Abstract
It has been approved for the clinical application of β-elemene to treat various cancers mainly brain tumors in China. In the present study, we found that β-elemene significantly inhibited the in vitro growth of human breast cancer cells by inducing apoptosis. In addition, β-elemene also induced the conversion of LC3-I into LC3-II as well as the formation of autolysosomes, indicating the activation of autophagy. Interestingly, inhibition of autophagy significantly potentiated the growth-inhibitory effect of β-elemene on breast cancer cells. In summary, β-elemene induced cytoprotective autophagy in human breast cancer cells in addition to apoptosis. Inhibition of autophagy significantly enhanced the cytotoxicity of β-elemene to human breast cancer cells. Therefore, combination of β-elemene with autophagy inhibitors could be a promising strategy for the treatment of breast cancer.
Keywords: β-elemene, breast cancer, apoptosis, autophagy
Introduction
Elemenes including α-, β-, γ-, and δ-elemene, are a group of natural chemical compounds extracted from a variety of medical herbs and plants such as Curcuma WenYuJin [1]. Elemenes contribute to the floral aromas of some plants, and are used as pheromones by some insects. Recently, β-elemene has attracted great interest because of its potent anti-proliferation effects toward some types of cancer cells [2-5]. Based on some small scale clinical trials with low quality in China, β-elemene has been approved by the State Food and Drug Administration of China for anti-cancer treatment. Up-to-now, β-ELE has been clinically used to treat leukemia and carcinomas in brain, breast, liver and other tissues.
It seems that β-elemene exerts its anti-cancer effect through multiple mechanisms. For example, β-elemene can induce apoptosis or cell cycle arrest [5-7], and reverse epithelial-mesenchymal transition (EMT) [8] and multidrug resistance (MDR) [9-11] in various cancer cells. Recently, it was frequently reported that autophagy could be induced by chemotherapeutic and targeted therapy drugs as a cytoprotection mechanism to count-act anti-cancer drugs [12-14]. In the present study, we found that β-elemene induced cytoprotective autophagy in breast cancer cells in addition to apoptosis. Inhibition of autophagy significantly enhanced the cytotoxicity of β-elemene, indicating that combination of β-elemene with autophagy inhibitors could be a promising strategy for cancer treatment.
Materials and methods
Reagents and antibodies
The chemicals such as β-elemene and Chlo-roquine were provided by J&K chemical Ltd. (Shanghai, China). The primary antibodies against microtubule-associated protein 1 light chain 3 (LC3) and GAPDH were bought from Santa Cruz.
Cell cultures
The breast cancer cell lines (Bcap37, MBA-MD-231) were bought from cell bank (Chinese Academy of Sciences). Monolayer culture of cancer cells was maintained in DMEM supplemented with 10% fetal bovine serum 10% fetal bovine serum, 100 units/mL penicillin, 100 μg/mL streptomycin and incubated at 5% CO2, 37°C, and 95% humidity.
Cell viability assay (MTS)
Cell growth was determined by CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay kit (Promega, Madison, WI, USA) according to the protocol provided. Cells were seeded into 96-well plates (cultured overnight for adherent cells) and treated with chemicals with different concentrations. After 24-h, 48-h, 72-h incubation, 20 μl MTS (5 mg/ml) was added into each well respectively. The absorbance was then measured using a model ELX800 Micro Plate Reader (Bio-Tek Instruments, Inc.) at 570 nm.
Electron microscopy
Treated cells were washed and fixed for 30 min in 2.5% glutaraldehyde. The samples were treated with 1.5% osmium tetroxide, dehydrated with acetone and embedded in Durcupan resin. Thin sections were poststained with lead citrate and examined in the TECNAI 10 electron microscope (Philips, Holland) at 60 kV.
Western blot analysis
Western blotting of LC3 was carried out as previously reported [13]. The protein was applied to a proper concentration of SDS-polyacrylamide gel, transferred to a PVDF membrane, and then detected by the proper primary and secondary antibodies before visualization with a chemiluminescence kit. Visualization was done with Image Quant LAS-4000 (Fujifilm, Tokyo, Japan) using image Multi-Gauge Software (Fujifilm, Tokyo, Japan).
Cell apoptosis analysis
The human breast cancer cell lines were seeded into 6-well plates (3 × 105/well) and treated with either the diluent control (DMSO) or ELE at various concentrations. At the end of incubation, the cells on each well were trypsinized, washed with PBS and stained with Annexin V-FITC Apoptosis Detection Kit (BD Pharmingen). The cell apoptosis was determined by the flow cytometry (Becton Dickinson, Mountain View, CA, USA).
Statistical analyses
Unless otherwise stated, data were expressed as the mean ± SD, and analyzed by Student’s t test.
Results
β-elemene inhibited viability of breast cancer cells
We started to evaluate the effect of β-elemene on breast cancer cells by evaluating the viability of breast cancer cells treated with MTS assay before and after β-elemene treatment. As shown in Figure 1A and 1B, β-elemene significantly inhibit the viability of Bcap37 and MBA-MD-231 cells in both time-and dose-dependent manners (Figure 1, P < 0.05, One-Way ANOVA test).
Figure 1.
