB1 did not significantly alter the fraction of apoptotic cells in SW480 cell line (Fig. 6B).
Figure 6. Suppression of CCNB1 induces apoptosis in p53-wt HCT116 cells. (A and B) Seventy-two hours after transfection, apoptosis assay was performed to determine the early apoptotic rate of HCT116 and SW480 cells. Data were presented as mean ± SD of three independent experiments. (C and D) Western blot analysis of P53 and Bax proteins in CCNB1-repressed HCT116 cells and tumor xenografts generated from HCT116-shCCNB1 cells. The data shown were mean ± SD of three individual experiments. (E and F) Western blot analysis of P53 and Bax proteins in CCNB1-repressed SW480 cells and tumor xenografts generated from SW480-shCCNB1 cells. The data shown were mean ± SD of three individual experiments.
A recent publication by Kreis et al. suggested that downregulation of CCNB1 increased p53 expression in human papillomavirus 16/18-infected cancer cells.6 It has been well known that HCT116 contains wild-type p53, while SW480 exhibits dysregulation of p53 function by p53 mutation. We, therefore, need to know whether the different effects of CCNB1 on apoptosis of HCT116 and SW480 cells were due to the p53 status. Thus, we checked the levels of p53 protein and its main cellular target, Bax, both in vitro and in vivo. Surprisingly, knocking down CCNB1 by specific siRNA in HCT116 cells increased both p53 and Bax protein levels (Fig. 6C). Similar effects on p53 and Bax levels were observed in the tumor xenografts generated from HCT116-shCCNB1 cells, compared with those from HCT116-shCtrl cells (Fig. 6D). As expected, ectopic downregulation of CCNB1 in the SW480 cells did not alter p53 or Bax protein levels either in vitro or in vivo (Fig. 6E and F). These data suggested that ectopic inhibition of CCNB1 might induce apoptosis enhancement through activating p53 and Bax in p53-wild HCT116 cells.
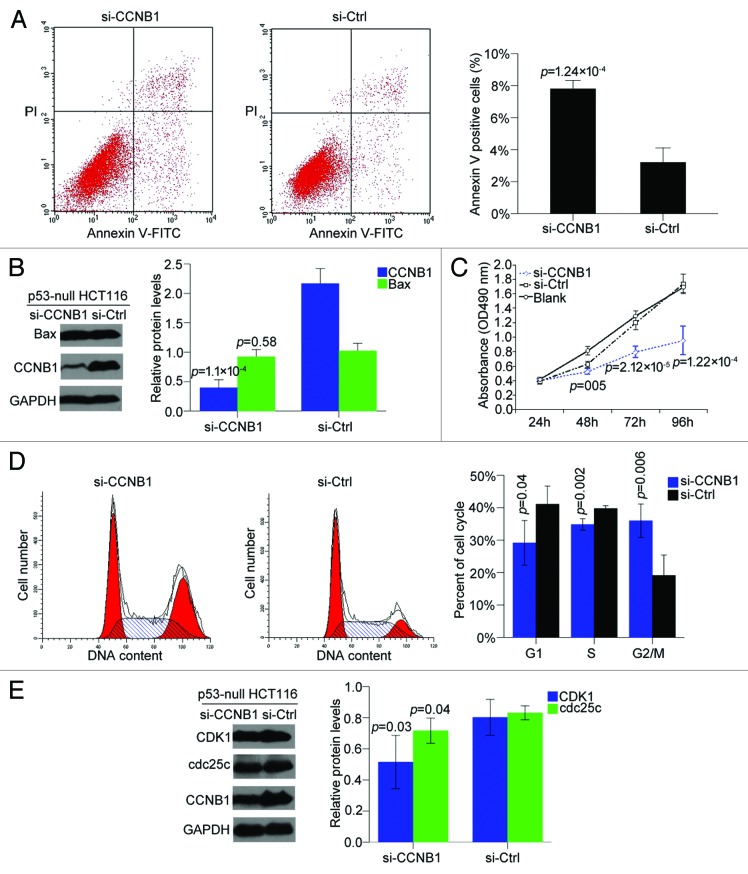
Inhibition of CCNB1 expression suppresses cell proliferation, blocks cell cycle progression, and induces apoptosis in p53-null HCT116 cells
To further confirm whether p53 contributed to the apoptosis induction of CCNB1 suppression, the effect of CCNB1 in apoptosis activity was also assessed in p53 -null HCT116 colon cancer cells. Interestingly, inhibition of CCNB1 provoked a robust apoptotic response in the p53-null HCT116 cells without any change of Bax protein level (Fig. 7A and B). We then evaluated cell growth and cell cycle in this cell line and found inhibition of CCNB1 suppressed cell proliferation and blocked cell cycle in G2/M phase by repressing cdc25c and CDK1 protein level (Fig. 7C–E), in line with the effects on HCT116 and SW480 cells. These above data demonstrated that ectopic inhibition of CCNB1 could activated p53, but did not require p53 for inducing the anticancer action in colorectal cancer cells.
