Abstract
Microtubule-associated protein light chain 3 (LC3) is a key mediator bridging autophagy, apoptosis and differentiation. However, its role and clinical significance in resectable esophageal squamous cell carcinoma (ESCC) is still scanty. The purpose of this study was to investigate the clinical significance of LC3 by immunohistochemistry in a group of patients with ESCC treated with surgical resection. Tissue microarray that included 253 surgically resected ESCC specimens was successfully generated for immunohistochemical evaluation. The clinical/prognostic significance of LC3 expression was analyzed statistically. The association of LC3 expression with the ESCC survival rate was assessed by Kaplan-Meier and Cox proportional-hazards regression. The results showed that the immunostaining of LC3 was distributed in cytoplasm and plasma-membrane. Significantly high LC3 expression was found in ESCC cells compared with that of normal esophageal epithelial cells. Patients with low expression of LC3 demonstrated higher overall survival compared with those with high expression of LC3 (mean of 71.1 months versus 55.5 months, P = 0.022). A similar result was observed for disease-free survival (mean of 68.7 months versus 51.8 months, P = 0.021). In subgroup analysis, LC3 expression could stratify pN0 patients with ESCC. Multivariate analysis showed that the level of LC3 expression was an independent prognostic factor in ESCC (RR = 1.407, P = 0.049). This paper shows high level of LC3 suggests poor prognosis for resectable ESCC patients.
Keywords: Esophageal cancer, immunohistochemistry, LC3, surgery
Introduction
Esophageal squamous cell carcinoma (ESCC) is one of the most aggressive malignancies of the digestive tract [1,2]. Surgical resection is still as the mainstay strategy employed for operable ESCC. Despite the great advances has been achieved in multimodal therapy, its five-year survival rate remains unsatisfactory [3-5]. Discovering suitable biomarkers will probably be a key to monitoring cancer recurrence or screening high risk population of ESCC, giving information on the need for adjuvant or neoadjuvant therapy.
LC3, microtubule-associated protein light chain 3, was originally identified as a protein that co-purifies with large microtubule associated with MAP1A and MAP1B from rat brain [6]. LC3 is an autophagosomal orthologue of yeastAtg8, with approximately 30% amino acid homology with Atg8 [7,8]. It exists in two forms, LC3-I and LC3-II (molecular weight, 18 and 16kD, respectively), localized in the cytoplasm. LC3 is now widely used as a specific molecular marker to monitor autophagosome formation. Up-regulation of LC3 expression was observed in the presence of various stresses such as genomic injury, hypoxia, viral/bacterial infection, starvation [9].
Currently, numerous studies have been investigated on the role of autophagy in cancer development and cancer treatment. Accumulating data provide evidence that autophagy is involved in tumor suppressor pathways. Beclin-1, an essential mediator of autophagy, was confirmed as a tumor suppressor in heterozygous mouse models [10,11], and intensive expression of Beclin-1 was found in breast, colorectal, and gastric cancers [12-14]. Furthermore, higher expression of Beclin-1 has shown favorable survival than the lower ones in ESCC patients [15]. However, other studies supported the idea that autophagy enhances tumor progression and protects cancer cells from anticancer therapies. As tumors grow, autophagy may contribute to cancer cells survival under nutrient deprivation and hypoxia conditions [16-19]. Knockdown of autophagy, in combination with tamoxifen or 4-hydroxy-tamoxifen (4-OH-T), resulted in decreased cell viability of estrogen receptor-positive breast cancer cells [20,21]; Inhibition of autophagy along with irradiation lead to enhanced cytotoxicity of radiotherapy in resistant cancer cells [22]. Thus, the role of autophagy in tumor suppression and development is still controversial.
LC3, a specific marker of autophagy, has been examined in gastrointestinal cancers [23], but the evidence related to survival is still scanty in ESCC. Therefore, we performed this study to evaluate and explore the possible relation between the LC3 expression and prognosis in a large cohort of ESCC (253 cases) by immunohistochemistry.
