Abstract
Inhibitors of isoprenylcysteine carboxylmethyltransferase (Icmt) are promising anti-cancer agents, as modification by Icmt is an essential component of the protein prenylation pathway for a group of proteins that includes Ras GTPases. Cysmethynil, a prototypical indole-based inhibitor of Icmt, effectively inhibits tumor cell growth. However, the physical properties of cysmethynil, such as its low aqueous solubility, make it a poor candidate for clinical development. A novel amino-derivative of cysmethynil with superior physical properties and marked improvement in efficacy, termed compound 8.12, has recently been reported. We report here that Icmt −/− mouse embryonic fibroblasts (MEFs) are much more resistant to compound 8.12-induced cell death than their wild-type counterparts, providing evidence that the anti-proliferative effects of this compound are mediated through an Icmt specific mechanism. Treatment of PC3 prostate and HepG2 liver cancer cells with compound 8.12 resulted in pre-lamin A accumulation and Ras delocalization from the plasma membrane, both expected outcomes from inhibition of the Icmt-catalyzed carboxylmethylation. Treatment with compound 8.12 induced cell cycle arrest, autophagy and cell death, and abolished anchorage-independent colony formation. Consistent with its greater in vitro efficacy, compound 8.12 inhibited tumor growth with greater potency than cysmethynil in a xenograft mouse model. Further, a drug combination study identified synergistic antitumor efficacy of compound 8.12 and the epithelial growth factor receptor (EGFR)-inhibitor gefitinib, possibly through enhancement of autophagy. This study establishes compound 8.12 as a pharmacological inhibitor of Icmt that is an attractive candidate for further preclinical and clinical development.
Keywords: Icmt, Ras, cysmethynil, compound 8.12, autophagy, anti-proliferation, gefitinib, synergy
Introduction
Protein prenylation has been known for many years as an important post-translational processing event for proteins containing a so-called CaaX motif at their C-terminus (CaaX-proteins).1 The CaaX motif can be found in a wide variety of proteins, ranging from small G proteins such as members of the Ras, Rho, and Rac families to structural proteins such as nuclear lamins.2 The CaaX motif consists of a cysteine residue fourth from the C-terminus, followed by two generally aliphatic amino acids; the last position can accommodate a wide range of amino acids.3 Protein prenylation is a three-step process that often targets CaaX proteins to cellular membranes where they function. The first step involves the addition of an isoprenyl group to the cysteine residue in the CaaX motif. This cytoplasmic process can either be farnesylation or geranylgeranylation, catalyzed by protein farnesyltransferase (FTase) or protein geranylgeranyltransferase-1 (GGTase-1), respectively.4 This prenylation step is crucial for the function of CaaX proteins.2 The second step of this pathway, which takes place in the endoplasmic reticulum, involves the cleavage of the C-terminal tripeptide aaX by the CaaX endoprotease Rce1, exposing the isoprenylated cysteine at the C-terminal.5
Icmt is a highly-conserved endoplasmic reticulum-resident protein consisting of 8 transmembrane domains and a cytoplasmic catalytic domain.6,7 Icmt catalyzes the last step of the prenylation pathway for CaaX-proteins, where it uses S-adenosyl-l-methionine (AdoMet) as the methyl donor for the carboxylate group at the exposed C-terminal cysteine through an esterification reaction.8 For membrane-targeted CaaX-proteins such as the Ras-family of proteins, the methyl group at the C-terminal neutralizes the negative charge on the cysteine residue and increases its hydrophobicity, thus increasing its affinity to the plasma membrane.2,7 In mice, disruption of the Icmt gene is embryonically lethal at embryonic day 10.5–11.5.9 Mouse embryonic fibroblasts (MEFs) derived from conditional Icmt knockout mice are resistant to oncogenic transformation by activated forms of Kras and Braf,10 and conditional deletion of Icmt ameliorated myeloproliferation phenotypes in a mouse model of Kras-induced cancer.11
Several CaaX-proteins, such as members of the Ras and Rho families, among others, have been implicated in tumorigenesis and tumor progression. Activating Ras mutations can be found in a third of all human cancers and more in some cancer types,12,13 making the Ras pathway an attractive target for cancer therapy development. Direct Ras inhibition has been a difficult task primarily due to the biochemical properties of the GTPase,14 and any inhibitor identified may only be limited to one subtype of Ras (e.g., Kras) and one specific type of activating mutation (e.g., G12C).15 To circumvent this challenge, much effort has been put into the development of molecules that target tumorigenic CaaX proteins, particularly Ras, through the inhibition of their posttranslational prenylation process.12 Inhibitors targeting the enzyme required for the first step of Ras prenylation, FTase, has proceeded to clinical trials with only limited success; one potential reason being that geranylgeranylation can replace farnesylation to activate some forms of Ras in the absence of FTase activity.16,17 Thus, Icmt has emerged as an anticancer target, since both farnesylated and geranylgeranylated CaaX-proteins, including the Ras and Rho family proteins, are processed by the same enzyme.
An indole-based inhibitor of Icmt, cysmethynil, was discovered via an in vitro screening of a diverse chemical library.18 Cysmethynil treatment has been shown to inhibit growth and induce cell cycle arrest in cancer cells of different tumor origins, including colon, liver and prostate, and elicit cell death in a process that involves autophagy induction.18,19 Cysmethynil treatment has also been shown to induce Ras mislocalization from the plasma membrane.18 The functions of RhoA and Rac1, which are CaaX-proteins involved in cytoskeletal reorganization and cell migration, were also attenuated in cells after treatment with cysmethynil, suggesting that Icmt inhibition could also negatively impact cancer metastasis.20,21 Despite its specificity and effectiveness in affecting tumor cell growth and biological functions in cells, cysmethynil may not be a good candidate for drug development because of its high lipophilicity, low water solubility, and affinity to plasma proteins.22
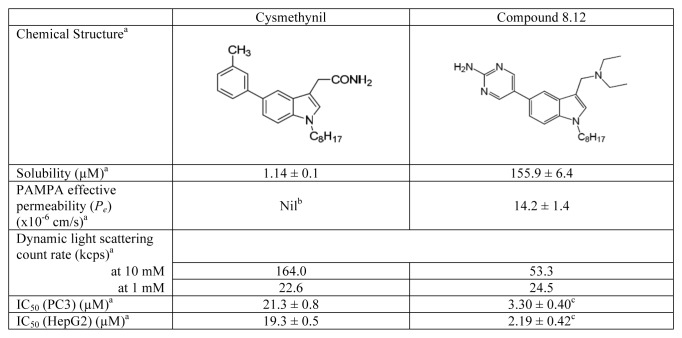
In studies aimed at modifying the indole core of cysmethynil to improve its “drug-likeness” while retaining its activity against Icmt, we found that amino-substituents at position 3 of the indole ring produced analogs with improved activities in Icmt inhibition and cell growth inhibition.22 By making further modifications to the functional group at position 5 of the indole ring, compound 8.12 was discovered which exhibits superior physical properties compared with cysmethynil. Compound 8.12 is more water soluble and has better cell permeability (Scheme 1).23 Most notably, IC50 values for inhibition of both HepG2 and PC3 cell growth for compound 8.12 are almost 10-fold lower than that of cysmethynil (Scheme 1).23
Scheme 1. Chemical structures and physical properties of cysmethynil and compound 8.12. aThese values are obtained from our previous report,23 unless otherwise indicated. bValue could not be determined due to cysmethynil’s low water solubility. cThese IC50 values are determined with the conditions described in Materials and Methods in this study. PC3, n = 16; HepG2, n = 12. PAMPA, parallel artificial membrane permeability assay.