Figure 7. Inhibition of CCNB1 suppresses cell proliferation, blocks cell cycle progression, and induces apoptosis in p53-null HCT116 cells. (A and B) Seventy-two hours after transfection, apoptosis assay was performed to determine the early apoptotic rate of p53-null HCT116 cells, and western blot was to detect Bax protein level. (C) At 24, 48, 72, and 96 h after transfection, cell proliferation was determined by MTT assay. (D and E) Seventy-two hours after transfection, cells were collected, stained with PI, and cell cycle was analyzed with FACS. Protein levels of cdc25c and CDK1 were determined by western blot. Data were presented as mean ± SD of three independent experiments.
Discussion
As one of the major health problems, colorectal cancer causes unbearable morbility and mortality worldwild.1 Aberrant cell cycle regulation has been considered as one of the essential characteristics of cancer cells including colorectal cancer cells.17 Targeting the deregulated cell cycle has been suggested as a practical strategy to check uncontrolled proliferation in cancer cells. Here we report on the actions of a key regulator of G2/M checkpoint, CCNB1 in promoting colorectal cancer development both in vitro and in vivo.
The CCNB1-CDK1 complex (so-called mitosis promoting factor, MPF) is required for orderly G2/M transition.18 It has been known that the activity of CDK1 is usually increased in cancer cells. Accordingly, pursuits for the inhibitors, targeting the activity of CDK1, has been the intense area of research for last two decades.19 CCNB1, the regulatory subunit of MPF, could specifically increases the activity of CDK1, further leading to a strong proliferation promotion in a variety of cancer cells.10,20 Studies have also revealed that CCNB1 is highly expressed in various primary tumors,5-9 including colorectal cancer.21 These data highlight the potential pivotal roles of CCNB1 both in the development and progression of malignancy. However, convincing data of the functional characteristics of CCNB1 in colorectal cancer are still lacking. Thus, we first examined CCNB1 expression in human colorectal cancer tumor samples compared with paired normal mucosa and found CCNB1 was overexpressed in tumors compared with normal tissues. Then, we used both transient and stable gene silencing of CCNB1 approaches in two colorectal cancer cell lines, HCT116 and SW480. Consistent with clinical data, knockdown of CCNB1 expression remarkably suppressed the proliferation and tumorigenesis in colorectal cancer cells tested in vitro and in vivo.
Few studies describe the regulation of CCNB1 specifically. A recent study suggested that CCNB1 was an efficacy-predicting biomarker for Chk1 inhibitors across different types of cancers.16 Chk1 is an essential kinase in governing cell cycle G1/S, S, and G2/M phase checkpoints and determining cellular responses to DNA damage.22-26 In contrast to the well-known utility of Chk1 inhibitors in sensitizing tumors to chemotherapy agents,23,27,28 Chk1 overexpression or downexpression also occurs in some types of tumors, including breast cancer, ovarian cancer, cervical cancer, and neuroblastoma.22,29-33 In our study, we found Chk1 mRNA and protein levels were upregulated in colorectal cancer tissues compared with paired normal tissues, and positively correlated with CCNB1 expression. We hypothesized that CCNB1 might serve as a Chk1-induced oncogene in colorectal cancer. Therefore, we assessed the repression of Chk1 by specific siRNA on CCNB1 expression and cell proliferation in colorectal cancer cells. A significant decrease in CCNB1 protein level and cell growth was observed in both HCT116 and SW480 cancer cells. Collectively, these results suggested that Chk1 might be a right regulator of CCNB1 in colorectal cancer cells.
