Abstract
Background and aim: Protein Kinase (casein kinase 2, CK2) is a pleiotropic serine-threonine kinase that is frequently dysregulated in many human tumors; microRNAs (miRNAs) are a class of small noncoding RNAs which play important roles in human cancers. This study aimed to investigate the role of CK2 and miRNA expression in breast cancer. Methods: Casein kinase 2 in MCF-7 breast cancer cell line was inhibited by the CK2 inhibitor TBB (4,5,6,7-tetrabromobenzotriazole), then cell proliferation was studied using MTT assay, and cell cycle distribution and apoptosis were detected by flow cytometry. The alteration of microRNAs expression profile was determined by microarray technology, followed by RT-PCR confirmation. Results: Here, we for the first time showed that inhibition of CK2 in MCF-7 breast cancer cells causes suppressed cell growth, which was related with dysregulation of the miRNA profile and altered expression. CK2 inhibition induced the up-regulated expression of 17 miRNAs and 10 down-regulated microRNAs which contributed to the impaired growth, inhibited cell cycle progress and increased apoptosis of MCF-7 cells by a CK2 inhibitor. Conclusions: These findings highlight the potential role of dysregulated miRNA expression regulated by CK2 in breast cancer.
Keywords: TBB, casein kinase 2, microRNA, breast cancer
Introduction
Breast cancer is the leading cause of cancer death for women worldwide. It is estimated that 232,620 new cases and 39,970 deaths in 2011 in the United States [1]. Despite advances in early detection and treatment of breast cancer, our understanding of the molecular mechanism that governs the progression of breast cancer remains fragmentary.
Casein kinase 2 (CK2) is a ubiquitous eukaryotic serine and threonine protein kinase which consists of two catalytic subunits α and (or) α′ and two regulatory subunits β [2]. As a heterologous tetramer, CK2 has three isoforms, including α2β2, α′2β2 or αα′β2. It has been proved that CK2 plays a pivotal role in the regulation of cellular survival, growth, proliferation as well as apoptosis through phosphorylating a variety of substrates [3]. CK2 is involved in processes such as proliferation, apoptosis and embryonic development, and is found to be overexpressed in human cancer cell lines and solid cancer tissues [4].
MicroRNAs (miRNAs) are a class of evolutionarily conserved small non-coding RNAs that control gene expression by targeting mRNAs for translational repression or cleavage [5]. Studies have shown that miRNAs have critical functions in regulating cellular differentiation, proliferation and apoptosis and deregulation of miRNAs often alter the normal cell growth and development, leading to a variety of disorders including human cancer [6-8]. Aberrant expression of miRNAs has been well documented in a variety of human cancers including breast cancer [9]. Recent findings have shown that miR-21, miR-29b-2 and miR-27a/b were upregulated, while miR-125b, miR-145, miR-10b, miR-155 and miR-17-5p were downregulated in breast cancers compared with normal tissues [10].
In the present study, we analyzed the role of CK2 and its correlation with miRNA in human breast cancer cell line MCF-7 cells, and the study for the first time shows that inhibition of CK2 in MCF-7 cells induced changes in cell viability, cell cycle and apoptosis and these changes correlated with the deregulation of miRNA expression.
Materials and methods
Cell culture
The human breast cancer cell line MCF-7 was obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Cells were maintained in DMEM (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) in a humidified incubator at 37°C under a 5% CO2 atmosphere.
TBB treatment
The CK2 inhibitor 4,5,6,7-tetrabromo-2-azabenzimidazole (TBB) was purchased from the Sigma-Aldrich Company (Sigma, St. Louis, MO, USA). TBB was dissolved in 100% dimethylsulphoxide (DMSO; Sigma) to make a stock solution of 10 mM, which was then diluted in culture medium to obtain the desired concentrations of 50, 100 and 200 μM. DMSO diluted in culture medium at the final percentage of 0.05% without TBB was designated as 0 μM. Untreated cells were incubated in the culture medium without any additives. Except for the cell proliferation assay, cells were treated with or without TBB for 48 hours, after which total RNA or protein was extracted and flow cytometry was carried out.