In the current study, we further investigated the biological consequences of treating cancer cells with compound 8.12, and determined its pharmacokinetic parameters as well as anti-tumor effects in vivo. In cell-based systems, compound 8.12 has similar biological activity to cysmethynil in terms of Ras mislocalization and pre-lamin A accumulation, as well as cell cycle arrest induction and autophagy initiation. Additionally, compound 8.12 was also much more effective than cysmethynil in inhibiting tumor growth in vivo, and displayed promising synergistic activity with approved anti-tumor agent gefitinib in multiple cancer cell lines. The pharmacokinetic parameters of compound 8.12 suggested that it can be given in vivo with realistic dosing regimens and without significant toxicity, and its synergistic interaction with gefitinib suggests that compound 8.12 can potentially be developed together with tyrosine kinase-inhibitors as novel drug combinations in cancer therapy.
Results
Compound 8.12 negatively affects cellular processes that require carboxylmethylation of prenylated CaaX-proteins
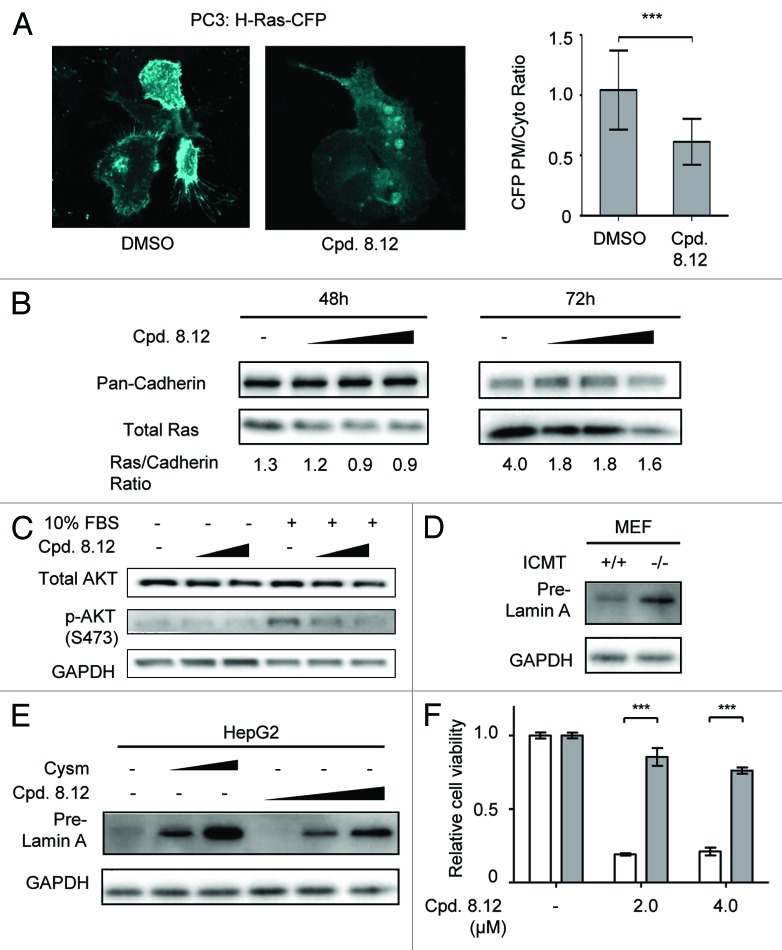
One of the major effects of pharmacological inhibition of Icmt is the mislocalization of Ras from the plasma membrane,18 as carboxylmethylation of the prenylated cysteine is important for proper plasma membrane localization of Ras.10,24 Indeed, compound 8.12 was found to cause Ras mislocalization. PC3 cells expressing cyan fluorescent protein (CFP)-tagged Hras were treated with either compound 8.12 or dimethyl sulfoxide (DMSO) control. In DMSO-treated cells, CFP fluorescence was readily observed at the periphery of the cells, indicating plasma membrane localization of the CFP-tagged Hras. In contrast, in compound 8.12-treated cells, CFP fluorescence in the cell periphery diminished, indicating mislocalization of CFP-Hras (Fig. 1A, left). Image analysis of populations of control and compound 8.12-treated cells provided quantitative data showing that plasma membrane to cytoplasmic ratio for CFP fluorescence was significantly reduced by compound 8.12 treatment (Fig. 1A, right). In addition, treating PC3 cells with increasing concentrations of compound 8.12 resulted in a dose-dependent reduction in the level of endogenous Ras in the plasma membrane fraction (Fig. 1B). Since proper Ras localization is required for downstream signaling, we investigated the impact of compound 8.12 on phosphoinositide-3-kinase (PI3K) signaling stimulated by fetal bovine serum (FBS) after serum starvation. Compound 8.12 treatment resulted in decreased Akt activation in response to serum, indicating that signaling downstream of Ras was negatively regulated by compound 8.12 (Fig. 1C). Icmt loss-of-function will interfere with pre-lamin A’s proteolytic cleavage to mature lamin A, resulting in the accumulation of pre-lamin A.25,26 In Icmt−/− MEFs, level of pre-lamin A is higher compared with Icmt+/+ MEFs, indicating accumulation of pre-lamin A (Fig. 1D). Notably, compound 8.12 treatment also resulted in a dose-dependent pre-lamin A accumulation in HepG2 cells (Fig. 1E). This provided further evidence supporting the impact of compound 8.12 on Icmt-mediated processes.
Figure 1. Compound 8.12 negatively affects cellular processes that require carboxylmethylation of a prenylated CaaX-protein. (A) CFP-tagged Hras was introduced into PC3 cells by electroporation; 48 h after transfection, the cells were treated with 1.6 µM compound 8.12 for 24 h. CFP fluorescence in transfected cells was observed using confocal microscopy, and all pictures were taken using a 63X oil immersion lens with identical optical settings throughout a single experiment. The ratio of plasma membrane: cytoplasmic CFP-tagged Hras was calculated using ImageJ (NIH), and the results plotted as mean ± SD. Control: n = 68; compound 8.12 treated: n = 50; P ≤ 0.001 (***). (B) Compound 8.12 treatment reduces plasma membrane-associated endogenous Ras. PC3 cells were treated with either DMSO control or increasing concentrations of compound 8.12 (2.0, 2.4, and 2.8 µM for 48 h, 2.4, 3.2, and 3.6 µM for 72 h). The membrane fraction was isolated and analyzed for total endogenous Ras using pan-Ras antibody; pan-cadherin was used as a loading control. The values below the immunoblots represent the ratios of total Ras to pan-cadherin in the samples. (C) Impact of compound 8.12 treatment on Akt activation. MEFs were maintained in 1% FBS together with either vehicle (–), 1.6 or 1.8 µM compound 8.12 for 72 h before stimulation with 10% FBS for 15 min. Immunoblot analysis was performed using antibodies to total AKT and phospho-AKT (S473); GAPDH expression was probed as a loading control. (D) Icmt+/+ and Icmt−/− MEFs are harvested and analyzed for pre-lamin A levels. (E) Impact of compound 8.12 treatment on pre-lamin A processing. HepG2 cells were treated with vehicle only, 18 or 21 µM cysmethynil, or 1.6, 2.4, or 3.2 µM of compound 8.12 for 48 h. Cells were harvested and processed for immunoblot analysis using antibodies against pre-lamin A; GAPDH was probed as a loading control. All data shown above are from a single experiment that has been repeated at least twice with similar results. (F) To demonstrate Icmt specificity of compound 8.12, Icmt+/+ MEFs (white bars) and Icmt−/− MEFs (gray bars) were treated with vehicle only (−), 2.0 or 4.0 µM of compound 8.12 for 48 h. Cell viability was determined as described in Materials and Methods. Viability of each cell type is reported relative to that treated with vehicle only (cell viability = 1) and is plotted as mean ± normalized SD P ≤ 0.001 (***). Data shown are from a single experiment that has been repeated three times with similar results.