Materials and methods
Patient selection
This study was approved by the medical ethics committee of Sun Yat-Sen University Cancer Center. Two hundred and sixty-five primary ESCC patients who underwent surgery at the Department of Thoracic surgery, Cancer Center, Sun Yat-Sen University, between October 2000 and April 2007 were eligible for enrollment in the study. The histologic grade and clinical stage of the tumors were defined according to the 7th edition of the TNM classification of the International Union Against Cancer [24]. The cases selected in this study fulfilled the following criteria: (a) newly diagnosed cancer of the esophagus without previous treatment; (b) histologically confirmed primary thoracic ESCC; (c) no distant metastases, including supraclavicular or celiac lymph nodes metastases; (d) underwent a complete surgical resection (R0) at our cancer center; (e) adequate clinical information and follow-up data were available. Patients with a non-curative resection (R1) or died from postoperative complications were excluded from the study. Patients with neoadjuvant or adjuvant therapy were also excluded. In our cancer center, patients with ESCC that has invaded the airway or major vessels (such as the thoracic aorta) or accompanied by visceral metastasis are not indicated for surgery. The tumor specimens and paracancerous samples were obtained as paraffin blocks from the Bank of Tumor Source at our cancer center. Clinical data were obtained from hospital records after surgery. All the patients were followed up in May 2010 to determine their current status.
Tissue microarray construction
Tumor tissue samples from 265 ESCC cases were collected, fixed in formalin, and embedded in paraffin. H&E-stained sections from a single random block from each patient were reviewed by a senior pathologist (R-Z Luo) to define representative tumor regions. Two targeted core samples of each specimen were obtained using a tissue array instrument (ALPHELYS Minicore instruments, France). Briefly, tissue cylinders with a diameter of 10 mm were punched and arrayed on a recipient paraffin block. Sections (5 μm) of the tissue array (recipient) block were cut and placed on class slides. After the exclusion of cores with inadequate tissue following sectioning and tissuetransfer, the final immunohistochemical analyses included cores from 253 ESCC cases. Each of the 253 different ESCC cases contributed to the biomarker analyses. Among the 253 cases, formalin-fixed paracancerous normalesophageal tissues were available for 56 cases, which served as controls. The microarray for the normal esophageal tissues was constructed according to the same method described above.
Immunohistochemistry (IHC) staining and assessment
IHC staining was performed using TMA sections that were rehydrated via a graded alcohol series. Endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide for 15 minutes. For antigen retrieval, the TMA slides were boiled in tris(hydroxymethyl) aminomethane-EDTA buffer (pH 8.0) in a pressure cooker for 20 min. Nonspecific binding was blocked with 10% normal goat serum for 20 min. The TMA slides were incubated with rabbit anti-LC3 antibody (NB100-2220, 1:400 dilution, Novus) for 12 hours at 4°C in a moist chamber. Subsequently, the slides were sequentially incubated with biotinylated rabbit anti-mouse immunoglobulin antibody at a concentration of 1:100 for 30 min at 37°C and then with a streptavidin-peroxidase conjugate for 30 min at 37°C and 3’-3’diaminobenzidine as the chromogen substrate. The nucleus was counterstained using Meyer’s hematoxylin. The negativecontrol was obtained by replacing the primary antibody with normal rabbit IgG. Positive expression of LC3 in ESCC and normal esophageal mucosa cells exhibited a primarily cytoplasm pattern (Figure 1). Internal positive and negative controls, including normal squamous mucosa of the esophagus from non-cancer patients, were utilized as available to further support the staining patterns. Two independent observers (R-Z Luo and M Li) blinded to the clinicopathological information performed the immunoreactivity score (IRS) for LC3 expression. The staining results were scored based on the following criteria: (a) percentage of positive tumor cells in the tumor tissue: zero (0%), 1 (1%-25%), 2 (26%-50%), 3 (51%-75%), and 4 (76%-100%); (b) signal intensity: zero (no signal), 1 (weak), 2 (moderate), 3 (marked). IRS was calculated by multiplying the score for the percentage of positive cells by the intensity score (range of 0 to 12). The average IRS for each case was assigned as the staining result for the patient. The specimens were rescored if the difference between the scores determined by the two pathologists was greater than 3 [25]. The median IRS was defined as the cut-off value; IRS greater than this value was considered high and, otherwise, low.
Figure 1.