To better understand how deregulation of CCNB1 impacts colorectal cancer cell growth, we showed that knockdown of CCNB1 could induce cell cycles arrest in both HCT116 and SW480 cells. CCNB1 silencing mediated G2/M arrest was associated with a decrease in the protein levels of cdc25c and CDK1. Progression through G2/M phase involves the association of CDK1 with CCNB1.18 During G2 phase, the CDK1/CCNB1 complex is inactive. At the onset of mitosis, the complex is actived by the phosphatase cdc25c. CDK1/CCNB1 can then phosphorylate cdc25c, further activating it and initiating a positive feedback loop.34 In our study, we found that suppression of CCNB1 reduces the levels of CDK1 as well as cdc25c both in cell culture and xenograft tissues, and thereby possibly causes the G2/M phase arrest. In addition, we also showed that the molecular mechanism by which repression of CCNB1 retarded colorectal cancer cell growth was due, at least in part, to acceleration of apoptosis, upregulation of p53 and its downstream effector, Bax, in the p53-wild HCT116 cells silencing of CCNB1. Even in HCT116 cells grown as tumor xenograft in nude mice, increased levels of p53 and Bax were also observed in tumors from HCT116-shCCNB1 cells, thereby implying that reduction of apoptosis may be a contributing factor in the CCNB1-mediated promoted tumor growth. Previous studies revealed that the p53 signaling was required for apoptosis progression. In addition, activation of p53 directly induces Bax expression, and thereby promoting cell apoptosis. Mutation of p53 leads to a loss of normal function of the wild-type protein. In our study, despite significant CCNB1 down-expression, we could not observe any apoptosis induction and p53 protein change of p53-mutant SW480 cells tested in vitro and in vivo. However, when we further evaluated the effects of ectopic suppression of CCNB1 by transfection with CCNB1 siRNA on cell apoptosis in p53-null HCT116 cells, we found that inhibition of CCNB1 also provoked a robust apoptotic response without any change of Bax protein level in p53-null HCT116 cells. The molecular mechanism of CCNB1 in apoptosis pathway seems much more complicated than we expected, and much more studies should be done to explore the possible mechanism in the future. In addition, MTT and cell cycle analysis showed that inhibition of CCNB1 could suppress cell proliferation, blocked cell cycle in G2/M phase by repressing cdc25c and CDK1 in the p53-null HCT116 cells. Our data suggested that CCNB1 could suppress p53, but did not require p53 for inducing the anticancer action in colorectal cancer cells.
In summary, our findings suggest an oncogenic role for CCNB1 in colorectal cancer. Molecular analyses showed that CCNB1 is activated by Chk1 overexpression and inhibition of CCNB1 induces cell cycle arrest in different colorectal cancer cell lines through modulating the expression of G2/M cell cycle regulators. Also, suppression of CCNB1 induces apoptosis in certain colorectal cancer cells. Overall, understanding the precise roles played by CCNB1 in the development of colorectal cancer will help our understanding of the biology of this tumor type, and may also allow the development of a novel therapeutic approach against colorectal cancer based on inhibition of CCNB1.
Materials and Methods
Tissue collection
Fresh primary colorectal cancer specimens and paired non-cancerous colorectal tissues from 30 patients during Mar 2012–Sep 2012 were provided by Tumor Tissue Bank of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine. The study was approved by the Hospital Ethical Committee and specimens were obtained with informed consent. All tissue samples were immediately snap-frozen in liquid nitrogen and stored at –80 °C until use.
RNA isolation and quantitative real-time RT-PCR (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen) following the manufacturer’s instructions, and cDNA was synthesized with the PrimeScript RT reagent Kit (TaKaRa Otsu). qRT-PCR analyses for mRNA were performed using SYBR Premix Ex Taq (TaKaRa) on an ABI 7500HT system (ABI). GAPDH was identified as a suitable internal control, and fold changes were calculated through relative quatifucation (2−ΔCT). The primers used are listed in Table 1.