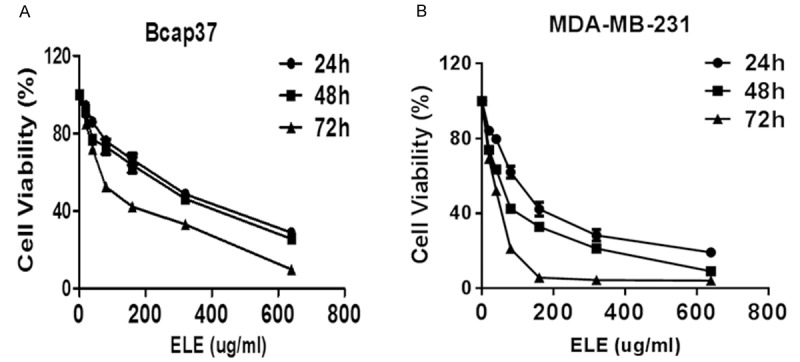
β-elemene inhibits the viability of breast cancer cells. Bcap37 (A) and MDA-MB-231 (B) cells were treated with varied concentration of β-elemene for different times as indicated. The cell viability was determined by MTS assay. The statistical significance was analyzed by One-Way ANOVA test. ELE: β-elemene.
β-elemene induced apoptosis of breast cancer cells
Some cancer cells became floating after the exposure to β-elemene (Figure 2A and 2B), indicating that β-elemene may induce the apoptosis of breast cancer cells. Indeed, PI and Annexin V double staining results confirm the induction of apoptosis by β-elemene (Figure 2C and 2D). However, there are still many live cells left, indicating that β-elemene may induce other changes in addition to the apoptosis of breast cancer cells.
Figure 2.
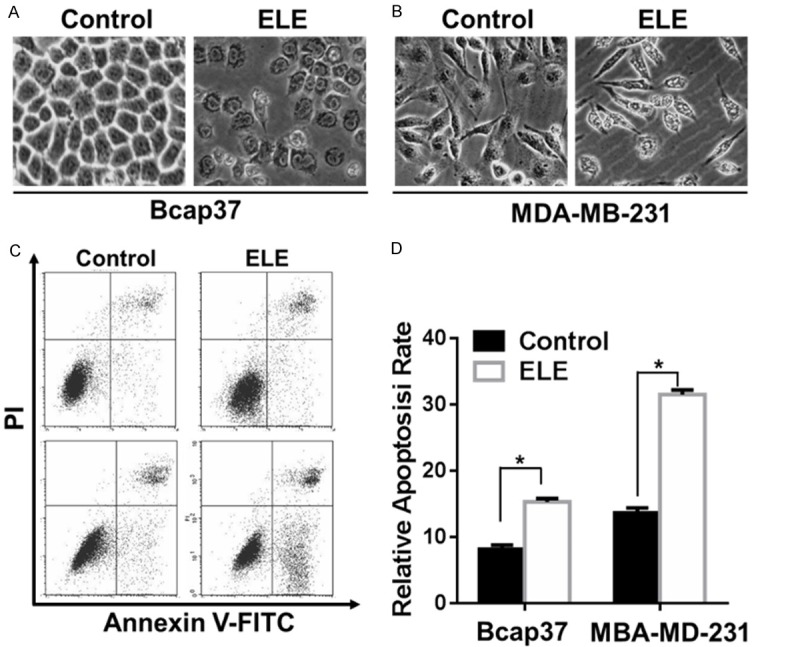
ELE induces the apoptosis of breast cancer cells. A. The morphological changes of Bcap37and MDA-MB-231 cells treated with β-elemene (320 μg/ml) for 24 hours were observed under light microscopy. B and C. The apoptosis of Bcap37and MDA-MB-231 cells treated with β-elemene (320 μg/ml) for 24 hours were analyzed by PI and Annexin V staining. The statistical significance was analyzed by Student’s t test. The asterisks indicate statistical significance (P < 0.05). ELE: β-elemene.
β-elemene induced autophagy in breast cancer cells
Therefore, we asked whether autophagy was induced after β-elemene treatment. First, we evaluated the conversion of LC3-I into LC3-II before and after β-elemene treatment by western blotting analysis. As shown in Figure 3, β-elemene can induce the switch of LC-I to LC-II in a dose-dependent manner in both Bcap37 and MBA-MD-231 cells, indicating that autophagy might be activated by β-elemene (Figure 3A and 3B). Next, we confirmed the activation of autophagy by electronic microscopy examination. As shown in Figure 3C, typical autophagosomes were presented in β-elemene-treated cells rather than untreated cells, confirming the induction of autophagy by β-elemene.
Figure 3.
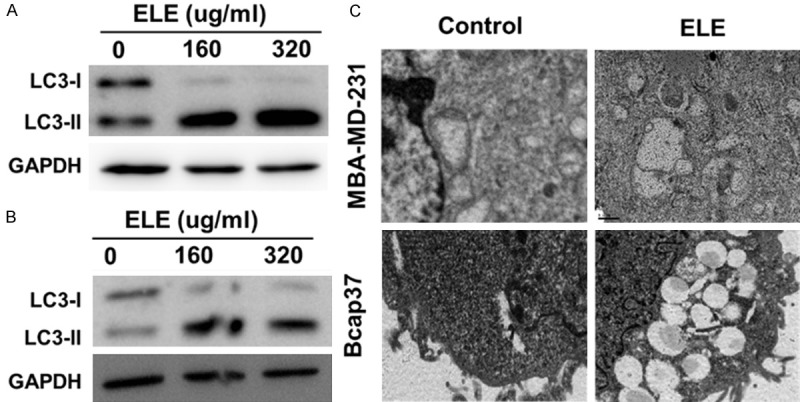
ELE activates autophagy in breast cancer cells. LC3 in Bcap37 (A) and MDA-MB-231 (B) cells treated with indicated concentration for 24 hours were detected by western blotting. (C) The changes in ultra-microstructure of Bcap37and MDA-MB-231 cells treated with β-elemene (320 μg/ml) for 24 hours were observed under electron microscopy. ELE: β-elemene.