The impact of compound 8.12 on cell viability is specifically due to Icmt inhibition
To assess whether the impact of compound 8.12 on cell viability is Icmt-dependent, we treated Icmt+/+ and Icmt−/− MEFs with various concentrations of the drug. Icmt+/+ MEFs were significantly more sensitive to compound 8.12 treatment compared with Icmt−/− MEFs. The viability of Icmt+/+ MEFs was markedly reduced with compound 8.12 treatment, whereas that of Icmt−/− MEFs treated with the same concentrations of compound 8.12 decreased only slightly (Fig. 1F). These data indicate that the primary target of compound 8.12 in MEFs is indeed Icmt, and that the pharmacological impact of the inhibitor on cell viability is Icmt-dependent.
Compound 8.12 induces cell cycle arrest and inhibits anchorage-independent growth in HepG2 and PC3 cells
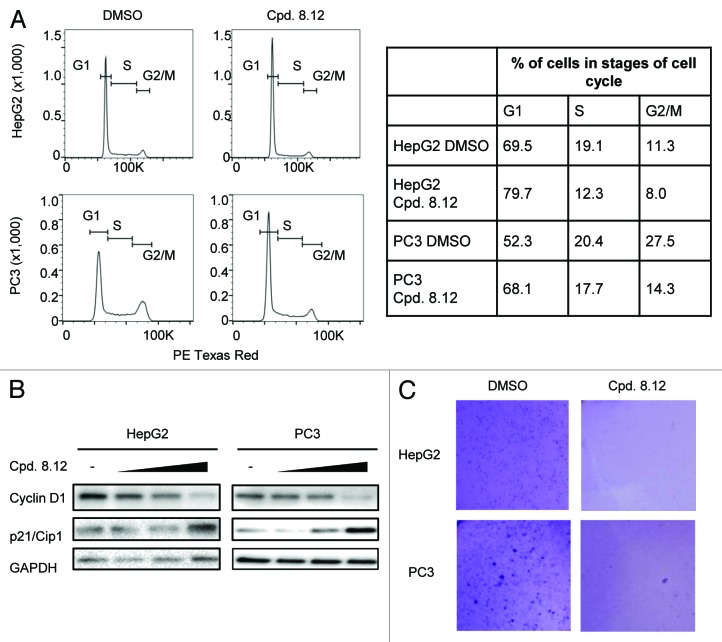
We previously reported that Icmt inhibition can induce cell cycle arrest, cell death and loss of anchorage-independent growth in a number of cancer cell lines.18,19,27 Therefore, compound 8.12 was evaluated for its anti-proliferative property. DNA content analysis, by flow cytometry, of HepG2 and PC3 cells showed an increased proportion of cells in the G1 phase of the cell cycle after compound 8.12 treatment (Fig. 2A). Consistently, immunoblot analysis of the level of cyclin D1, a marker for cell proliferation, decreased with compound 8.12 treatment while the level of p21/Cip1, a marker of G1-arrest, correspondingly increased (Fig. 2B). To evaluate compound 8.12’s ability in attenuating tumorigenesis, HepG2 and PC3 cells were suspended in agar and cultured under anchorage-independent conditions. Cells treated with increasing concentrations of compound 8.12 formed fewer and smaller colonies, and no colony could be observed at 0.8 µM and 1.6 µM of compound 8.12 treatment for HepG2 and PC3 cells, respectively (Fig. 2C).
Figure 2. Treatment with compound 8.12 induces G1-arrest in HepG2 and PC3 cells. (A) HepG2 and PC3 cells were treated for 24 h with 1.6 or 3.6 µM compound 8.12, respectively, and subjected to DNA content analysis by flow cytometry (left panel). The right panel presents quantification in percentages of the cells in different stages of the cell cycle in both cell lines. (B) HepG2 and PC3 cells were treated with vehicle only, 1.2, 1.6, and 2.0 µM (HepG2) or 1.2, 2.4, and 3.6 µM (PC3) of compound 8.12 for 24 h, and the cell lysates were subjected to immunoblot analysis using antibodies to cyclin D1 and p21/Cip1. GAPDH expression was probed as a loading control. Data shown are from a single experiment that has been repeated four times with similar results. (C) Proliferative potential of HepG2 and PC3 cells in the absence or presence of compound 8.12 (HepG2, 0.8 µM; PC3, 1.6 µM) was assessed by the soft agar anchorage-independent clonogenic assay as described in Materials and Methods. Pictures were taken using an Olympus SZX16® Research Stereo Microscope with 10× magnification. Data shown are from a single experiment that has been repeated twice with similar results.
Compound 8.12 induces autophagy, reducing cancer cell viability
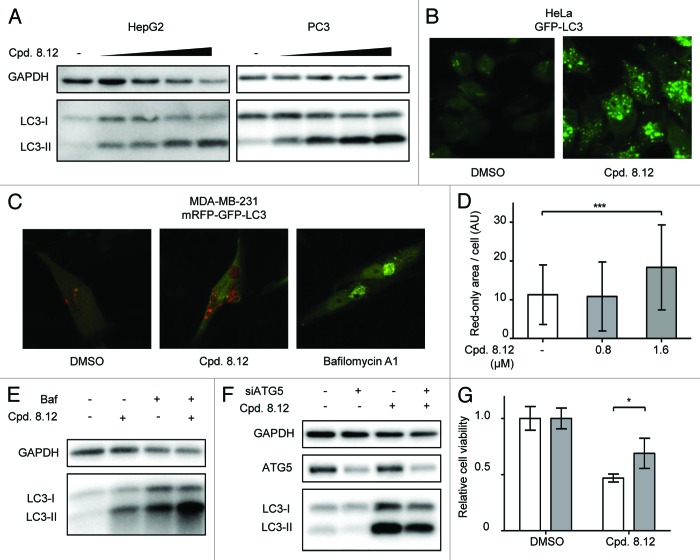
Autophagy induction is one of the major effects of pharmacological Icmt inhibition.19,27 This can be readily detected as an increase in the level of LC3-II, which is the lipidated form of LC3 localized to autophagosomes. Following compound 8.12 treatment, HepG2 and PC3 cells showed a dose-dependent increase in LC3-II levels, indicative of autophagy induction (Fig. 3A). Increased autophagosomes were also observed in HeLa cells stably transfected with green fluorescent protein (GFP)-tagged LC3,28 wherein compound 8.12 treatment resulted in a dose-dependent increase in GFP-positive puncta in the cells (Fig. 3B).