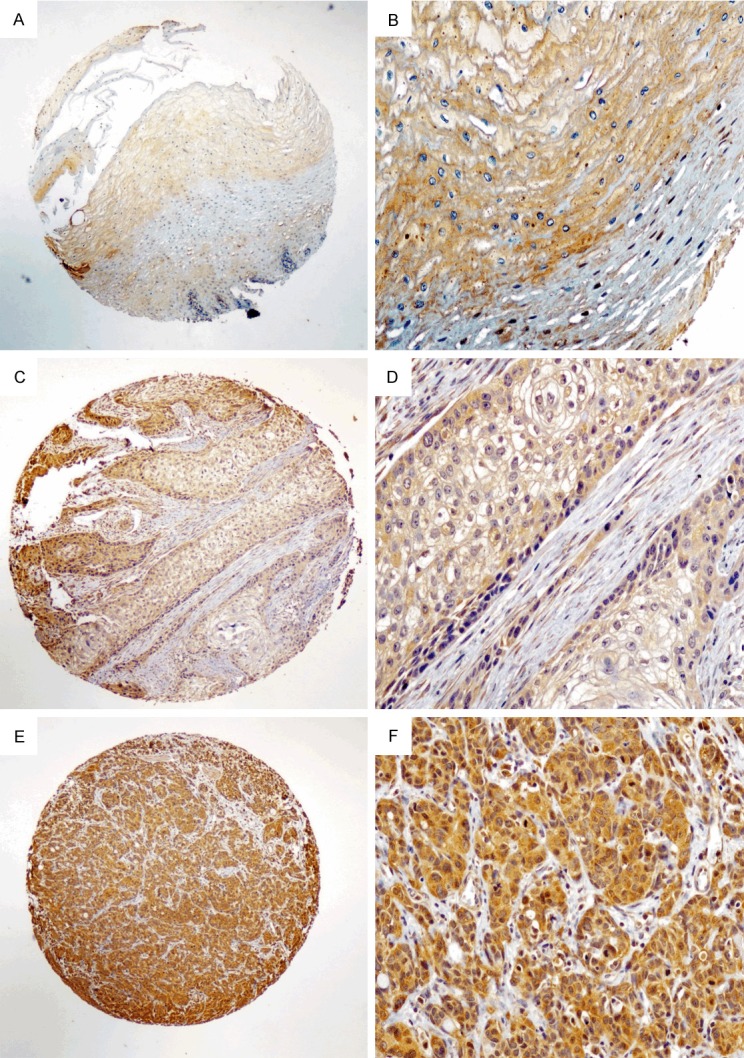
LC3 expression by immunohistochemical staining. (A, B) Normal esophageal mucosa demonstrated low expression of LC3 protein in the cytoplasm of all esophageal squamous cells (magnification: A, ×40, B, ×200). (C, D) An ESCC case demonstrating a low expression level of p300 (magnification: C, ×40, D, ×200). (E, F) High expression level of LC3 detected in ESCC (magnification: E, ×40, F, ×200).
Statistical analysis
Statistical analysis was performed using SPSS software (standard version 16.0, SPSS, Chicago, IL, USA). The correlation between LC3 expression and clinicopathological features was assessed using Pearson’s χ2 test. Disease-free survival (DFS) was defined as the time from surgery to regional relapse or distant metastasis. Overall survival (OS) was defined as the time from surgery to death. DFS and OS were assessed using the Kaplan-Meier method and compared by the log-rank test. Multivariate survival analysis was performed for all of the parameters that were significant in the univariate analysis using the Cox regression model. A two-sided probability value of less than 0.05 was considered statistically significant.
Results
Patient characteristics
Seventy females and one-hundred and eighty-three males, aged from 32 to 80 years (median 58.0 years), were included in the study. The clinicopathological characteristics of the 253 patients are listed in Table 1.
Table 1.