Inhibition of autophagy promotes growth-inhibitory effect of β-elemene
To explore the relevance of autophagy in β-elemene-induced growth inhibition, we determined the effect of β-elemene on breast cancer cells before and after the suppression of autophagy. While autophagy inhibitor chloroquine (CQ) alone had no significant effect on breast cancer cells, the inhibitory effect of β-elemene on cell viability was significantly enhanced in both breast cancer cell lines s after CQ treatment (Bcap 37 in Figure 4A and MBA-MD-231 in Figure 4D). Consistently, apoptosis induced by β-elemene were increased after CQ treatment (Bcap 37 Figure 4B, 4C, and MBA-MD-231 in Figure 4D, 4E), indicating that autophagy in response to ELE treatment played a cyto-protective role to help cancer cells survive under unfavorable conditions.
Figure 4.
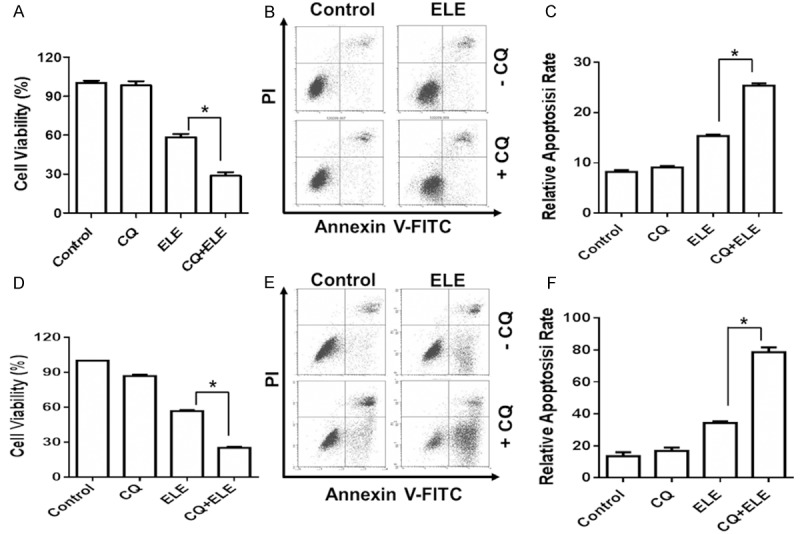
Autophagy inhibition enhances the sensitivity of breast cancer cells to ELE. The viability of Bcap37 (A) and MDA-MB-231 (D) cells treated with or without Chloroquine (10 μm) and β-elemene (320 μg/ml) were analyzed by MTS assay. The apoptosis of Bcap37 (B and C) and MDA-MB-231 (E and F) cells treated with or without Chloroquine (10 μm) and β-elemene (320 μg/ml) were analyzed by PI and Annexin V staining. The statistical significance was analyzed by Student’s t test. The asterisks indicate statistical significance (P < 0.05). ELE: β-elemene; CQ: Chloroquine.
Discussion
Although β-elemene has been approved by the State Food and Drug Administration of China and widely used for the clinical management of patients with various cancers mainly brain tumors, the mechanism of the anti-cancer effect of β-elemene remains largely unknown. In addition, it would be interesting to know whether β-elemene can improve the clinical efficacy of commonly used treatment modalities such as chemotherapy and targeted therapy. In this study, we found that β-elemene inhibited the viability of breast cancers in vitro in a dose- and time-dependent manner (Figure 1). Such an inhibition can be attributed to the induction of both apoptosis (Figure 2) and cell cycle arrest (data not shown). These data indicates that β-elemene could be used potentially for the treatment of patients with breast cancer, which is one of the most commonly diagnosed cancer and the first leading cause of cancer-related deaths in women [15].
Interestingly, we found β-elemene induced autophagy in addition to apoptosis (Figure 3). Autophagy is morphologically characterized by the appearance of “double-membrane” vacuoles (autophagosomes) in the cytoplasm. In addition, the mammalian homologue of LC3, a homologue of Apg8p essential for autophagy in yeast, is found to be a specific biochemical marker for autophagy. Newly synthesized LC3 termed LC3-1 is evenly distributed throughout the cytoplasm. Upon induction of autophagy, some LC3-I is converted into LC3-II, which is tightly bound to the autophagosomal membranes forming ring-shaped structures in the cytosol and degraded after the maturation of autophagosomes. We found that β-elemene indeed induced the conversion of LC3-II from LC3-I (Figure 3B). The accumulation of LC3-II can result from the blockage of the autophagy flux so that the degradation of LC3-II was hampered [16]. However, the analysis of ultrastructure by electron microscopy revealed that mature autolysosomes formed in breast cancer cells treated with β-elemene, indicating that β-elemene indeed activates rather than inhibits autophagy. Although it was designated as programmed cell death type II, autophagy was recently found to promote cellular survival under unfavorable stress conditions such as deprivation of amino acids, revealing a new role of autophagy in cancer development. In consistence, we found that inhibition of autophagy induced by β-elemene could potentiate the anti-cancer effect of β-elemene. Therefore, β-elemene-induced autophagy functions as a stress response to protect cells from apoptosis. A number of cancer therapeutics including DNA-damaging chemotherapeutics, endocrine therapies such as tamoxifen and radiation therapy have been found to induce autophagy in vitro and in vivo [17-19]. Recently, it was found that autophagy can be activated and protected tumor cells from targeted therapies, such as the imatinib mesylate in Philadelphia chromosome-positive cells [20], trastuzumab in breast cancer [21], Src family kinase inhibitors in prostate cancer [22], proteasome inhibitors in prostate cancer [23]. All of these studies indicate that the combination of anti-autophagy strategies could potentially improve the clinical efficacy of current anti-cancer regiments including β-elemene.