Figure 3. Compound 8.12 increases cellular autophagy. (A) HepG2 and PC3 cells were treated with either vehicle only, 0.2, 0.4, 0.8, and 1.6 µM (HepG2) or 0.4, 0.8, 1.6, and 2.0 µM (PC3) of compound 8.12 for 48 h, whereupon cells were harvested and lysates subject to immunoblot analysis for LC3-II levels to detect autophagy induction. (B) HeLa cells stably expressing GFP-tagged LC3 were treated with vehicle only or 1.6 µM of compound 8.12 for 48 h, whereupon green fluorescent puncta were visualized by confocal microscopy. (C) MDA-MB-231 cells stably expressing tandem-tagged mCherry-GFP-LC3 were treated with vehicle only, 1.6 µM of compound 8.12 or with 25 nM of bafilomycin A1, for 48 h. The extent of colocalization of red and green puncta was analyzed by confocal microscopy. (D) Quantification of the area of red-only puncta in MDA-MB-231 cells expressing tandem-tagged mCherry-GFP-LC3 after treatment with vehicle only, 0.8 µM or 1.6 µM compound 8.12 was performed using ImageJ software as described in Materials and Methods. P ≤ 0.001 (***). All pictures were taken using a 63× oil immersion lens with identical optical settings throughout a single experiment. (E) PC3 cells were treated with 1.6 µM of compound 8.12, 25 nM of bafilomycin A1, or both, for 48 h, whereupon the cells were harvested and lysates were subjected to immunoblotting using an antibody against LC3. (F) HepG2 cells were transfected with siRNA targeting ATG5 (siATG5) or control siRNA and then treated with 1.2 µM compound 8.12 for 48 h. Cells were harvested and lysates subject to immunoblot analysis using an antibody to LC3. (G) The viability of HepG2 cells subjected to ATG5 knockdown was assessed with treatment using vehicle only or 2.4 µM compound 8.12 for 48 h using the MTS assay as described in Materials and Methods. P ≤ 0.05 (*). White bars: control siRNA; gray bars: siATG5. Relative cell viability was calculated as described in Figure 1. All data shown are from single experiments that have been repeated at least twice with similar results.
The increase in LC3-II and the accumulation of autophagosomes could be explained by either an increase in autophagy initiation or a reduction of autophagic flux. To evaluate whether compound 8.12 impacts on autophagic flux, MDA-MB-231 cells stably expressing LC3 tandem-tagged with mouse red fluorescent protein (mRFP) and GFP were treated with compound 8.12. As autophagy is initiated and LC3-II becomes integrated onto autophagosomes, the mRFP and GFP signals colocalizes in the autophagosomes. However, as acidic autophagolysosomes form when autophagy progresses, the GFP signal is quenched by the acidic environment, while the more stable mRFP signal remains.29 Therefore the intensity and quantity of the fluorescent vesicles and the colocalization status of the green and red fluorescence measures both the initiation and progression of autophagy process. Treatment of the cells expressing the tandem-tagged LC3 with compound 8.12 for 48 h resulted in an increase in the total area of red-only puncta in cells, indicating that autophagy was driven to completion upon drug treatment, whereas treatment with proton pump inhibitor bafilomycin A1 resulted in perfect colocalization of red and green signals (Fig. 3C and D). Consistent with imaging study, when cells were subjected to concomitant treatment with compound 8.12 and bafilomycin A1, more LC3-II accumulation was observed compared with either compound 8.12 or bafilomycin treatment alone as shown by western blot (Fig. 3E), suggesting that compound 8.12 treatment increases both autophagy initiation and flux.
Our previous studies have provided evidence that Icmt inhibition could induce persistent autophagy that leads to cell demise. To investigate whether the induction of autophagy by compound 8.12 indeed impacted cell viability, HepG2 cells were transfected with small interfering ribonucleic acids (siRNAs) against an essential autophagy gene Atg530 and then subjected to compound 8.12 treatment. Atg5 knock-down in HepG2 cells resulted in a decrease in both the endogenous levels of LC3-II as well as the LC3-II upregulation induced by compound 8.12 treatment (Fig. 3F). Atg5 knock-down also partially rescued cell death induced by compound 8.12 treatment in HepG2 cells (Fig. 3G), suggesting that autophagy likely plays an important role in mediating compound 8.12-induced cell death. The incomplete extent of autophagy inhibition and rescuing by concurrent Atg5 knockdown is likely due to incomplete Atg5 knockdown in HepG2 cells, which proved to be variable in transfection efficiency.
Compound 8.12 effectively attenuates tumor growth in vivo
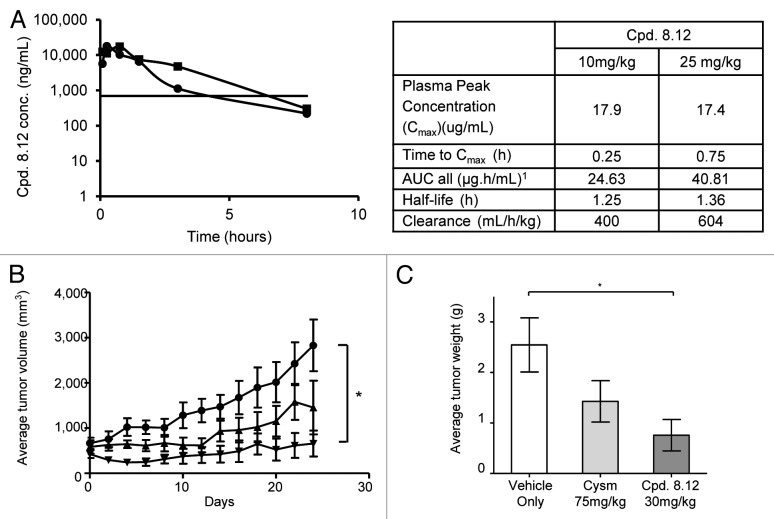
To determine the in vivo efficacy of compound 8.12, a HepG2 xenograft mouse model was employed. In preliminary studies, 6- to 8-wk-old Balb/c mice were dosed intraperitoneally in groups of two with compound 8.12 to determine its maximum tolerated dose (MTD). Cysmethynil has been reported to be well-tolerated up to 300 mg/kg19; in this study, compound 8.12 was found to be well-tolerated up to 50 mg/kg at 24 h without any morbidity. A pharmacokinetic study suggested that compound 8.12 requires a more frequent dosing regimen compared with cysmethynil to maintain a steady effective serum concentration, due to its shorter half-life (Fig. 4A).31
Figure 4. Compound 8.12 displays higher potency than cysmethynil in tumor growth inhibition in vivo. (A) Plasma concentrations of compound 8.12 in treated mice. Left: Compound 8.12 was administered at 10mg/kg (●) and 25 mg/kg (■) intraperitoneally, and plasma drug concentrations at time points up to 8 h were determined as described in Materials and Methods. The horizontal line indicates the plasma drug concentration that is approximately equivalent to the in vitro IC50 of compound 8.12 for HepG2 cells. Each data point was obtained from 3 animals (n = 3). Right: Table shows the pharmacokinetic parameters of compound 8.12. AUC, area under the time-concentration curve. (B) Mice bearing tumor xenografts were treated with: ●, vehicle only; ▲, cysmethynil 75 mg/kg; ▼, compound 8.12 30 mg/kg, and the volumes of the tumors were measured as described in Materials and Methods. The volumes of the tumors up to 24 d post-treatment were plotted as mean ± SEM P ≤ 0.05 (*). Each treatment group consists of 8 animals (n = 8). (C) The tumors from the study in (B) were harvested and weighed at the end of the study period; the mean ± SD is plotted as shown. P ≤ 0.05 (*). Similar experiments were conducted three times with similar results.
To determine the ability of compound 8.12 to impact tumor growth in vivo, compound 8.12 and cysmethynil were delivered intraperitoneally to the immunodeficient (SCID) mice bearing HepG2 xenografts on both flanks once a day and once every 2 d, respectively, for 24 d. Compared with mice treated with vehicle only, compound 8.12 given at 30 mg/kg markedly attenuated tumor growth throughout the experiment, with the average tumor volumes remain close to that at the beginning of the treatment course, while the control tumors grew rapidly (Fig. 4B). Compound 8.12 at 30 mg/kg was more effective in attenuating tumor growth than cysmethynil dosed at 75 mg/kg, however the difference was not significant (Fig. 4B). The weights of individual tumors harvested at the end of the experiment support a similar conclusion (Fig. 4C).