Relationship between LC3 expression and clinicopathological variables
| Variables | Case number | LC3 Expression (%) | ||
|---|---|---|---|---|
|
| ||||
| Low | High | P-valued | ||
| Age (years) | ||||
| ≤60 | 153 | 79 (51.6) | 74 (48.4) | 0.955 |
| >60 | 100 | 52 (52.0) | 48 (48.0) | |
| Gender | ||||
| Male | 183 | 97 (53.0) | 86 (47.0) | 0.528 |
| Female | 70 | 34 (48.6) | 36 (51.4) | |
| Surgery | ||||
| Standard | 155 | 82 (52.9) | 73 (47.1) | 0.653 |
| Three-incision | 98 | 49 (50.0) | 49 (50.0) | |
| Tumor location | ||||
| Upper | 13 | 6 (46.2) | 7 (53.8) | 0.902 |
| Middle | 174 | 90 (51.7) | 84 (48.3) | |
| Lower | 66 | 35 (53.0) | 31 (47.0) | |
| Histologic grade | ||||
| G1 | 58 | 32 (55.2) | 26 (44.8) | 0.155 |
| G2 | 163 | 78 (47.9) | 85 (52.1) | |
| G3 | 32 | 21 (65.6) | 11 (34.4) | |
| pT category | ||||
| 1 | 5 | 1 (20.0) | 4 (80.0) | 0.238 |
| 2 | 60 | 35 (58.3) | 25 (41.7) | |
| 3 | 185 | 93 (33.1) | 92 (66.9) | |
| 4 | 3 | 1 (33.3) | 2 (67.7) | |
| pN category | ||||
| 0 | 135 | 70 (51.9) | 65 (48.1) | 0.980 |
| 1/2/3 | 118 | 61 (51.7) | 57(48.3) | |
| pTNM stage | ||||
| I | 9 | 7 (77.8) | 2 (22.2) | 0.195 |
| II | 145 | 77 (53.1) | 68 (46.9) | |
| III | 99 | 47 (47.5) | 52 (52.5) | |
Pearson’s χ2 test;
standard: left thoracotomy, three-incision: thoracic-abdominal-cervical anastomosis; Fisher’s χ2 test.
Expression of LC3 in ESCC
In the present study, LC3 staining of ESCC tissue and normal esophageal mucosa revealed immunoreactivity primarily in cytoplasm within tumor cells. The protein expression of LC3 was examined by IHC in 253 cases of primary ESCC and in 56 cases of paracancerous esophageal mucosa. Using the criteria described above (median IRS of 8.0), high expression of LC3 was observed in 62.5% (122/253) of the ESCC. There was no significant correlation of LC3 expression with clinicopathological parameters, such as age, sex, tumor location, histologic grade, T status, N status and pathological stage.
LC3 expression and survival
Among the 253 ESCC patients, no patients were lost to follow-up. The median observation period was 63.0 months (4-115 months), and 135 patients were deceased and 118 were alive at the end of the follow-up. The 5-year DFS and OS for the entire cohort were 47.5% and 50.3%, with median survival times of 52.0 and 63.0 months, respectively.
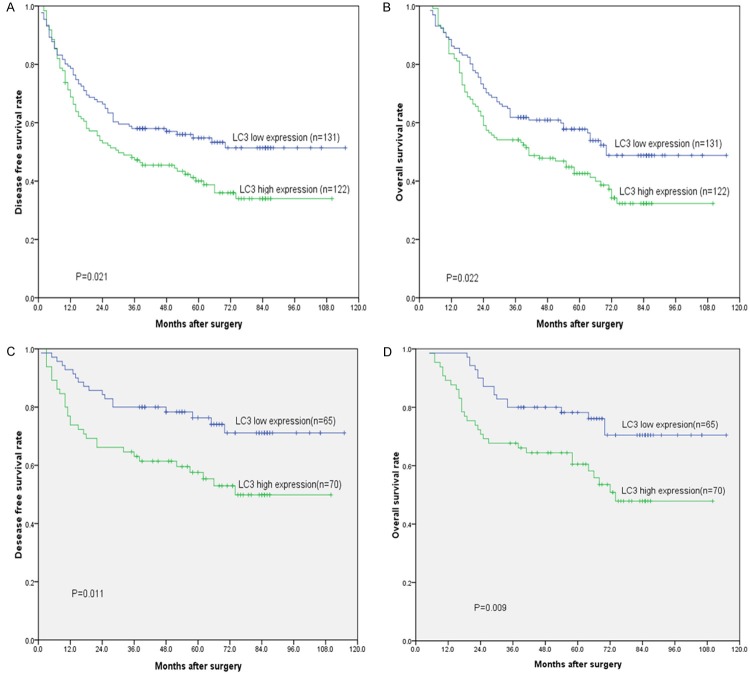
Patients with low expression of LC3 demonstrated longer OS compared with those with high expression of LC3 (mean of 71.1 months versus 55.5 months, P = 0.022, Figure 2, Table 3). A similar result was obtained for DFS (mean of 68.7 months versus 51.8 months, P = 0.021, Figure 2, Table 2). In the subgroup analysis, LC3 expression distinguished the DFS/OS well for pathological N0 patients (Table 2, P = 0.011/0.009), but not for pathological N1-3 patients (Table 2, P = 0.515/0.597).