Cell viability assay
Cell viability was measured by using MTT assay. Exponentially growing MCF-7 cells were digested by 0.25% trypsin for 1-2 min, and washed thrice with PBS. Then 5×103 cells in 100 μl culture medium were plated in 96-well plates followed 12 hours later by addition of TBB at the final concentration of 0, 50, 100 and 200 μM. At the indicated time points (24 h, 48 h), the medium was aspirated and each well was added with 100 μl serum-free DMEM and 10 μl tetrazolium compound MTT (Sigma) and incubated at 37°C for 4 h. Then the supernatant was discarded, 100 μl DMSO (Sigma) was applied to each well, and the plate was softly shaken for 10 min. Absorbance was measured at 450 nm with a reference wavelength of 630 nm on a spectrophotometer (Molecular Devices, Sunnyvale, CA). Cell viability was assessed as percent cell viability in terms of untreated control cells, which were determined for each concentration by use of the following equation: %viability = ODexperiment/ODcontrol×100%. Negative control cells were considered as 100% viable. All experiments were repeated in five times.
Cell cycle and apoptosis analysis by flow cytometry
After treatment of MCF-7 cells with TBB for 24 h, cells were harvested and immediately fixed in 75% ethanol at 4°C overnight, treated with 50 mg/L RNAse A (Sigma, St. Louis, MO, USA) for 30 min at 37°C, and stained with 50 mg/L PI (Sigma) for 10 min. Samples were then analyzed for their DNA content by a FACSAria Cell Cytometer (BD Biosciences, San Jose, CA, USA). Apoptosis analysis was performed by using a Annexin V-FITC KIT (Bender, Burlingame, CA, USA) according to the manufacturer’s instructions. The percentage of cells that were annexin V positive but PI negative was compared among the different treatment groups. Each experiment was performed in triplicate. The data were analyzed with CellQuest software (BD Biosciences).
CK2 activity detection
MCF-7 cells were with TBB at the concentration of 0, 50, 100 and 200 μM for 24 h, respectively. CK2 activity was determined using a CK2 kinase assay kit (MBL, Woburn, MA) according to the manufacturer’s instructions. Each experiment was performed in triplicate.
microRNA expression analysis
MCF-7 cells were treated with the DMSO or 100 μM for 24 h and total RNA was harvested using TRIzol (Invitrogen) and miRNeasy mini kit (QIAGEN) according to the manufacturer’s instructions. Total RNA (10 μg) was size fractionated (< 200 nucleotides) by using a mirVana kit (Ambion Inc., Austin, TX) and labeled using the miRCURY™ Hy3™/Hy5™ Power labeling kit and hybridized on the miRCURY™ LNA Array (v.16.0). The slides were scanned using the Axon GenePix 4000B microarray scanner. Scanned images were then imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. Data adjustments included data filtering, log2 transformation, and gene centering and normalization. The t test analysis was conducted between TBB-treated MCF-7 cells and control, and miRNA with P values < 0.05 were selected for cluster analysis. The clustering analysis was done using a hierarchical method and average linkage and Euclidean distance metrics [11].
Quantitative real-time PCR analysis for miRNA expression
MCF-7 cells were treated with the DMSO or 100 μM for 24 h and miRNA-enriched total RNA was extracted using the mirVana miRNA isolation kit (Ambion Inc., Austin, TX). Quantification of miRNAs was performed using TaqMan MicroRNA Assays (Applied Biosystems, Foster City, CA) following the manufacturer’s instructions. Reactions contained mirVana qRT-PCR Primer Sets (Ambion) specific for human miR-21, miR-29b-2, miR-27a/b, miR-125a/b, miR-145 and miR-205. U6 RNA was used for normalization of miRNA expression. Analysis was carried out using the comparative threshold cycle (Ct) method. The results are presented as fold change of each miRNA in the TBB-treated MCF-7 cells relative to DMSO-treated (control).
Statistical analysis
GraphPad Prism version 5.03 (GraphPad, San Diego, CA, USA) was used for all statistical analyses. All data are expressed as mean ± SEM. Differences between groups were analyzed by a 2-tailed Student’s paired t-test for single comparisons and by one-way ANOVA with LSD post-hoc test for multiple comparisons. Bonferroni’s correction was used to adjust for multiple comparisons. A P value < 0.05 was considered to be statistically significant.