Compound. 8.12 synergizes with gefitinib to reduce viability of some cancer cells
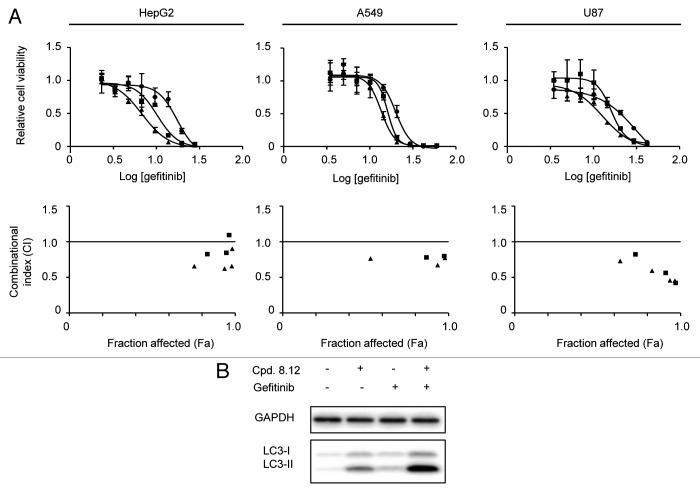
To further explore Icmt as a therapeutic target for cancer, we investigated whether Icmt inhibitors such as compound 8.12 are able to act synergistically with other agents used in cancer treatment. Such information could potentially allow future investigators to develop combination therapies with compound 8.12 that have increased efficacy with acceptable toxicity. Gefitinib, an EGFR-inhibitor currently used mainly in the treatment of non-small cell lung cancer,32 emerged from a screen of a small panel of drugs tested in combination with compound 8.12 in HepG2 cells. The combination of gefitinib with compound 8.12 was then tested in HepG2, A549 and U87 cell lines; for each of these lines the IC50 for gefitinib is above 10µM, a level characteristic of gefitinib resistance.33 Cells were treated with fixed concentrations of compound 8.12 that had no impact on viability as a single agent, combined with a wide range of gefitinib concentrations. The results were plotted in viability curves, and the combinational indexes (CIs) were calculated using CompuSync software.34 In the cell lines tested, compound 8.12 was able to synergize with gefitinib in reducing cell viability, as indicated by the left-ward shifts of the viability curves when the cells were treated with a range of gefitinib concentrations in a background concentration of compound 8.12 that alone had no impact on cell viability (Fig. 5A). Consistently, the CIs calculated from the cell viability data were mostly below 1.0 across all effect levels, supporting a synergistic relationship between compound 8.12 and gefitinib (Fig. 5A). Both Icmt inhibitors19,27 and gefitinib have been reported to induce autophagy in multiple cell lines,35-37 and our previous studies have established that autophagy induction mediated by Icmt inhibitors can lead to cell death.19,27 Therefore, we proceeded to investigate whether the combination of compound 8.12 and gefitinib would result in more autophagy induction than either agent alone. Indeed, LC3-II levels significantly increased in HepG2 cells treated with both compound 8.12 and gefitinib compared with the sum of the levels induced by either drug alone, suggesting a synergistic relationship in autophagy induction between compound 8.12 and gefitinib in HepG2 cells (Fig. 5B).
Figure 5. Compound 8.12 synergizes with gefitinib in cell killing in vitro. (A) HepG2, A549, and U87 cells were treated with the indicated range of concentrations of gefitinib in the presence of fixed concentrations of compound 8.12 that are unable to impact on the viability as a single agent. Relative cell viability was plotted against the log of gefitinib concentration, and combination indices (CI) were also plotted against the fraction affected (Fa) value. Details of experimental methods and CI calculation are described in Materials and Methods. ●, DMSO; ■, 0.8 µM compound 8.12; ▲, 1.2 µM compound 8.12. (B) HepG2 cells were treated with either vehicle only, 1.2 µM compound 8.12, 4 µM gefitinib, or both, for 48 h, whereupon cells were harvested and lysates subjected to immunoblot analysis using a LC3 antibody. Data shown are from a single experiment that has been repeated at least twice with similar results.
Discussion
Accumulating evidence on the biological effects of suppression of Icmt activity has supported the notion that this enzyme is a prime candidate for cancer therapy development. Inhibition of Icmt attracted attention initially as a way to reduce the activity of mutant Ras, infamous for its role in multiple human cancers. It is now recognized that multiple Icmt substrates are involved in tumorigenesis. The potential beneficial consequences of Icmt inhibition extend beyond cancer, as a recent study has demonstrated that reduction of Icmt activity rescues mice from symptomatic progression of progeria by inhibiting prelamin A carboxylmethylation.26 This study highlights the utility of targeting Icmt as a means to manipulate the functions of its substrates involved in pathological processes. Here, we report that compound 8.12, an amino-derivative of the prototypical indole-based Icmt inhibitor, cysmethynil, possesses significantly improved pharmacological properties and in vivo potency compared with the parent compound. As expected, treatment of cancer cells with compound 8.12 inhibits cell proliferation and induces autophagy and cell death in an Icmt-dependent fashion. Given its favorable pharmacokinetics profile, low toxicity and higher potency in tumor growth attenuation, compound 8.12 has the best overall properties among current Icmt inhibitors, and therefore holds significant promise as a potential candidate for drug development.
A subject of major interest in drug development is combination therapy. Cancer treatment regimens often consist of a combination of drugs.38 In principle, as drugs used in combination allow targeting of multiple molecular mechanisms within the tumor, they could produce higher rates of treatment response and lower rates of drug resistance, and potentially allow lower doses of each drug to reduce toxicity.39,40 In this study, compound 8.12 was found to act synergistically with gefitinib, an EGFR inhibitor, in inhibiting growth of multiple cancer cell lines. Gefitinib is indicated for non-small cell lung cancer as first-line therapy in patients with known EGFR-activating mutations; it has also been employed as a second-line agent for patients who have failed chemotherapy, albeit with limited efficacy.32 Interestingly, gefitinib had been reported to induce autophagy in breast cancer cells.37 A recent finding also suggested that non-small cell lung cancer cells with activating EGFR mutations that have acquired resistance to gefitinib harbored higher basal autophagy,41 providing additional evidence that autophagy plays a role in the antitumor effect of gefitinib. As Icmt inhibitors reduce cell viability via an autophagy-dependent mechanism, it is perhaps not surprising that compound 8.12 was able to synergize with gefitinib to enhance cell death in gefitinib-resistant cell lines such as HepG2. The combination of compound 8.12 and gefitinib may be able to promote autophagy to the point where the balance is tipped toward autophagic cell death.42 We observed synergistic enhancement of autophagy in HepG2 cells treated with low doses of both compound 8.12 and gefitinib, suggesting that detrimental levels of autophagy may indeed be the mechanism of synergy between these two drugs. Further, the synergy between compound 8.12 and gefitinib was not limited to HepG2 cells, as multiple cancer cell lines responded to the combination in a synergistic manner. Further delineation of mechanisms of synergy will be helpful both in the clinical application of gefitinib and in the development of Icmt inhibitors.
Although both genetic and pharmacologic suppression of Icmt has now been shown to impact on proliferation, autophagy and survival,10,18,19 the molecular mechanisms responsible for the biological effects observed have not been fully elucidated. The difficulty is in part due to the fact that Icmt acts on multiple CaaX-motif containing proteins that are implicated in cancer, and the exact contributions of these proteins in the tumorigenesis process have not been thoroughly defined. We have previously established that induction of autophagy occurs in multiple cancer cell lines treated with Icmt inhibitors; in addition, we have demonstrated that persistent autophagy induction from Icmt inhibition reduces cell survival.19,27 Similarly, compound 8.12-induced autophagy also detrimentally impacts on cell survival, as suppression of the autophagic process through ATG5 knockdown provided partial rescue from compound 8.12 treatment. The molecular mechanism(s) behind this phenomenon warrant additional study in the future; such information could provide additional insight into the mechanism of action of Icmt inhibitors in cancer cells, as well as potential clinical applications of not just Icmt inhibitors but also EGFR inhibitors.