In summary, β-elemene is a potential drug for breast cancer by including apoptosis. Inhibition of autophagy has the potential to improve the clinical efficacy of β-elemene for cancer treatment. However, how β-elemene simultaneously initiates apoptosis program and activates autophagy to maintain cell survival remains further studies.
Acknowledgements
This work was supported by Department of Health in Zhejiang Province (WKJ2012-2-013; 2013RCA029) and 973 projects of MOST (2012CB526600).
Disclosure of conflict of interest
None.
References
- 1.Edris AE. Anti-cancer properties of Nigella spp. essential oils and their major constituents, thymoquinone and beta-elemene. Curr Clin Pharmacol. 2009;4:43–46. doi: 10.2174/157488409787236137. [DOI] [PubMed] [Google Scholar]
- 2.Lu JJ, Dang YY, Huang M, Xu WS, Chen XP, Wang YT. Anti-cancer properties of terpenoids isolated from Rhizoma Curcumae--a review. J Ethnopharmacol. 2012;143:406–411. doi: 10.1016/j.jep.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Zhao J, Li QQ, Zou B, Wang G, Li X, Kim JE, Cuff CF, Huang L, Reed E, Gardner K. In vitro combination characterization of the new anticancer plant drug beta-elemene with taxanes against human lung carcinoma. Int J Oncol. 2007;31:241–252. [PubMed] [Google Scholar]
- 4.Tao L, Zhou L, Zheng L, Yao M. Elemene displays anti-cancer ability on laryngeal cancer cells in vitro and in vivo. Cancer Chemother Pharmacol. 2006;58:24–34. doi: 10.1007/s00280-005-0137-x. [DOI] [PubMed] [Google Scholar]
- 5.Wang G, Li X, Huang F, Zhao J, Ding H, Cunningham C, Coad JE, Flynn DC, Reed E, Li QQ. Antitumor effect of beta-elemene in non-small-cell lung cancer cells is mediated via induction of cell cycle arrest and apoptotic cell death. Cell Mol Life Sci. 2005;62:881–893. doi: 10.1007/s00018-005-5017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li QQ, Lee RX, Liang H, Zhong Y. Anticancer activity of beta-Elemene and its synthetic analogs in human malignant brain tumor cells. Anticancer Res. 2013;33:65–76. [PMC free article] [PubMed] [Google Scholar]
- 7.Li QQ, Wang G, Huang F, Li JM, Cuff CF, Reed E. Sensitization of lung cancer cells to cisplatin by beta-elemene is mediated through blockade of cell cycle progression: antitumor efficacies of beta-elemene and its synthetic analogs. Med Oncol. 2013;30:488. doi: 10.1007/s12032-013-0488-9. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Li Y, Zhang Y, Song J, Wang Q, Zheng L, Liu D. Beta-elemene blocks epithelial-mesenchymal transition in human breast cancer cell line MCF-7 through Smad3-mediated down-regulation of nuclear transcription factors. PLoS One. 2013;8:e58719. doi: 10.1371/journal.pone.0058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu HB, Li L, Fu J, Mao XP, Xu LZ. Reversion of multidrug resistance in a chemoresistant human breast cancer cell line by beta-elemene. Pharmacology. 2012;89:303–312. doi: 10.1159/000337178. [DOI] [PubMed] [Google Scholar]
- 10.Ding XF, Shen M, Xu LY, Dong JH, Chen G. 13,14-bis(cis-3,5-dimethyl-1-piperazinyl)-beta-elemene, a novel beta-elemene derivative, shows potent antitumor activities via inhibition of mTOR in human breast cancer cells. Oncol Lett. 2013;5:1554–1558. doi: 10.3892/ol.2013.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Mu XD, Li EZ, Luo Y, Song N, Qu XJ, Hu XJ, Liu YP. The Role of E3 Ubiquitin Ligase Cbl Proteins in beta-Elemene Reversing Multi-Drug Resistance of Human Gastric Adenocarcinoma Cells. Int J Mol Sci. 2013;14:10075–10089. doi: 10.3390/ijms140510075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han W, Pan H, Chen Y, Sun J, Wang Y, Li J, Ge W, Feng L, Lin X, Wang X, Wang X, Jin H. EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PLoS One. 2011;6:e18691. doi: 10.1371/journal.pone.0018691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Ding L, Wang X, Zhang J, Han W, Feng L, Sun J, Jin H, Wang XJ. Pterostilbene simultaneously induces apoptosis, cell cycle arrest and cyto-protective autophagy in breast cancer cells. Am J Transl Res. 2012;4:44–51. [PMC free article] [PubMed] [Google Scholar]