Results
TBB suppresses the growth of MCF-7 cells
To examine whether inhibition of CK2 would affect the viability of MCF-7 cells, cells were treated with varying concentrations of the CK2 inhibitor TBB for 24, 48 h and the cell viability was evaluated by MTT assay. As shown in Figure 1, increasing TBB concentration and treatment time resulted in a progressive inhibition of MCF-7 viability. The first significant reduction was observed at the concentration of 100 μM after incubating with TBB for 24 h, with an inhibition of 54.65% (P < 0.05). These results demonstrated that TBB suppressed growth of MCF-7 in a dose-dependent manner, suggesting TBB had a potent inhibitory effect on the growth of MCF-7 cells. The treatment time point of 48 h was therefore selected for further studies.
Figure 1.
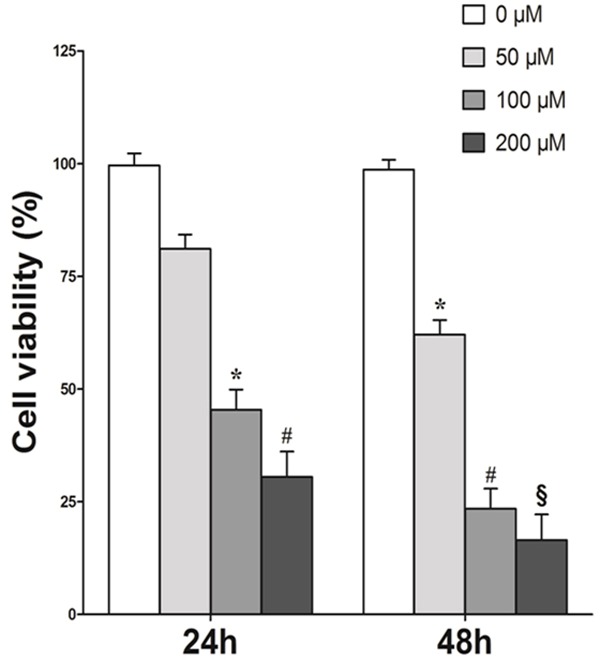
Growth inhibition of MCF-7 cells by the CK2 inhibitor TBB. MCF-7 cells (5000/well) were plated into 96-well plates. After incubation overnight, cells were treated with TBB with various concentrations for 24 and 48 hours. Cell viability was determined by MTT assay. Results are expressed as means ± SEM of five samples. *P < 0.05; #P < 0.01; §P < 0.001 versus control (0 μM).
TBB affects cell cycle and apoptosis of MCF-7 cells
We performed PI staining and flow cytometry to define the cell-cycle distribution of TBB-treated MCF-7 cells (Figure 2A-1). Treatment of MCF-7 cells with 0-200 μM of TBB for 24 h resulted in arrest in the G0/G1 phase and shortening of the S phase in a dose-dependent manner (Figure 2A-2). Apoptosis rate was determined by double staining of Annexin-V FITC and PI using flow cytometry assay. Apoptotic cells were determined as those cells that were annexin V positive, but PI negative (Figure 2B-1). The percentage of apoptotic cells increased with the elevated concentration of TBB as compared with the negative control after TBB treatment of MCF-7 cells for 24 hours (Figure 2B-2). The effects initially became significant at TBB concentrations of 100 μM or higher. These data were consistent with the previous results that 100 and 200 μM TBB treatment for 24 hours significantly inhibited the proliferation of MCF-7 cells. Taken together, these results suggest that decreased DNA synthesis (S phase) and increased apoptosis induced by TBB contributed to the impaired viability of MCF-7 cells treated with TBB.
Figure 2.