Table 1. Sequences of primers siRNA and shRNA.
| Primers, siRNA and shRNA | Sequence (5′ - 3′) |
|---|---|
| GAPDH forward | GACAGTCAGC CGCATCTTCT |
| GAPDH reverse | TTAAAAGCAG CCCTGGTGAC |
| CCNB1 forward | CCAAATCAGA CAGATGGAAA T |
| CCNB1 reverse | GCCAAAGTAT GTTGCTCGA |
| Chk1 forward | AAGCGTGCCG TAGACTGT |
| Chk1 reverse | TTATCCCTTT CATCCAAC |
| CCNB1 siRNA forward | GAAUUCUGCA CUAGUUCAA (dTdT) |
| CCNB1 siRNA reverse | UUGAACUAGU GCAGAAUUC (dTdT) |
| CCNB1 shRNA forward | CACCGAATTC TGCACTAGTT CAACGAATTG AACTAGTGCA GAATTC |
| CCNB1 shRNA reverse | AAAAGAATTC TGCACTAGTT CAATTCGTTG AACTAGTGCA GAATTC |
Cell culture and transfection
Human colorectal cancer cell lines, HCT116 (p53-wild HCT116), SW480, RKO, SW620, and HT29 were obtained from the American Type Culture Collection (ATCC). p53-null HCT116 was obtained from prof. Hongchuan, Jin (Department of Surgical Oncology, Sir Run Run Shaw Hospital). Two HCT116 cell lines were cultured in McCOY’S 5A and others were grown in RPMI 1640, all supplemented with 10% fetal bovine serum (FBS) (Gibco) at 37 °C and 5% CO2.
siRNA against Chk1 and scrambled siRNA negative control were synthesized by Ribobio. siRNA against CCNB1 and scrambled siRNA negative control were synthesized by bioneer. For transient transfection, siRNAs were transfected at a working concentration of 50 nM using lipid-based transfection reagents DharmaFECT1 (Dharmacon) according to the manufacturer’s guidelines. The transfection efficiency was detected by a cyc3-labled siRNA Ncontrol for monitoring the delivery into cancer cells. The relative expression of CCNB1 at 0, 24, 48, 72, and 96 h posttransfection was directly determered by qRT-PCR.
Western blot analysis
Total soluble proteins were prepared in RIPA buffer and assessed by immunoblot analysis. Cell lysates (20–50 μg) were subjected to 10% SDS-PAGE, transferred electrophoretically to PVDF membrances, and blotted with specific primary rabbit antibodies to CCNB1 (1:2000; Epitomics), Bax (1:2000; Epitomics), CDK1 (1:2000; Epitomics), and cdc25c (1:1000; Epitomics); and mouse antibodies to GAPDH (1:2000; Santa Cruz Laboratories), p53 (1:1000; Santa Cruz). The appropriate secondary antibodies conjugated to horseradish peroxidase (HRP) (Dawen Biotec) were then added and antigen-antibody complex was detected by enhanced chemiluminescence (ECL) (Biological Industries, Kibbutz Beit Haemek) reagent. GAPDH was used as a loading control.
MTT assay
Cell viability was identified by 3- (4,5- dimethylthiazol-2-yl)- 2, 5- diphenyl- tetrazolium bromide (MTT) assay. Twenty microliters of 5 mg/mL MTT solution (Sigma-Aldrich) was added into each test well 24, 48, 72, and 96 h respectively post-transfection, and incubated for 4 h. The supernatant was then removed, and 150 μL of dimethylsulfoxide (DMSO) (Sigma-Aldrich) was added to stop the reaction. Optical density (OD) was evaluated by measuring the absorbance at a wavelength of 490 nm on a a microplate spectrophotometer ELx800 (Bio-Tek). All experiments were performed in triplicate.
Cell cycle analysis
After incubation for 48 h post-transfection, cells were collected, washed with PBS, fixed with 70% ethanol, and stored overnight at 4 °C prior to staining. Cell cycle progression was then quantified by PI (20 μg/mL) (Biyotime) staining and subsequently analyzed with flow cytometry FACS EPICS (Beckman Coulter; Becton Dickinson), according to the manufacturer’s instructions. All experiments were repeated in triplicate.
Apoptosis assay
The apoptosis ratio was identified using the Annexin V-FITC Apoptosis Detection Kit (Biouniquer). Cells were harvested 72 h post-transfection, washed twice with PBS, and resuspended in binding buffer containing Annexin V-FITC and PI following the manufacturer’s protocol. The samples were analyzed by flow cytometry (Beckman Coulter; Becton Dickinson). The percentages of apoptotic cells from each group were then compared. Tests were repeated in triplicate.