- 14.Han W, Sun J, Feng L, Wang K, Li D, Pan Q, Chen Y, Jin W, Wang X, Pan H, Jin H. Autophagy inhibition enhances daunorubicin-induced apoptosis in K562 cells. PLoS One. 2011;6:e28491. doi: 10.1371/journal.pone.0028491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 16.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, Ahn HJ, Ait-Mohamed O, Ait-Si-Ali S, Akematsu T, Akira S, Al-Younes HM, Al-Zeer MA, Albert ML, Albin RL, Alegre-Abarrategui J, Aleo MF, Alirezaei M, Almasan A, Almonte-Becerril M, Amano A, Amaravadi R, Amarnath S, Amer AO, Andrieu-Abadie N, Anantharam V, Ann DK, Anoopkumar-Dukie S, Aoki H, Apostolova N, Arancia G, Aris JP, Asanuma K, Asare NY, Ashida H, Askanas V, Askew DS, Auberger P, Baba M, Backues SK, Baehrecke EH, Bahr BA, Bai XY, Bailly Y, Baiocchi R, Baldini G, Balduini W, Ballabio A, Bamber BA, Bampton ET, Banhegyi G, Bartholomew CR, Bassham DC, Bast RC Jr, Batoko H, Bay BH, Beau I, Bechet DM, Begley TJ, Behl C, Behrends C, Bekri S, Bellaire B, Bendall LJ, Benetti L, Berliocchi L, Bernardi H, Bernassola F, Besteiro S, Bhatia-Kissova I, Bi X, Biard-Piechaczyk M, Blum JS, Boise LH, Bonaldo P, Boone DL, Bornhauser BC, Bortoluci KR, Bossis I, Bost F, Bourquin JP, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady NR, Brancolini C, Brech A, Brenman JE, Brennand A, Bresnick EH, Brest P, Bridges D, Bristol ML, Brookes PS, Brown EJ, Brumell JH, Brunetti-Pierri N, Brunk UT, Bulman DE, Bultman SJ, Bultynck G, Burbulla LF, Bursch W, Butchar JP, Buzgariu W, Bydlowski SP, Cadwell K, Cahova M, Cai D, Cai J, Cai Q, Calabretta B, Calvo-Garrido J, Camougrand N, Campanella M, Campos-Salinas J, Candi E, Cao L, Caplan AB, Carding SR, Cardoso SM, Carew JS, Carlin CR, Carmignac V, Carneiro LA, Carra S, Caruso RA, Casari G, Casas C, Castino R, Cebollero E, Cecconi F, Celli J, Chaachouay H, Chae HJ, Chai CY, Chan DC, Chan EY, Chang RC, Che CM, Chen CC, Chen GC, Chen GQ, Chen M, Chen Q, Chen SS, Chen W, Chen X, Chen X, Chen X, Chen YG, Chen Y, Chen Y, Chen YJ, Chen Z, Cheng A, Cheng CH, Cheng Y, Cheong H, Cheong JH, Cherry S, Chess-Williams R, Cheung ZH, Chevet E, Chiang HL, Chiarelli R, Chiba T, Chin LS, Chiou SH, Chisari FV, Cho CH, Cho DH, Choi AM, Choi D, Choi KS, Choi ME, Chouaib S, Choubey D, Choubey V, Chu CT, Chuang TH, Chueh SH, Chun T, Chwae YJ, Chye ML, Ciarcia R, Ciriolo MR, Clague MJ, Clark RS, Clarke PG, Clarke R, Codogno P, Coller HA, Colombo MI, Comincini S, Condello M, Condorelli F, Cookson MR, Coombs GH, Coppens I, Corbalan R, Cossart P, Costelli P, Costes S, Coto-Montes A, Couve E, Coxon FP, Cregg JM, Crespo JL, Cronje MJ, Cuervo AM, Cullen JJ, Czaja MJ, D’Amelio M, Darfeuille-Michaud A, Davids LM, Davies FE, De Felici M, de Groot JF, de Haan CA, De Martino L, De Milito A, De Tata V, Debnath J, Degterev A, Dehay B, Delbridge LM, Demarchi F, Deng YZ, Dengjel J, Dent P, Denton D, Deretic V, Desai SD, Devenish RJ, Di Gioacchino M, Di Paolo G, Di Pietro C, Diaz-Araya G, Diaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dikic I, Dinesh-Kumar SP, Ding WX, Distelhorst CW, Diwan A, Djavaheri-Mergny M, Dokudovskaya S, Dong Z, Dorsey FC, Dosenko V, Dowling JJ, Doxsey S, Dreux M, Drew ME, Duan Q, Duchosal MA, Duff K, Dugail I, Durbeej M, Duszenko M, Edelstein CL, Edinger AL, Egea G, Eichinger L, Eissa NT, Ekmekcioglu S, El-Deiry WS, Elazar Z, Elgendy M, Ellerby LM, Eng KE, Engelbrecht AM, Engelender S, Erenpreisa J, Escalante R, Esclatine A, Eskelinen EL, Espert L, Espina V, Fan H, Fan J, Fan QW, Fan Z, Fang S, Fang Y, Fanto M, Fanzani A, Farkas T, Farre JC, Faure M, Fechheimer M, Feng CG, Feng J, Feng Q, Feng Y, Fesus L, Feuer R, Figueiredo-Pereira ME, Fimia GM, Fingar DC, Finkbeiner S, Finkel T, Finley KD, Fiorito F, Fisher EA, Fisher PB, Flajolet M, Florez-McClure ML, Florio S, Fon EA, Fornai F, Fortunato F, Fotedar R, Fowler DH, Fox HS, Franco