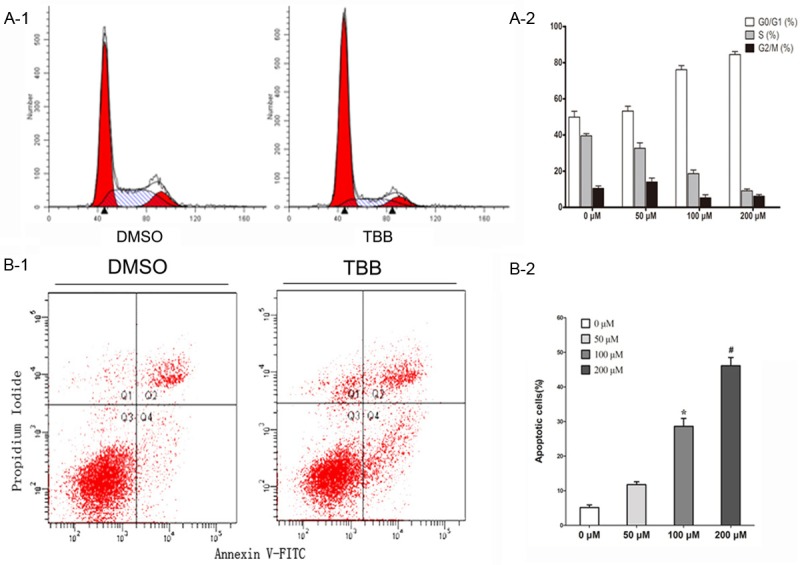
TBB affects apoptosis and the cell cycle of MCF-7 cells. MCF-7 cells were treated with TBB at various concentrations for 24 hours. A-1. Cell cycle distribution of MCF-7 cells was examined by PI staining and flow cytometry synchronized by serum starvation. A-2. Treatment of MCF-7 cells with 0-200 μM of TBB for 24 h resulted in arrest in the G0/G1 phase and shortening of the S phase in a dose-dependent manner. B-1. Each was performed in triplicate. P < 0.05 and #P < 0.01 versus control (0 μM TBB). Apoptosis of MCF-7 cells treated with TBB for 24 hours was detected by annexin V and PI staining using FACS analysis. B-2. The percentage of cells that were annexin V positive but PI negative was compared among different groups. Results visualized as a representative experiment (above) or means ± SEM of 3 experiments (below). *P < 0.05 and #P < 0.01 versus control (0 μM TBB).
Confirmation of the inhibition of CK2 activity by TBB in MCF-7 cells
MCF-7 cells treated with various concentrations of TBB for 24 h were assessed for the CK2 activity.
As shown in Figure 3, TBB treatment greatly reduced the CK2 activity in a dose-dependent manner. A significant reduction was observed at 100 and 200 μM of TBB treatment. These data were consistent with the previous results that treatment with 100 and 200 μM TBB for 24 h significantly inhibited the growth and promoted the apoptosis of MCF-7 cells.
Figure 3.
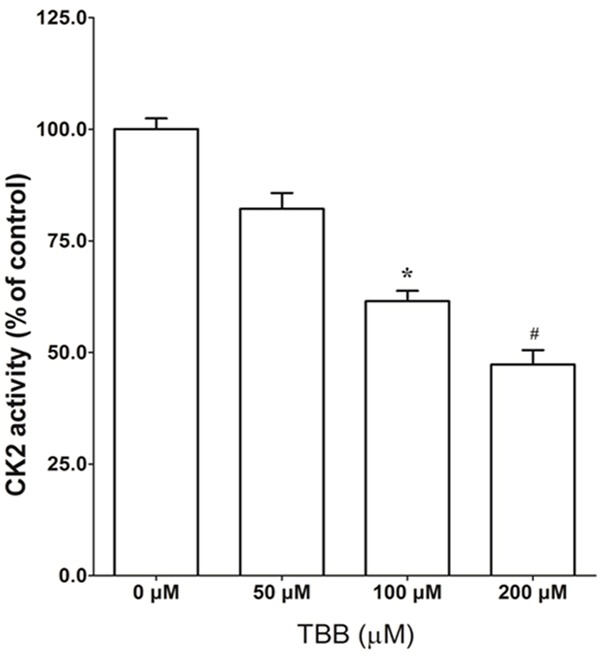
Effect of TBB on CK2 activity in MCF-7 cells. MCF-7 cells were treated with TBB at the indicated concentration for 24 hours and cell lysates (5 μg) were used for measuring CK2 activity as described in Materials and methods. The data (mean ± SEM) are expressed as percent CK2 activity compared with DMSO (0 μM) treated control cells. *P < 0.05 and #P < 0.01 versus control.