Construction and transduction of lentiviral vectors encoding CCNB1 shRNA
For constant suppression of CCNB1 expression in HCT116 and SW480 cells, a lentiviral vector encoding CCNB1 shRNA (pL/shRNA/F-CCNB1) was constructed for knockdown of CCNB1. The sequences of CCNB1 shRNA were shown in Table 1. For cell infection, viral supernatants were supplemented with 5 μg/mL polybrene and incubated with cells for 12 h. HCT116 and SW480 cells were transduced by the lentiviral particles followed by blasticidine selection (4 μg/mL and 6 μg/mL, respectively) for 10 d. The cells stably expressing shRNA were confirmed by performing qRT-PCR and western blot analysis.
Animal model experiments
All animal studies were performed in accordance with the rules and guidelines concerning the use and care of laboratory animals and approved by the Animal Care Committee of Zhejiang University. For in vivo tumor growth assay, 4-wk-old BALB/c nude mice were purchased from Shanghai Laboratory Animal Center (SLAC) and housed in a dedicated SPF facility at Laboratory Animal Center of Zhejiang University. Equal numbers of cells stably infected with pL/shRNA/F-CCNB1 or pL/shRNA/F (5 × 106 cells/injection for HCT116 and SW480, respectively) were resuspended in 100 μL PBS, and injected subcutaneously in the left-side axilla of each animal, respectively (n = 6/each group). The diameters of tumors were measured once a week with precision calipers. Tumor mass (xenograft) volume was calculated according to the following formula: volume = [(tumor length) × (tumor width)2] / 2. About 4 wk after xenograft, mice were sacrificed and tumors were removed, weighed, and photographed. Tissue samples were fixed overnight in 10% paraformaldehyde, embedded in paraffin, and sectioned for histological studies and western blot analysis.
Immunohistochemistry analysis
The tumor xenografts embedded in paraffin were sectioned of 4 μm and stained with hematoxylin and eosin according to standard protocols. Sections were further under IHC staining using the Envision method. In brief, a unique band was obtained when the selected antibody in western blot analysis was used, thus the same anti-CCNB1 antibody was used for IHC analysis.
Sections were deparaffinized in xylene and rehydrated with ethanol. Next, hydrated autoclave pretreatment was performed by boiling the samples in 10 mM citrate buffer for 2 min. Endogen peroxidase was quenched with 50 μL of 3% hydrogen peroxidase for 10 min at room temperature (RT). Slides were then incubated with the primary anti-CCNB1 antibody at a dilution of 1:10 000 in Tris-buffered solution at RT for 1 h followed by incubation with Dako Envision Peroxidase (Dako Diagnostica) for 1 h at RT. The antibody staining was visualized with DAB (Dako Diagnostica). The section slides were counterstained with Mayer’s hematoxylin, dehydrated and mounted. Controls without incubation with primary antibody were processed. Stained sections were evaluated by Image pro-plus 6.0 (Media Cybernetics) and the average score for each slide was used for statistical analysis.
Statistical analysis
Statistical calculations were performed using SPSS 19.0 (WPSS Ltd). Results were presented as mean ± SE/SD. Relative quantification of mRNA expression was calculated with the 2−ΔCT method. Differences between colorectal cancer and normal colorectal tissues were assessed by the nonparametric Mann–Whitney U test. For other experiments in vitro and in vivo, differences between two groups were evaluated using the Student t test. All P values were two-sides and P values < 0.05 were considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Professor Xing Xie and Weiguo Lu for their helpful suggestions and comments of this manuscript. We would also like to acknowledge continuous financial support by grants from the International Scientific and Technological Cooperation Projects (2012DFA30410) and the National Science-technology Support Plan Projects of China (2012BAI14B06).