R, Frankel LB, Fransen M, Fuentes JM, Fueyo J, Fujii J, Fujisaki K, Fujita E, Fukuda M, Furukawa RH, Gaestel M, Gailly P, Gajewska M, Galliot B, Galy V, Ganesh S, Ganetzky B, Ganley IG, Gao FB, Gao GF, Gao J, Garcia L, Garcia-Manero G, Garcia-Marcos M, Garmyn M, Gartel AL, Gatti E, Gautel M, Gawriluk TR, Gegg ME, Geng J, Germain M, Gestwicki JE, Gewirtz DA, Ghavami S, Ghosh P, Giammarioli AM, Giatromanolaki AN, Gibson SB, Gilkerson RW, Ginger ML, Ginsberg HN, Golab J, Goligorsky MS, Golstein P, Gomez-Manzano C, Goncu E, Gongora C, Gonzalez CD, Gonzalez R, Gonzalez-Estevez C, Gonzalez-Polo RA, Gonzalez-Rey E, Gorbunov NV, Gorski S, Goruppi S, Gottlieb RA, Gozuacik D, Granato GE, Grant GD, Green KN, Gregorc A, Gros F, Grose C, Grunt TW, Gual P, Guan JL, Guan KL, Guichard SM, Gukovskaya AS, Gukovsky I, Gunst J, Gustafsson AB, Halayko AJ, Hale AN, Halonen SK, Hamasaki M, Han F, Han T, Hancock MK, Hansen M, Harada H, Harada M, Hardt SE, Harper JW, Harris AL, Harris J, Harris SD, Hashimoto M, Haspel JA, Hayashi S, Hazelhurst LA, He C, He YW, Hebert MJ, Heidenreich KA, Helfrich MH, Helgason GV, Henske EP, Herman B, Herman PK, Hetz C, Hilfiker S, Hill JA, Hocking LJ, Hofman P, Hofmann TG, Hohfeld J, Holyoake TL, Hong MH, Hood DA, Hotamisligil GS, Houwerzijl EJ, Hoyer-Hansen M, Hu B, Hu CA, Hu HM, Hua Y, Huang C, Huang J, Huang S, Huang WP, Huber TB, Huh WK, Hung TH, Hupp TR, Hur GM, Hurley JB, Hussain SN, Hussey PJ, Hwang JJ, Hwang S, Ichihara A, Ilkhanizadeh S, Inoki K, Into T, Iovane V, Iovanna JL, Ip NY, Isaka Y, Ishida H, Isidoro C, Isobe K, Iwasaki A, Izquierdo M, Izumi Y, Jaakkola PM, Jaattela M, Jackson GR, Jackson WT, Janji B, Jendrach M, Jeon JH, Jeung EB, Jiang H, Jiang H, Jiang JX, Jiang M, Jiang Q, Jiang X, Jiang X, Jimenez A, Jin M, Jin S, Joe CO, Johansen T, Johnson DE, Johnson GV, Jones NL, Joseph B, Joseph SK, Joubert AM, Juhasz G, Juillerat-Jeanneret L, Jung CH, Jung YK, Kaarniranta K, Kaasik A, Kabuta T, Kadowaki M, Kagedal K, Kamada Y, Kaminskyy VO, Kampinga HH, Kanamori H, Kang C, Kang KB, Kang KI, Kang R, Kang YA, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kanthasamy A, Karantza V, Kaushal GP, Kaushik S, Kawazoe Y, Ke PY, Kehrl JH, Kelekar A, Kerkhoff C, Kessel DH, Khalil H, Kiel JA, Kiger AA, Kihara A, Kim DR, Kim DH, Kim DH, Kim EK, Kim HR, Kim JS, Kim JH, Kim JC, Kim JK, Kim PK, Kim SW, Kim YS, Kim Y, Kimchi A, Kimmelman AC, King JS, Kinsella TJ, Kirkin V, Kirshenbaum LA, Kitamoto K, Kitazato K, Klein L, Klimecki WT, Klucken J, Knecht E, Ko BC, Koch JC, Koga H, Koh JY, Koh YH, Koike M, Komatsu M, Kominami E, Kong HJ, Kong WJ, Korolchuk VI, Kotake Y, Koukourakis MI, Kouri Flores JB, Kovacs AL, Kraft C, Krainc D, Kramer H, Kretz-Remy C, Krichevsky AM, Kroemer G, Kruger R, Krut O, Ktistakis NT, Kuan CY, Kucharczyk R, Kumar A, Kumar R, Kumar S, Kundu M, Kung HJ, Kurz T, Kwon HJ, La Spada AR, Lafont F, Lamark T, Landry J, Lane JD, Lapaquette P, Laporte JF, Laszlo L, Lavandero S, Lavoie JN, Layfield R, Lazo PA, Le W, Le Cam L, Ledbetter DJ, Lee AJ, Lee BW, Lee GM, Lee J, Lee JH, Lee M, Lee MS, Lee SH, Leeuwenburgh C, Legembre P, Legouis R, Lehmann M, Lei HY, Lei QY, Leib DA, Leiro J, Lemasters JJ, Lemoine A, Lesniak MS, Lev D, Levenson VV, Levine B, Levy E, Li F, Li JL, Li L, Li S, Li W, Li XJ, Li YB, Li YP, Liang C, Liang Q, Liao YF, Liberski PP, Lieberman A, Lim HJ, Lim KL, Lim K, Lin CF, Lin FC, Lin J, Lin JD, Lin K, Lin WW, Lin WC, Lin YL, Linden R, Lingor P, Lippincott-Schwartz J, Lisanti MP, Liton PB, Liu B, Liu CF, Liu K, Liu L, Liu QA, Liu W, Liu YC, Liu Y, Lockshin RA, Lok CN, Lonial S, Loos B, Lopez-Berestein G, Lopez-Otin C, Lossi L, Lotze MT, Low P, Lu B, Lu B, Lu B, Lu Z, Luciano F, Lukacs NW, Lund AH, Lynch-Day MA, Ma Y, Macian F, MacKeigan JP, Macleod KF, Madeo F, Maiuri L, Maiuri MC, Malagoli D, Malicdan MC, Malorni W, Man N, Mandelkow EM, Manon S, Manov I, Mao K, Mao X, Mao Z, Marambaud P, Marazziti D, Marcel YL, Marchbank K, Marchetti P, Marciniak