TBB regulates the expression profile of miRNAs in MCF-7 cells
To test whether TBB affected the miRNA expression of MCF-7 cells, miRNA microarray was performed to analyze the miRNA expression profiles of MCF-7 cells 24 h after treatment with either TBB (100 μM) or vehicle control DMSO. We identified 27 miRNA genes (17 up-regulated and 10 down-regulated) that were differentially expressed (P < 0.05) in the MCF-7 cells treated with TBB compared with vehicle control (Table 1). Cluster analysis based on differentially expressed miRNA, generated a tree with clear distinction between TBB and control groups (Figure 4A). The results obtained by miRNA microarray analysis were independently confirmed by the qR-TPCR. We analyzed the status of differentially expressed miR-21, miR-29b-2, miR-27a/b, miR-125a/b, miR-145 and miR-205 genes in MCF-7 cells treated with TBB or vehicle control. The qRT-PCR confirmed the data obtained by microarray analysis (Figure 4B). These results suggest that inhibition of CK2 by TBB alters the expressions of these microRNAs, which may be related with the suppressed growth of MCF-7 cells.
Table 1.
The effect of TBB on miRNA expression in MCF-7 cells
| MicroRNA | Fold change | P-value | |
|---|---|---|---|
| Upregulated | hsa-miR-24 | 3.31 | 0.030 |
| hsa-miR-181a | 5.60 | 0.031 | |
| hsa-miR-101 | 4.82 | 0.002 | |
| hsa-miR-3607-3p | 11.86 | 0.038 | |
| hsa-miR-320a | 3.74 | 0.002 | |
| hsa-miR-15a | 10.57 | 0.027 | |
| hsa-miR-205 | 4.47 | 0.013 | |
| hsa-miR-331-3p | 4.52 | 0.004 | |
| hsa-miR-145 | 4.89 | 0.016 | |
| hsa-miR-125b | 9.99 | 0.009 | |
| hsa-miR-125a | 8.93 | 0.002 | |
| hsa-miR-3607-5p | 10.63 | 0.048 | |
| hsa-miR-93 | 8.81 | 0.046 | |
| hsa-miR-652 | 4.75 | 0.038 | |
| hsa-miR-191 | 26.94 | 0.047 | |
| hsa-miR-23b | 3.70 | 0.003 | |
| hsa-miR-27-b | 2.21 | 0.048 | |
| Downregulated | hsa-miR-634 | 0.22 | 0.002 |
| hsa-miR-29b-2 | 0.12 | 0.019 | |
| hsa-miR-21 | 0.20 | 0.012 | |
| hsa-miR-593 | 0.35 | 0.027 | |
| hsa-miR-302c | 0.08 | 0.016 | |
| hsa-miR-625 | 0.33 | 0.043 | |
| hsa-miR-27-a | 0.33 | 0.021 | |
| hsv1-miR-H14-3p | 0.16 | 0.001 | |
| ebv-miR-BART8 | 0.30 | 0.019 | |
| hsa-miR-3195 | 0.18 | 0.016 |
MCF-7 cells were treated with 100 μM TBB or DMSO for 24 h and microRNA expression array analysis was performed. Only those microRNAs whose expression differences with a P-value of less than 0.05 among TBB-treated and DMSO treated cells were presented.
Figure 4.
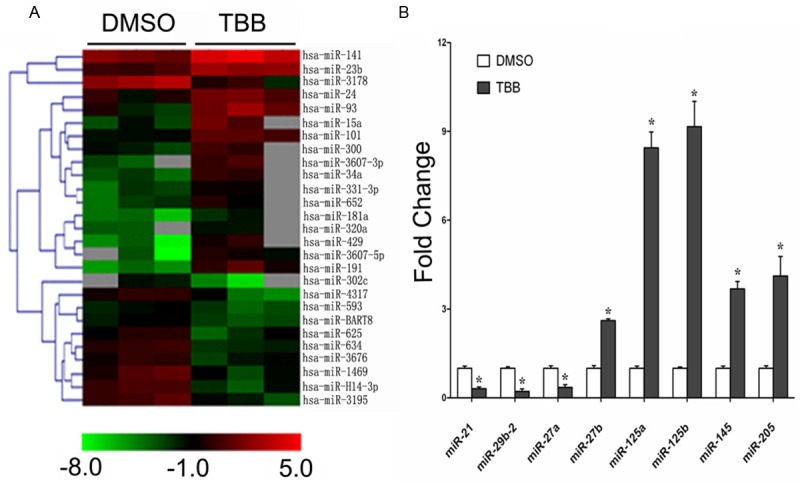
MicroRNA dysregulation in the MCF-7 human breast adenocarcinoma cells treated with 100 μM TBB for 24 h. A. Hierarchical clustering of the differentially expressed miRNA genes (as determined by ANOVA) in the MCF-7 cells after being treated with 100 μM TBB or DMSO for 24 h. Rows, miRNA; columns, independent biological replicates. Red indicates high expression and green indicates low expression. Each miRNA listed is significantly differentially expressed (P < 0.05) between TBB and control (DMSO). B. qRT-PCR analysis of the differentially expressed miRNA in the TBB and DMSO MCF-7 cells. Values are fold increase compared to DMSO for each miRNA. Values are the average of three means ± SEM. *P < 0.01, TBB versus control.