Similar to other targeted therapies, Icmt targeted therapy will likely not be equally effective against all cancers. Therefore stratification of Icmt-sensitive and Icmt-resistant cancers will be critically important for clinical development of these compounds. Genetic deletion of Icmt obliterated the ability of oncogenic Kras-G12V and Braf-V599E to transform MEF cells,10 and reduced the tumorigenicity of oncogenic Kras-G12D-driven myeloproliferative disease in mice.11 Pharmacological inhibition of Icmt with small molecule inhibitors, including cysmethynil and compound 8.12, led to inhibition of proliferation, tumorigencity, and cell death in multiple cancer cell lines and xenograft models.18,19,32 Despite these promising results, Icmt inhibition showed no efficacy in a mouse model of Kras-G12D driven pancreatic cancer.43 The divergent responses to Icmt suppression further underscore the importance of understanding the molecular signaling circuitry of different cancer cells and molecular stratification of responsive and resistant cells. Icmt functions and the effects of Icmt inhibition will almost assuredly be dependent on cellular contexts, which clearly require further delineation in order to uncover the full potential of Icmt inhibition as a cancer therapeutic strategy. Careful study of the differences in the molecular signatures between resistant and sensitive cells may not only shed light on Icmt function in cancer cells and the molecular mechanisms behind the effects caused by Icmt inhibition, but also enable future investigations to identify patient populations suitable for Icmt inhibitor therapy.
Materials and Methods
Materials and cell culture
Cells (HepG2, PC3, A549, and U87) were obtained from American Type Culture Collection. HeLa cells stably expressing GFP-tagged LC3, MDA-MB-231 cells stably expressing tandem tagged mRFP-GFP-LC3 as well as wild-type and Icmt-null MEFs were previously described.9,10,28,29 Cells were maintained in Dulbecco’s minimal essential medium (DMEM) with 5 or 10% FBS (v/v) and penicillin (100 U/mL)/streptomycin (100 µg/mL), all obtained from GIBCO/Life Technologies, under standard conditions. Cysmethynil was prepared by the Duke Small Molecule Synthesis Facility; compound 8.12 was synthesized as described previously23 and gefitnib (G-4408) was obtained from LC Laboratories. Antibodies for pan-cadherin (4068), Cyclin D1 (DCS6, 2926), p21 Waf1/Cip1 (12D1, 2947), LC3B (2775), GAPDH (14C10, 2118), and Atg5 (2630), were all from Cell Signaling Technology. The antibody for Ras (Ras10, MA1-012) was from Pierce Scientific; this antibody is able to recognize H-, N-, and Kras.44 The antibody for lamin A (C20; sc-6214) obtained from Santa Cruz Biotechnology has been validated for the specific detection of pre-lamin A.45 Plasmids expressing CFP-tagged Hras (pECFP-Hras) were kind gifts from Dr Won Do Heo; siATG5 and control siRNA were obtained from Sigma-Aldrich.
Cell fractionation and plasma membrane protein isolation
Plasma membrane fraction was isolated from PC3 cells using the Plasma Membrane Protein Extraction Kit (K268-50) from BioVision according to the manufacturer’s protocol.
Transient transfection and immunoblot analysis
pECFP-Hras was electroporated into PC3 cells with 300 V/1000 µF using a Gene Pulser Xcell™ (Bio-Rad), and control siRNA/siATG5 were transfected into HepG2 cells using Lipofectamine 2000 (Invitrogen). For immunoblot analysis, cells were harvested and lysed by sonication in RIPA buffer (Pierce) supplemented with phosphatase and protease inhibitors (Roche). Lysates were resolved by sodium dodecyl sulfate-PAGE (SDS-PAGE) and immunoblot analysis performed using standard protocols.
Cell viability assay
Cells were seeded in 96-well culture plates at a density of 2500 cells per well and allowed to settle overnight. Cells were then incubated in 200 µL of drug- or vehicle-containing 5% FBS supplemented DMEM for a further 48 h. A tetrazolium-based colorimetric cell viability assay was used to measure cell viability (CellTiter 96® AQueous One Solution Cell Proliferation Assay, Promega) per manufacturer’s protocol. Cell viability data was analyzed using the GraphPad Prism software; combinational index for each combinational treatment was calculated from the fraction-affected value using Compusyn software (ComboSyn, Inc.), with an index value smaller than 1.0 indicating synergism.34
Anchorage-independent clonogenic assay
A bottom layer of 1% noble agar (Sigma-Aldrich) in 5% FBS-supplemented DMEM with the appropriate drug concentrations was placed in each well of a 24-well culture plate. PC3 and HepG2 wells were suspended in a mixture of 0.4% noble agar in DMEM with 5% FBS and the appropriate drug concentrations at a final density of 5000 cells per 400 µL. This mixture was placed over the bottom agar layer. After setting at room temperature, 5% FBS-supplemented DMEM with the appropriate drug concentrations was added to each well. The drug-containing cell culture media was changed every 2 d, and the cultures were incubated for 3–4 wk before fixing with 4% paraformaldehyde and staining using 0.5% crystal violet.
Flow cytometry and cell cycle analysis
PC3 and HepG2 cells were seeded at 75 000 cells per well in a 6-well culture plate and allowed to settle overnight, and cells were treated with either DMSO-only or appropriate concentrations of compound 8.12 for 24 h. Both the culture media and cells for each condition were collected and cells pelleted by centrifugation. Seventy percent ethanol was added drop-wise to the resultant cell pellet with gentle vortexing, and cells were fixed at 4 °C overnight. Fixed cells were stained with 100 µg/mL propidium iodide + 100 µg/mL ribonuclease A in the dark for at least 30 min at room temperature. Flow Cytometry was performed using a BD LSRFortessa Cell Analyzer (BD BioSciences) and cell cycle analysis performed using FlowJo software (Tree Star Inc.).
Confocal microscopy and image analysis
Cells were seeded at 30 000 cells per well directly on glass coverslips placed in each well of a 24-well culture plate and allowed to settle overnight in 10% FBS-supplemented DMEM. Cells were then treated with the appropriate concentrations of compound 8.12 or DMSO-only, fixed with 4% paraformaldehyde and the coverslips mounted onto glass slides. Confocal microscopy was performed using a Zeiss LSM 710 Confocal Microscope and the resultant images analyzed using ImageJ (NIH). The total area of red puncta in mRFP-GFP-LC3- expressing MDA-MB-231 cells was calculated using the Green and Red Puncta Colocalization Macro for the modified version of Image J designed for the purpose of this kind of analysis (Shiwarski et al.46; Pampliega, et al.47).