SJ, Marcondes M, Mardi M, Marfe G, Marino G, Markaki M, Marten MR, Martin SJ, Martinand-Mari C, Martinet W, Martinez-Vicente M, Masini M, Matarrese P, Matsuo S, Matteoni R, Mayer A, Mazure NM, McConkey DJ, McConnell MJ, McDermott C, McDonald C, McInerney GM, McKenna SL, McLaughlin B, McLean PJ, McMaster CR, McQuibban GA, Meijer AJ, Meisler MH, Melendez A, Melia TJ, Melino G, Mena MA, Menendez JA, Menna-Barreto RF, Menon MB, Menzies FM, Mercer CA, Merighi A, Merry DE, Meschini S, Meyer CG, Meyer TF, Miao CY, Miao JY, Michels PA, Michiels C, Mijaljica D, Milojkovic A, Minucci S, Miracco C, Miranti CK, Mitroulis I, Miyazawa K, Mizushima N, Mograbi B, Mohseni S, Molero X, Mollereau B, Mollinedo F, Momoi T, Monastyrska I, Monick MM, Monteiro MJ, Moore MN, Mora R, Moreau K, Moreira PI, Moriyasu Y, Moscat J, Mostowy S, Mottram JC, Motyl T, Moussa CE, Muller S, Muller S, Munger K, Munz C, Murphy LO, Murphy ME, Musaro A, Mysorekar I, Nagata E, Nagata K, Nahimana A, Nair U, Nakagawa T, Nakahira K, Nakano H, Nakatogawa H, Nanjundan M, Naqvi NI, Narendra DP, Narita M, Navarro M, Nawrocki ST, Nazarko TY, Nemchenko A, Netea MG, Neufeld TP, Ney PA, Nezis IP, Nguyen HP, Nie D, Nishino I, Nislow C, Nixon RA, Noda T, Noegel AA, Nogalska A, Noguchi S, Notterpek L, Novak I, Nozaki T, Nukina N, Nurnberger T, Nyfeler B, Obara K, Oberley TD, Oddo S, Ogawa M, Ohashi T, Okamoto K, Oleinick NL, Oliver FJ, Olsen LJ, Olsson S, Opota O, Osborne TF, Ostrander GK, Otsu K, Ou JH, Ouimet M, Overholtzer M, Ozpolat B, Paganetti P, Pagnini U, Pallet N, Palmer GE, Palumbo C, Pan T, Panaretakis T, Pandey UB, Papackova Z, Papassideri I, Paris I, Park J, Park OK, Parys JB, Parzych KR, Patschan S, Patterson C, Pattingre S, Pawelek JM, Peng J, Perlmutter DH, Perrotta I, Perry G, Pervaiz S, Peter M, Peters GJ, Petersen M, Petrovski G, Phang JM, Piacentini M, Pierre P, Pierrefite-Carle V, Pierron G, Pinkas-Kramarski R, Piras A, Piri N, Platanias LC, Poggeler S, Poirot M, Poletti A, Pous C, Pozuelo-Rubio M, Praetorius-Ibba M, Prasad A, Prescott M, Priault M, Produit-Zengaffinen N, Progulske-Fox A, Proikas-Cezanne T, Przedborski S, Przyklenk K, Puertollano R, Puyal J, Qian SB, Qin L, Qin ZH, Quaggin SE, Raben N, Rabinowich H, Rabkin SW, Rahman I, Rami A, Ramm G, Randall G, Randow F, Rao VA, Rathmell JC, Ravikumar B, Ray SK, Reed BH, Reed JC, Reggiori F, Regnier-Vigouroux A, Reichert AS, Reiners JJ Jr, Reiter RJ, Ren J, Revuelta JL, Rhodes CJ, Ritis K, Rizzo E, Robbins J, Roberge M, Roca H, Roccheri MC, Rocchi S, Rodemann HP, Rodriguez de Cordoba S, Rohrer B, Roninson IB, Rosen K, Rost-Roszkowska MM, Rouis M, Rouschop KM, Rovetta F, Rubin BP, Rubinsztein DC, Ruckdeschel K, Rucker EB 3rd, Rudich A, Rudolf E, Ruiz-Opazo N, Russo R, Rusten TE, Ryan KM, Ryter SW, Sabatini DM, Sadoshima J, Saha T, Saitoh T, Sakagami H, Sakai Y, Salekdeh GH, Salomoni P, Salvaterra PM, Salvesen G, Salvioli R, Sanchez AM, Sanchez-Alcazar JA, Sanchez-Prieto R, Sandri M, Sankar U, Sansanwal P, Santambrogio L, Saran S, Sarkar S, Sarwal M, Sasakawa C, Sasnauskiene A, Sass M, Sato K, Sato M, Schapira AH, Scharl M, Schatzl HM, Scheper W, Schiaffino S, Schneider C, Schneider ME, Schneider-Stock R, Schoenlein PV, Schorderet DF, Schuller C, Schwartz GK, Scorrano L, Sealy L, Seglen PO, Segura-Aguilar J, Seiliez I, Seleverstov O, Sell C, Seo JB, Separovic D, Setaluri V, Setoguchi T, Settembre C, Shacka JJ, Shanmugam M, Shapiro IM, Shaulian E, Shaw RJ, Shelhamer JH, Shen HM, Shen WC, Sheng ZH, Shi Y, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shintani T, Shirihai OS, Shore GC, Sibirny AA, Sidhu SB, Sikorska B, Silva-Zacarin EC, Simmons A, Simon AK, Simon HU, Simone C, Simonsen A, Sinclair DA, Singh R, Sinha D, Sinicrope FA, Sirko A, Siu PM, Sivridis E, Skop V, Skulachev VP, Slack RS, Smaili SS, Smith DR, Soengas MS, Soldati T, Song X, Sood AK, Soong TW, Sotgia F, Spector SA, Spies CD, Springer W, Srinivasula SM, Stefanis L, Steffan JS, Stendel R, Stenmark H, Stephanou