Discussion
Casein kinase 2 (CK2) is a highly conserved and ubiquitous protein Ser/Thr kinase localized in the nuclear and cytoplasmic compartments in the eukaryotic cell [2]. Over-expression of CK2 has been documented in various cancers including breast cancer [12,13]. Recently a very selective cell-permeant inhibitor of CK2, 4,5,6,7-tetrabromobenzotriazole (TBB) has been available for investigating the biological functions of this kinase [14]. Downregulation of CK2 activity by means of chemical inhibition, as well as antisense oligodeoxynucleotide, RNAi, or overexpression of exogenous kinase-inactive CK2 can induce apoptosis in cancer cells [15,16]. In this work, we have exploited TBB to gain information about the role of CK2 in MCF-7 cells. We found that inhibition of CK2 activity in MCF-7 cells significantly impaired cell proliferation, inhibited cell cycle progression and promoted apoptosis. These data suggest that CK2 is required for the survival of MCF-7 cells.
MicroRNAs (miRNAs) are an evolutionarily conserved class of pleiotropically acting small RNAs that regulates gene expression post-transcriptionally via sequence-specific interactions with the 3’ untranslated regions (UTRs) of cognate mRNA targets [17]. Deregulation of microRNA expression has been observed in various tumor types, including breast cancer [18]. miRNAs can function as tumor suppressors or oncogenes, depending on whether they specifically target oncogenes or tumor suppressor genes [19]. In this regard, tumor suppressive miRNAs are usually underexpressed in tumors. For instance, miR-125a/b, miR-145 and miR-205 are downregulated in breast cancer. In contrast, oncogenic miRNAs, such as mir-21, mir-29b-2, mir-27a/b, are overexpressed in breast tumors [9]. In this study, we found that CK2 inhibition by the inhibitor TBB significantly altered microRNA expression profile of MCF-7 breast cancer cells: the expression of oncogenic miRNAs, such as mir-21, mir-29b-2, mir-27a were suppressed, while suppressive miRNAs mir-27b, miR-125a/b, miR-145 and miR-205 were overexpressed. These data demonstrate that dysregulation of miRNA expression is associated with inhibited growth induced by CK2 inhibition in MCF-7 cells. Specifically, miR-21 was reported to be consistent and significant increased expression in breast cancer cell lines and human tissue when compared with normal cells and tissues [20]. The tumor-suppressor gene programmed cell death (PDCD)4 was target for miR-21, which downregulates PDCD4 at the mRNA and protein level [21]. In the present study, inhibition of CK2 leads to the decreased expression of miR-21, resulting in enhancing expression of PDCD4, which thus caused the increased apoptosis of MCF-7 cells by CK2 inhibition. In addition, miR-27a is upregulated in breast cancer, and target ZBTB10, which represses the specificity protein 1 (SP1) transcription factor that plays a role in the G0–G1 to S phase progression in breast cancer cells. Our data suggest that CK2 inhibition caused the suppressed cell cycle progression of MCF-7 cells is related with the decreased expression of miR-27a.
Although biological functions of microRNAs are being studied extensively, studies about mechanism controlling their expression are rarely explored. Our data suggest that CK2 plays an important role in regulation of microRNAs expression in breast cancer cell line MCF-7 cells. Considering CK2 can phosphorylates a great number of substrates such as transcriptional factors, the possible mechanisms of CK2-regulated expression of microRNAs including CK2-inducible expression of mRNA-encoding genes that harbor microRNA genes in their intronic regions, CK2 regulates the expression of transcription factors that control the expression of microRNAs. For instance, it is reported that p53 can bind to distinct microRNA promoter elements and modulate microRNA gene expression [22], and p53 was one of phosphorylated substrates of CK2 [23], thus it is fully possible that CK2 would affect microRNAs expression via phosphorylating p53. However, further studies need to be carried out to confirm this assumption.