Determination of maximum tolerated dose and pharmacokinetic parameters of compound 8.12 and cysmethynil
Animal studies were performed with compliance to protocols approved by the Institutional Animal Care and Use Committee. To determine the MTD of compound 8.12, pairs of 6- to 8-wk-old wild-type Balb/c mice were given either vehicle (ethanol, PEG 400 and 5% dextrose [Sigma-Aldrich] in the ratio 1:6:3) or the test drug dissolved in vehicle intraperitoneally and observed for 45 min for signs of acute toxicity. If no sign of acute toxicity was observed for a particular dosage, a higher dosage was given to the next pair of mice. Compound 8.12 was tested for toxicity up to 100 mg/kg. Mice were observed for any sign of toxicity for a further 24 h. Pharmacokinetic parameters were then determined for compound 8.12 up to 48 h. Compound 8.12 was administered intraperitoneally to 12 Balb/c mice, and blood was collected for analysis from 3 mice at each time point up to 48 h. Blood was collected twice from each group of animals; the first collection was done from the submandibular vein, while the second collection was performed through terminal cardiac puncture following anesthesia. Plasma prepared from the blood samples were analyzed for compound 8.12 using the Prominence UFLC high-speed liquid chromatography system (Shimadzu Scientific Instruments) coupled to a QTRAP 3200 triple quadruple mass spectrometer (Applied Biosystems). Pharmacokinetic parameters were calculated with Standard Version 5.0.1 of the WinNonlin® software (Standard Version 5.0.1; Pharsight) using non-compartmentalized analysis for per os dosing.
Tumor xenograft studies
A total of 1 × 107 HepG2 cells suspended in 40% Matrigel (BD BioSciences) were injected subcutaneously into both flanks of 6- to 8-wk-old female SCID mice. When the tumors reached 100–200 mm3, either vehicle only, 75 mg/kg cysmethynil (once every two days), or 30 mg/kg compound 8.12 (daily) was administered to the mice intraperitoneally over a total of 24 d. The animals’ general appearance, body weights, and tumor sizes were monitored every 2 d. Tumors were measured using the standard clipper method, and the tumor volumes were calculated using the formula of (length × width × width) ÷ 2. The animals were euthanized on the 24th day and the tumors removed and weighed.
Disclosure of Potential Conflicts of Interest
P.M.R., T.Y., M.L.G., P.J.C., and M.W. are listed on a patent filing covering the Icmt inhibitor used in this study; other authors have no conflict of interest.
Grant Support
The study is supported by grants from Singapore Biomedical Research Council and Singapore National Medical Research Council.
Glossary
Abbreviations:
- AdoMet
S-adenosyl-l-methionine
- CFP
cyan fluorescent protein
- DMEM
Dulbecco’s Minimal Essential Medium
- DMSO
dimethyl sulfoxide
- EGFR
epithelial growth factor receptor
- FBS
fetal bovine serum
- FTase
farnesyltransferase
- Icmt
isoprenylcysteine carboxylmethyltransferase
- GFP
green fluorescent protein
- GGTase-1
geranylgeranyltransferase-1
- MEFs
mouse embryonic fibroblasts
- mRFP
mouse red fluorescent protein
- MTD
maximum tolerated dose
- PEG
polyethylene glycol
- PI3K
phosphoinositide-3-kinase
- SCID
severe combined immunodeficiency
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- siRNA
small interfering ribonucleic acid
References
- 1.Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer. 2011;11:775–91. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao J, Liao J, Yang GY. CAAX-box protein, prenylation process and carcinogenesis. Am J Transl Res. 2009;1:312–25. [PMC free article] [PubMed] [Google Scholar]
- 3.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol. 2012;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casey PJ, Seabra MC. Protein prenyltransferases. J Biol Chem. 1996;271:5289–92. doi: 10.1074/jbc.271.10.5289. [DOI] [PubMed] [Google Scholar]
- 5.Otto JC, Kim E, Young SG, Casey PJ. Cloning and characterization of a mammalian prenyl protein-specific protease. J Biol Chem. 1999;274:8379–82. doi: 10.1074/jbc.274.13.8379. [DOI] [PubMed] [Google Scholar]
- 6.Dai Q, Choy E, Chiu V, Romano J, Slivka SR, Steitz SA, Michaelis S, Philips MR. Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J Biol Chem. 1998;273:15030–4. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- 7.Wright LP, Court H, Mor A, Ahearn IM, Casey PJ, Philips MR. Topology of mammalian isoprenylcysteine carboxyl methyltransferase determined in live cells with a fluorescent probe. Mol Cell Biol. 2009;29:1826–33. doi: 10.1128/MCB.01719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, Kulkarni K, Manolaridis I, Zhang Z, Dodd RB, Mas-Droux C, Barford D. Mechanism of isoprenylcysteine carboxyl methylation from the crystal structure of the integral membrane methyltransferase ICMT. Mol Cell. 2011;44:997–1004. doi: 10.1016/j.molcel.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 9.Bergo MO, Leung GK, Ambroziak P, Otto JC, Casey PJ, Gomes AQ, Seabra MC, Young SG. Isoprenylcysteine carboxyl methyltransferase deficiency in mice. J Biol Chem. 2001;276:5841–5. doi: 10.1074/jbc.C000831200. [DOI] [PubMed] [Google Scholar]
- 10.Bergo MO, Gavino BJ, Hong C, Beigneux AP, McMahon M, Casey PJ, Young SG. Inactivation of Icmt inhibits transformation by oncogenic K-Ras and B-Raf. J Clin Invest. 2004;113:539–50. doi: 10.1172/JCI200418829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahlstrom AM, Cutts BA, Liu M, Lindskog A, Karlsson C, Sjogren AK, Andersson KM, Young SG, Bergo MO. Inactivating Icmt ameliorates K-RAS-induced myeloproliferative disease. Blood. 2008;112:1357–65. doi: 10.1182/blood-2007-06-094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baines AT, Xu D, Der CJ. Inhibition of Ras for cancer treatment: the search continues. Future Med Chem. 2011;3:1787–808. doi: 10.4155/fmc.11.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slebos RJ, Hoppin JA, Tolbert PE, Holly EA, Brock JW, Zhang RH, Bracci PM, Foley J, Stockton P, McGregor LM, et al. K-ras and p53 in pancreatic cancer: association with medical history, histopathology, and environmental exposures in a population-based study. Cancer Epidemiol Biomarkers Prev. 2000;9:1223–32. [PubMed] [Google Scholar]
- 14.John J, Sohmen R, Feuerstein J, Linke R, Wittinghofer A, Goody RS. Kinetics of interaction of nucleotides with nucleotide-free H-ras p21. Biochemistry. 1990;29:6058–65. doi: 10.1021/bi00477a025. [DOI] [PubMed] [Google Scholar]
- 15.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–51. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James GL, Goldstein JL, Brown MS. Polylysine and CVIM sequences of K-RasB dictate specificity of prenylation and confer resistance to benzodiazepine peptidomimetic in vitro. J Biol Chem. 1995;270:6221–6. doi: 10.1074/jbc.270.11.6221. [DOI] [PubMed] [Google Scholar]
- 17.Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, Bishop WR, Pai JK. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem. 1997;272:14459–64. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 18.Winter-Vann AM, Baron RA, Wong W, dela Cruz J, York JD, Gooden DM, Bergo MO, Young SG, Toone EJ, Casey PJ. A small-molecule inhibitor of isoprenylcysteine carboxyl methyltransferase with antitumor activity in cancer cells. Proc Natl Acad Sci U S A. 2005;102:4336–41. doi: 10.1073/pnas.0408107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M, Tan W, Zhou J, Leow J, Go M, Lee HS, Casey PJ. A small molecule inhibitor of isoprenylcysteine carboxymethyltransferase induces autophagic cell death in PC3 prostate cancer cells. J Biol Chem. 2008;283:18678–84. doi: 10.1074/jbc.M801855200. [DOI] [PubMed] [Google Scholar]