Author Contributions
Y.F. conceived and performed experiments and drafted the article. H.Y., X.L., and J.X. conceived and performed partial experiments. X.C. designed the study, supervised experiments, and revised the article. All authors were involved in writing the paper and had final approval of the submitted and published versions.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Migliore L, Migheli F, Spisni R, Coppedè F. Genetics, cytogenetics, and epigenetics of colorectal cancer. J Biomed Biotechnol. 2011;2011:792362. doi: 10.1155/2011/792362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krek W, Nigg EA. Differential phosphorylation of vertebrate p34cdc2 kinase at the G1/S and G2/M transitions of the cell cycle: identification of major phosphorylation sites. EMBO J. 1991;10:305–16. doi: 10.1002/j.1460-2075.1991.tb07951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–4. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 5.Niméus-Malmström E, Koliadi A, Ahlin C, Holmqvist M, Holmberg L, Amini RM, Jirström K, Wärnberg F, Blomqvist C, Fernö M, et al. Cyclin B1 is a prognostic proliferation marker with a high reproducibility in a population-based lymph node negative breast cancer cohort. Int J Cancer. 2010;127:961–7. doi: 10.1002/ijc.25091. [DOI] [PubMed] [Google Scholar]
- 6.Kreis NN, Sanhaji M, Krämer A, Sommer K, Rödel F, Strebhardt K, Yuan J. Restoration of the tumor suppressor p53 by downregulating cyclin B1 in human papillomavirus 16/18-infected cancer cells. Oncogene. 2010;29:5591–603. doi: 10.1038/onc.2010.290. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida T, Tanaka S, Mogi A, Shitara Y, Kuwano H. The clinical significance of Cyclin B1 and Wee1 expression in non-small-cell lung cancer. Ann Oncol. 2004;15:252–6. doi: 10.1093/annonc/mdh073. [DOI] [PubMed] [Google Scholar]
- 8.Nozoe T, Korenaga D, Kabashima A, Ohga T, Saeki H, Sugimachi K. Significance of cyclin B1 expression as an independent prognostic indicator of patients with squamous cell carcinoma of the esophagus. Clin Cancer Res. 2002;8:817–22. [PubMed] [Google Scholar]
- 9.Kedinger V, Meulle A, Zounib O, Bonnet ME, Gossart JB, Benoit E, Messmer M, Shankaranarayanan P, Behr JP, Erbacher P, et al. Sticky siRNAs targeting survivin and cyclin B1 exert an antitumoral effect on melanoma subcutaneous xenografts and lung metastases. BMC Cancer. 2013;13:338. doi: 10.1186/1471-2407-13-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Androic I, Krämer A, Yan R, Rödel F, Gätje R, Kaufmann M, Strebhardt K, Yuan J. Targeting cyclin B1 inhibits proliferation and sensitizes breast cancer cells to taxol. BMC Cancer. 2008;8:391. doi: 10.1186/1471-2407-8-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng L, Du J, Zhou Q, Cheng B, Li J, Zhang D, Ling C. Identification of cyclin B1 and Sec62 as biomarkers for recurrence in patients with HBV-related hepatocellular carcinoma after surgical resection. Mol Cancer. 2012;11:39. doi: 10.1186/1476-4598-11-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Innocente SA, Lee JM. p53 is a NF-Y- and p21-independent, Sp1-dependent repressor of cyclin B1 transcription. FEBS Lett. 2005;579:1001–7. doi: 10.1016/j.febslet.2004.12.073. [DOI] [PubMed] [Google Scholar]
- 13.Yu M, Zhan Q, Finn OJ. Immune recognition of cyclin B1 as a tumor antigen is a result of its overexpression in human tumors that is caused by non-functional p53. Mol Immunol. 2002;38:981–7. doi: 10.1016/S0161-5890(02)00026-3. [DOI] [PubMed] [Google Scholar]
- 14.O’Connor MJ, Martin NM, Smith GC. Targeted cancer therapies based on the inhibition of DNA strand break repair. Oncogene. 2007;26:7816–24. doi: 10.1038/sj.onc.1210879. [DOI] [PubMed] [Google Scholar]