In summary, our data demonstrate that CK2 inhibition would suppress the growth and enhance the apoptosis of breast cancer cells by altering the miRNA expression, providing a substantial basis for exploring the role of CK2 in maintaining microRNAs properties as well as deepening our understanding the biological functions of CK2. Moreover, further study of the functions of these CK2-associated microRNAs will clarify the exact mechanisms.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (No. 82172240) and Shanghai Science Committee Foundation (STCSM No. 10411964700).
Disclosure of conflict of interest
No conflicts of interest are involved in this manuscript.
Abbreviations
- CK2
Casein Kinase 2
- TBB
4,5,6,7-Tetrabromobenzotriazole
- miRNA
microRNA.
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Hanif IM, Shazib MA, Ahmad KA, Pervaiz S. Casein Kinase II: an attractive target for anti-cancer drug design. Int J Biochem Cell Biol. 2010;42:1602–5. doi: 10.1016/j.biocel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 3.Pinna LA. Protein kinase CK2: a challenge to canons. J Cell Sci. 2002;115:3873–8. doi: 10.1242/jcs.00074. [DOI] [PubMed] [Google Scholar]
- 4.Munstermann U, Fritz G, Seitz G, Lu YP, Schneider HR, Issinger OG. Casein kinase II is elevated in solid human tumours and rapidly proliferating non-neoplastic tissue. Eur J Biochem. 1990;189:251–7. doi: 10.1111/j.1432-1033.1990.tb15484.x. [DOI] [PubMed] [Google Scholar]
- 5.Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–61. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 7.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–7. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 9.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 10.Lowery AJ, Miller N, McNeill RE, Kerin MJ. MicroRNAs as prognostic indicators and therapeutic targets: potential effect on breast cancer management. Clin Cancer Res. 2008;14:360–5. doi: 10.1158/1078-0432.CCR-07-0992. [DOI] [PubMed] [Google Scholar]
- 11.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tawfic S, Yu S, Wang H, Faust R, Davis A, Ahmed K. Protein kinase CK2 signal in neoplasia. Histol Histopathol. 2001;16:573–82. doi: 10.14670/HH-16.573. [DOI] [PubMed] [Google Scholar]
- 13.Landesman-Bollag E, Romieu-Mourez R, Song DH, Sonenshein GE, Cardiff RD, Seldin DC. Protein kinase CK2 in mammary gland tumorigenesis. Oncogene. 2001;20:3247–57. doi: 10.1038/sj.onc.1204411. [DOI] [PubMed] [Google Scholar]
- 14.Sarno S, Reddy H, Meggio F, Ruzzene M, Davies SP, Donella-Deana A, Shugar D, Pinna LA. Selectivity of 4,5,6,7-tetrabromobenzotriazole, an ATP site-directed inhibitor of protein kinase CK2 (‘casein kinase-2’) FEBS Lett. 2001;496:44–8. doi: 10.1016/s0014-5793(01)02404-8. [DOI] [PubMed] [Google Scholar]
- 15.Di Maira G, Brustolon F, Bertacchini J, Tosoni K, Marmiroli S, Pinna LA, Ruzzene M. Pharmacological inhibition of protein kinase CK2 reverts the multidrug resistance phenotype of a CEM cell line characterized by high CK2 level. Oncogene. 2007;26:6915–26. doi: 10.1038/sj.onc.1210495. [DOI] [PubMed] [Google Scholar]
- 16.Pagano MA, Andrzejewska M, Ruzzene M, Sarno S, Cesaro L, Bain J, Elliott M, Meggio F, Kazimierczuk Z, Pinna LA. Optimization of protein kinase CK2 inhibitors derived from 4,5,6,7-tetrabromobenzimidazole. J Med Chem. 2004;47:6239–47. doi: 10.1021/jm049854a. [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 19.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–91. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–33. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 22.Breving K, Esquela-Kerscher A. The complexities of microRNA regulation: mirandering around the rules. Int J Biochem Cell Biol. 2010;42:1316–29. doi: 10.1016/j.biocel.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 23.Meek DW, Simon S, Kikkawa U, Eckhart W. The p53 tumour suppressor protein is phosphorylated at serine 389 by casein kinase II. EMBO J. 1990;9:3253–60. doi: 10.1002/j.1460-2075.1990.tb07524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