Figure 2.
Disease-free survival (DFS) and overall survival (OS) curves for esophageal squamous cell carcinoma patients according to LC3 expression status. A, B: DFS and OS curves: patients with low and high expression levels of LC3. C, D: DFS and OS curves: patients with low and high expression levels of LC3 at stage N0.
Table 3.
Results of the univariate and multivariate survival analyses for OS according to the Cox regression model
| RR | 95% CI | P-value | |
|---|---|---|---|
| Univariate survival analysis | |||
| Age (≤60 vs. >60) | 1.142 | 0.812-1.606 | 0.444 |
| Gender (male vs. female) | 1.262 | 0.858-1.855 | 0.237 |
| Tumor locationg | 0.909 | 0.649-1.273 | 0.579 |
| Surgery (left thoracotomy vs. three incision) | 0.142 | 0.812-1.606 | 0.444 |
| Histologic gradeh | 1.297 | 0.971-1.731 | 0.078 |
| T categoryi | 1.470 | 1.010-2.140 | 0.044 |
| N category (0 vs. 1/2/3) | 3.465 | 2.414-4.972 | <0.001 |
| LC3 expression (high vs. low) | 1.479 | 1.053-2.077 | 0.024 |
| Multivariate survival analysis | |||
| T category | 1.389 | 0.963-2.005 | 0.079 |
| N category (1/2/3 vs. 0) | 3.388 | 2.358-4.868 | <0.001 |
| LC3 expression (high vs. low) | 1.407 | 1.001-1.977 | 0.049 |
Tumor location: upper vs. middle vs. lower;
Histologic grade: G1 vs. G2 vs. G3;
T category: T1 vs. T2 vs. T3 vs. T4;
RR: relative risk; CI: confidence interval.
Table 2.
Kaplan-Meier survival analysis (log-rank test) according to LC3 expression in ESCC patients
| Variable | Case number | DFS (months) | OS (months) | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Mean | Median | P-value | Mean | Median | P-value | ||
| Total | |||||||
| Low expression | 131 | 68.7 | NR | 0.021 | 71.1 | 70.0 | 0.022 |
| High expression | 122 | 51.8 | 30.0 | 55.5 | 40.0 | ||
| pT1-2 | |||||||
| Low expression | 37 | 68.2 | NR | 0.194 | 70.9 | NR | 0.243 |
| High expression | 28 | 57.3 | 57.0 | 62.1 | 66.0 | ||
| pT3-4 | |||||||
| Low expression | 94 | 66.3 | NR | 0.074 | 68.7 | 70.0 | 0.061 |
| High expression | 94 | 42.6 | 26.0 | 46.3 | 39.0 | ||
| pN0 | |||||||
| Low expression | 70 | 90.4 | NR | 0.011 | 92.1 | NR | 0.009 |
| High expression | 65 | 67.2 | 74.0 | 70.0 | 74.0 | ||
| pN1-3 | |||||||
| Low expression | 61 | 37.3 | 18.0 | 0.515 | 39.6 | 26.0 | 0.597 |
| High expression | 57 | 29.7 | 16.0 | 35.0 | 24.0 | ||
| Histologic grade | |||||||
| G1 | |||||||
| Low expression | 32 | 63.8 | 70.0 | 0.447 | 66.0 | 70.0 | 0.540 |
| High expression | 26 | 59.5 | 51.0 | 63.0 | 64.0 | ||
| G2-3 | |||||||
| Low expression | 99 | 68.1 | NR | 0.033 | 70.6 | 70.0 | 0.029 |
| High expression | 96 | 42.5 | 26.0 | 46.5 | 39.0 | ||
ESCC: esophageal squamous cell carcinoma; DFS: disease-free survival; OS: overall survival; NR: not reached.