- 20.Cushman I, Casey PJ. Role of isoprenylcysteine carboxylmethyltransferase-catalyzed methylation in Rho function and migration. J Biol Chem. 2009;284:27964–73. doi: 10.1074/jbc.M109.025296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cushman I, Casey PJ. RHO methylation matters: a role for isoprenylcysteine carboxylmethyltransferase in cell migration and adhesion. Cell Adh Migr. 2011;5:11–5. doi: 10.4161/cam.5.1.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Go ML, Leow JL, Gorla SK, Schüller AP, Wang M, Casey PJ. Amino derivatives of indole as potent inhibitors of isoprenylcysteine carboxyl methyltransferase. J Med Chem. 2010;53:6838–50. doi: 10.1021/jm1002843. [DOI] [PubMed] [Google Scholar]
- 23.Ramanujulu PM, Yang T, Yap SQ, Wong FC, Casey PJ, Wang M, Go ML. Functionalized indoleamines as potent, drug-like inhibitors of isoprenylcysteine carboxyl methyltransferase (Icmt) Eur J Med Chem. 2013;63:378–86. doi: 10.1016/j.ejmech.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Bergo MO, Leung GK, Ambroziak P, Otto JC, Casey PJ, Young SG. Targeted inactivation of the isoprenylcysteine carboxyl methyltransferase gene causes mislocalization of K-Ras in mammalian cells. J Biol Chem. 2000;275:17605–10. doi: 10.1074/jbc.C000079200. [DOI] [PubMed] [Google Scholar]
- 25.Davies BS, Fong LG, Yang SH, Coffinier C, Young SG. The posttranslational processing of prelamin A and disease. Annu Rev Genomics Hum Genet. 2009;10:153–74. doi: 10.1146/annurev-genom-082908-150150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ibrahim MX, Sayin VI, Akula MK, Liu M, Fong LG, Young SG, Bergo MO. Targeting isoprenylcysteine methylation ameliorates disease in a mouse model of progeria. Science. 2013;340:1330–3. doi: 10.1126/science.1238880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang M, Hossain MS, Tan W, Coolman B, Zhou J, Liu S, Casey PJ. Inhibition of isoprenylcysteine carboxylmethyltransferase induces autophagic-dependent apoptosis and impairs tumor growth. Oncogene. 2010;29:4959–70. doi: 10.1038/onc.2010.247. [DOI] [PubMed] [Google Scholar]
- 28.Zhu WL, Hossain MS, Guo DY, Liu S, Tong H, Khakpoor A, Casey PJ, Wang M. A role for Rac3 GTPase in the regulation of autophagy. J Biol Chem. 2011;286:35291–8. doi: 10.1074/jbc.M111.280990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–60. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 30.Yang YP, Liang ZQ, Gu ZL, Qin ZH. Molecular mechanism and regulation of autophagy. Acta Pharmacol Sin. 2005;26:1421–34. doi: 10.1111/j.1745-7254.2005.00235.x. [DOI] [PubMed] [Google Scholar]
- 31.Wang M, Khoo YM, Zhou J, Casey P, Lee HS. A high-performance liquid chromatography method for the quantification of cysmethynil, an inhibitor of isoprenylcysteine carboxylmethyl transferase, in mouse plasma. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:553–7. doi: 10.1016/j.jchromb.2008.12.067. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi K, Hagiwara K. Epidermal growth factor receptor (EGFR) mutation and personalized therapy in advanced nonsmall cell lung cancer (NSCLC) Target Oncol. 2013;8:27–33. doi: 10.1007/s11523-013-0258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono M, Hirata A, Kometani T, Miyagawa M, Ueda S, Kinoshita H, Fujii T, Kuwano M. Sensitivity to gefitinib (Iressa, ZD1839) in non-small cell lung cancer cell lines correlates with dependence on the epidermal growth factor (EGF) receptor/extracellular signal-regulated kinase 1/2 and EGF receptor/Akt pathway for proliferation. Mol Cancer Ther. 2004;3:465–72. [PubMed] [Google Scholar]
- 34.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–81. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 35.Han W, Pan H, Chen Y, Sun J, Wang Y, Li J, Ge W, Feng L, Lin X, Wang X, et al. EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PLoS One. 2011;6:e18691. doi: 10.1371/journal.pone.0018691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng Y, Zhang Y, Zhang L, Ren X, Huber-Keener KJ, Liu X, Zhou L, Liao J, Keihack H, Yan L, et al. MK-2206, a novel allosteric inhibitor of Akt, synergizes with gefitinib against malignant glioma via modulating both autophagy and apoptosis. Mol Cancer Ther. 2012;11:154–64. doi: 10.1158/1535-7163.MCT-11-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dragowska WH, Weppler SA, Wang JC, Wong LY, Kapanen AI, Rawji JS, Warburton C, Qadir MA, Donohue E, Roberge M, et al. Induction of autophagy is an early response to gefitinib and a potential therapeutic target in breast cancer. PLoS One. 2013;8:e76503. doi: 10.1371/journal.pone.0076503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol. 2012;30:679–92. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- 39.Delbaldo C, Michiels S, Syz N, Soria JC, Le Chevalier T, Pignon JP. Benefits of adding a drug to a single-agent or a 2-agent chemotherapy regimen in advanced non-small-cell lung cancer: a meta-analysis. JAMA. 2004;292:470–84. doi: 10.1001/jama.292.4.470. [DOI] [PubMed] [Google Scholar]
- 40.Zimmermann GR, Lehár J, Keith CT. Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov Today. 2007;12:34–42. doi: 10.1016/j.drudis.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Sakuma Y, Matsukuma S, Nakamura Y, Yoshihara M, Koizume S, Sekiguchi H, Saito H, Nakayama H, Kameda Y, Yokose T, et al. Enhanced autophagy is required for survival in EGFR-independent EGFR-mutant lung adenocarcinoma cells. Lab Invest. 2013;93:1137–46. doi: 10.1038/labinvest.2013.102. [DOI] [PubMed] [Google Scholar]
- 42.Zhai B, Hu F, Jiang X, Xu J, Zhao D, Liu B, Pan S, Dong X, Tan G, Wei Z, et al. Inhibition of akt reverses the acquired resistance to sorafenib by switching protective autophagy to autophagic cell death in hepatocellular carcinoma. Mol Cancer Ther. 2014;13:1589–98. doi: 10.1158/1535-7163.MCT-13-1043. [DOI] [PubMed] [Google Scholar]
- 43.Court H, Amoyel M, Hackman M, Lee KE, Xu R, Miller G, Bar-Sagi D, Bach EA, Bergö MO, Philips MR. Isoprenylcysteine carboxylmethyltransferase deficiency exacerbates KRAS-driven pancreatic neoplasia via Notch suppression. J Clin Invest. 2013;123:4681–94. doi: 10.1172/JCI65764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamer PJ, Trimpe KL, Pullano T, Ng S, LaVecchio JA, Petit DA, DeLellis R, Wolfe H, Carney WP. Production and characterization of anti-RAS p21 monoclonal antibodies. Hybridoma. 1990;9:573–87. doi: 10.1089/hyb.1990.9.573. [DOI] [PubMed] [Google Scholar]
- 45.Ragnauth CD, Warren DT, Liu Y, McNair R, Tajsic T, Figg N, Shroff R, Skepper J, Shanahan CM. Prelamin A acts to accelerate smooth muscle cell senescence and is a novel biomarker of human vascular aging. Circulation. 2010;121:2200–10. doi: 10.1161/CIRCULATIONAHA.109.902056. [DOI] [PubMed] [Google Scholar]
- 46.Shiwarski DJ, Dagda RK, Chu CT. Red and Green Puncta Colocalization Macro. 2014
- 47.Pampliega O, Orhon I, Patel B, Sridhar S, Díaz-Carretero A, Beau I, Codogno P, Satir BH, Satir P, Cuervo AM. Functional interaction between autophagy and ciliogenesis. Nature. 2013;502:194–200. doi: 10.1038/nature12639. [DOI] [PMC free article] [PubMed] [Google Scholar]