- 15.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–9. doi: 10.1016/S1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 16.Xiao Z, Xue J, Gu WZ, Bui M, Li G, Tao ZF, Lin NH, Sowin TJ, Zhang H. Cyclin B1 is an efficacy-predicting biomarker for Chk1 inhibitors. Biomarkers. 2008;13:579–96. doi: 10.1080/13547500802063240. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Sánchez I, Dynlacht BD. New insights into cyclins, CDKs, and cell cycle control. Semin Cell Dev Biol. 2005;16:311–21. doi: 10.1016/j.semcdb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Malumbres M, Pevarello P, Barbacid M, Bischoff JR. CDK inhibitors in cancer therapy: what is next? Trends Pharmacol Sci. 2008;29:16–21. doi: 10.1016/j.tips.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Yuan J, Yan R, Krämer A, Eckerdt F, Roller M, Kaufmann M, Strebhardt K. Cyclin B1 depletion inhibits proliferation and induces apoptosis in human tumor cells. Oncogene. 2004;23:5843–52. doi: 10.1038/sj.onc.1207757. [DOI] [PubMed] [Google Scholar]
- 21.Grabsch H, Lickvers K, Hansen O, Takeno S, Willers R, Stock W, Gabbert HE, Mueller W. Prognostic value of cyclin B1 protein expression in colorectal cancer. Am J Clin Pathol. 2004;122:511–6. doi: 10.1309/54H4Q88A1UBBWPTE. [DOI] [PubMed] [Google Scholar]
- 22.Albiges L, Goubar A, Scott V, Vicier C, Lefèbvre C, Alsafadi S, Commo F, Saghatchian M, Lazar V, Dessen P, et al. Chk1 as a new therapeutic target in triple-negative breast cancer. Breast. 2014;23:250–8. doi: 10.1016/j.breast.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 23.McNeely S, Beckmann R, Bence Lin AK. CHEK again: revisiting the development of CHK1 inhibitors for cancer therapy. Pharmacol Ther. 2014;142:1–10. doi: 10.1016/j.pharmthera.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Hunter T. Roles of Chk1 in cell biology and cancer therapy. Int J Cancer. 2014;134:1013–23. doi: 10.1002/ijc.28226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caparelli ML, O’Connell MJ. Regulatory motifs in Chk1. Cell Cycle. 2013;12:916–22. doi: 10.4161/cc.23881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson R, Eastman A. The cancer therapeutic potential of Chk1 inhibitors: how mechanistic studies impact on clinical trial design. Br J Clin Pharmacol. 2013;76:358–69. doi: 10.1111/bcp.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engelke CG, Parsels LA, Qian Y, Zhang Q, Karnak D, Robertson JR, Tanska DM, Wei D, Davis MA, Parsels JD, et al. Sensitization of pancreatic cancer to chemoradiation by the Chk1 inhibitor MK8776. Clin Cancer Res. 2013;19:4412–21. doi: 10.1158/1078-0432.CCR-12-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gadhikar MA, Sciuto MR, Alves MV, Pickering CR, Osman AA, Neskey DM, Zhao M, Fitzgerald AL, Myers JN, Frederick MJ. Chk1/2 inhibition overcomes the cisplatin resistance of head and neck cancer cells secondary to the loss of functional p53. Mol Cancer Ther. 2013;12:1860–73. doi: 10.1158/1535-7163.MCT-13-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cole KA, Huggins J, Laquaglia M, Hulderman CE, Russell MR, Bosse K, Diskin SJ, Attiyeh EF, Sennett R, Norris G, et al. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc Natl Acad Sci U S A. 2011;108:3336–41. doi: 10.1073/pnas.1012351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verlinden L, Vanden Bempt I, Eelen G, Drijkoningen M, Verlinden I, Marchal K, De Wolf-Peeters C, Christiaens MR, Michiels L, Bouillon R, et al. The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor /progesterone receptor /HER-2 breast carcinomas. Cancer Res. 2007;67:6574–81. doi: 10.1158/0008-5472.CAN-06-3545. [DOI] [PubMed] [Google Scholar]
- 31.Ehlén Å, Nodin B, Rexhepaj E, Brändstedt J, Uhlén M, Alvarado-Kristensson M, Pontén F, Brennan DJ, Jirström K. RBM3-regulated genes promote DNA integrity and affect clinical outcome in epithelial ovarian cancer. Transl Oncol. 2011;4:212–21. doi: 10.1593/tlo.11106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J, Li Y, Wang F, Wang X, Cheng B, Ye F, Xie X, Zhou C, Lu W. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene. 2013;32:976–87. doi: 10.1038/onc.2012.121. [DOI] [PubMed] [Google Scholar]
- 33.Gao Q, Huang X, Tang D, Cao Y, Chen G, Lu Y, Zhuang L, Wang S, Xu G, Zhou J, et al. Influence of chk1 and plk1 silencing on radiation- or cisplatin-induced cytotoxicity in human malignant cells. Apoptosis. 2006;11:1789–800. doi: 10.1007/s10495-006-9421-4. [DOI] [PubMed] [Google Scholar]
- 34.Izumi T, Maller JL. Elimination of cdc2 phosphorylation sites in the cdc25 phosphatase blocks initiation of M-phase. Mol Biol Cell. 1993;4:1337–50. doi: 10.1091/mbc.4.12.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]