A, Stern ST, Sternberg C, Stork B, Stralfors P, Subauste CS, Sui X, Sulzer D, Sun J, Sun SY, Sun ZJ, Sung JJ, Suzuki K, Suzuki T, Swanson MS, Swanton C, Sweeney ST, Sy LK, Szabadkai G, Tabas I, Taegtmeyer H, Tafani M, Takacs-Vellai K, Takano Y, Takegawa K, Takemura G, Takeshita F, Talbot NJ, Tan KS, Tanaka K, Tanaka K, Tang D, Tang D, Tanida I, Tannous BA, Tavernarakis N, Taylor GS, Taylor GA, Taylor JP, Terada LS, Terman A, Tettamanti G, Thevissen K, Thompson CB, Thorburn A, Thumm M, Tian F, Tian Y, Tocchini-Valentini G, Tolkovsky AM, Tomino Y, Tonges L, Tooze SA, Tournier C, Tower J, Towns R, Trajkovic V, Travassos LH, Tsai TF, Tschan MP, Tsubata T, Tsung A, Turk B, Turner LS, Tyagi SC, Uchiyama Y, Ueno T, Umekawa M, Umemiya-Shirafuji R, Unni VK, Vaccaro MI, Valente EM, Van den Berghe G, van der Klei IJ, van Doorn W, van Dyk LF, van Egmond M, van Grunsven LA, Vandenabeele P, Vandenberghe WP, Vanhorebeek I, Vaquero EC, Velasco G, Vellai T, Vicencio JM, Vierstra RD, Vila M, Vindis C, Viola G, Viscomi MT, Voitsekhovskaja OV, von Haefen C, Votruba M, Wada K, Wade-Martins R, Walker CL, Walsh CM, Walter J, Wan XB, Wang A, Wang C, Wang D, Wang F, Wang F, Wang G, Wang H, Wang HG, Wang HD, Wang J, Wang K, Wang M, Wang RC, Wang X, Wang X, Wang YJ, Wang Y, Wang Z, Wang ZC, Wang Z, Wansink DG, Ward DM, Watada H, Waters SL, Webster P, Wei L, Weihl CC, Weiss WA, Welford SM, Wen LP, Whitehouse CA, Whitton JL, Whitworth AJ, Wileman T, Wiley JW, Wilkinson S, Willbold D, Williams RL, Williamson PR, Wouters BG, Wu C, Wu DC, Wu WK, Wyttenbach A, Xavier RJ, Xi Z, Xia P, Xiao G, Xie Z, Xie Z, Xu DZ, Xu J, Xu L, Xu X, Yamamoto A, Yamamoto A, Yamashina S, Yamashita M, Yan X, Yanagida M, Yang DS, Yang E, Yang JM, Yang SY, Yang W, Yang WY, Yang Z, Yao MC, Yao TP, Yeganeh B, Yen WL, Yin JJ, Yin XM, Yoo OJ, Yoon G, Yoon SY, Yorimitsu T, Yoshikawa Y, Yoshimori T, Yoshimoto K, You HJ, Youle RJ, Younes A, Yu L, Yu L, Yu SW, Yu WH, Yuan ZM, Yue Z, Yun CH, Yuzaki M, Zabirnyk O, Silva-Zacarin E, Zacks D, Zacksenhaus E, Zaffaroni N, Zakeri Z, Zeh HJ 3rd, Zeitlin SO, Zhang H, Zhang HL, Zhang J, Zhang JP, Zhang L, Zhang L, Zhang MY, Zhang XD, Zhao M, Zhao YF, Zhao Y, Zhao ZJ, Zheng X, Zhivotovsky B, Zhong Q, Zhou CZ, Zhu C, Zhu WG, Zhu XF, Zhu X, Zhu Y, Zoladek T, Zong WX, Zorzano A, Zschocke J, Zuckerbraun B. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 18.Hoyer-Hansen M, Jaattela M. Autophagy: an emerging target for cancer therapy. Autophagy. 2008;4:574–580. doi: 10.4161/auto.5921. [DOI] [PubMed] [Google Scholar]
- 19.Lomonaco SL, Finniss S, Xiang C, Decarvalho A, Umansky F, Kalkanis SN, Mikkelsen T, Brodie C. The induction of autophagy by gamma-radiation contributes to the radioresistance of glioma stem cells. Int J Cancer. 2009;125:717–722. doi: 10.1002/ijc.24402. [DOI] [PubMed] [Google Scholar]
- 20.Bellodi C, Lidonnici MR, Hamilton A, Helgason GV, Soliera AR, Ronchetti M, Galavotti S, Young KW, Selmi T, Yacobi R, Van Etten RA, Donato N, Hunter A, Dinsdale D, Tirro E, Vigneri P, Nicotera P, Dyer MJ, Holyoake T, Salomoni P, Calabretta B. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest. 2009;119:1109–1123. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vazquez-Martin A, Oliveras-Ferraros C, Menendez JA. Autophagy facilitates the development of breast cancer resistance to the anti-HER2 monoclonal antibody trastuzumab. PLoS One. 2009;4:e6251. doi: 10.1371/journal.pone.0006251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Z, Chang PC, Yang JC, Chu CY, Wang LY, Chen NT, Ma AH, Desai SJ, Lo SH, Evans CP, Lam KS, Kung HJ. Autophagy Blockade Sensitizes Prostate Cancer Cells towards Src Family Kinase Inhibitors. Genes Cancer. 2010;1:40–49. doi: 10.1177/1947601909358324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu K, Dunner K Jr, McConkey DJ. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene. 2010;29:451–462. doi: 10.1038/onc.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]