Univariate analysis using Cox’s proportional hazard model showed that the following parameters correlated significantly with DFS and OS: T category, N category, and LC3 expression (Table 2). When the above parameters were included in multivariate analysis, the results suggested that T category, N category, and LC3 expression were independent factors that affected OS (Table 3).
Discussion
LC3 is an autophagasomal orthologue of yeast autophagy-related gene 8 (Atg8), introduction of autophagy by various stresses such as starvation, hypoxia, stimulates up-regulation of LC3 expression. In order to investigate the role of LC3 in ESCC, we evaluated LC3 expression in ESCC tissues using high throughput tissue microarray. Consistent with studies in several other tumor entities, including esophageal squamous cell carcinoma, gastric cancer and colorectal cancer [23], our results showed that a significant percentage of cells in the esophageal cancer mucosa demonstrated positive staining for LC3 compared with those in noncancerous esophageal mucosa. This may due to basal autophagy plays an important role in maintaining homeostasis in normal tissue [26,27].
Increasing evidence indicates that autophagy plays an important role in cancer development. LC3, as a specific molecular biomarker of autophagy, also has been involved in carcinogenesis [28,29]. In the present study, no significantcorrelation was observed between clinicopathological parameters and LC3 expression statistically. Nevertheless, high expression of LC3 in ESCC has shown shorter survival than the ones of low expression. Similar results were also reported in melanoma [29]. The lack of prognostic significance of LC3 was also reported in other surgical series of the patients with ESCC [23], this discrepancy is not surprising in light of studies with the difference of the sample enrolled.
Surgical resection can be considered as the standard treatment for patients with local ESCC. However, the problem how to identify the patients who could benefit from surgery is still unresovled. In the present study, elevated expression of LC3 was found to be an unfavorable prognostic factor in ESCC patients. High expression of LC3 was one of the most important predictors of poor DFS and OS in the multivariate analysis. This result was similar to the previous study reported on melanoma [29]. Therefore, we could conclude that LC3 is closely correlated with clinical outcome in human ESCC.
How LC3 promotes progression of ESCC is elusive. One possibility may that LC3 upregulation may represent an adaptive cellular mechanism directed to overcome uncontrolled proliferation and metabolic stress such as hypoxia and nutrient deprivation. Another possible mechanism is that the relatively poor blood supply in esophageal mucosa, increased expression of LC3 in cancer cells is more likely to sustain survival at this situation [30]. The third potential mechanism may relate to activation of positive regulator of apoptosis such as Bcl-2/ induced autophagy [31]. In future, identification the underlined mechanism would be helpful to designing ESCC patient-tailored therapy.
LC3 expression could be used to stratify DFS and OS in different subsets of patients, especially in pN0 stage patients, but not in stage pN1-3. This finding was supported by the previous study in ESCC [23], which suggest that LC3 is closely associated with the early phase of tumorigenesis in ESCC, but not with advanced stage. Therefore, the determination of LC3 expression by IHC could be used for these patients, who are more likely to experience disease recurrence or progression after surgical resection. In addition, adjuvant therapies should be recommended to these patients with higher expression of LC3 in pN0 stage.
There are some limitations in this study. Firstly, the samples selected from a single institution, and the number of samples enrolled may not enough for subgroup analysis. Subsequently, this is a retrospective study. Last, the informationon chemotherapy or radiotherapy is inadequate to draw a conclusion about the potential role of LC3 expression to therapeutic sensitivity.
In conclusion, the present study determined the prognostic value of LC3 in the ESCC patients treated with surgical resection. LC3, as detected by IHC, may serve as a novel molecular marker for the prognosis of ESCC patients treated with surgical resection. Further studies are needed to evaluate the role of LC3 and clinical application in the treatment of ESCC.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Hongo M, Nagasaki Y, Shoji T. Epidemiology of esophageal cancer: Orient to Occident. Effects of chronology, geography and ethnicity. J Gastroenterol Hepatol. 2009;24:729–735. doi: 10.1111/j.1440-1746.2009.05824.x. [DOI] [PubMed] [Google Scholar]
- 3.Tepper J, Krasna MJ, Niedzwiecki D, Hollis D, Reed CE, Goldberg R, Kiel K, Willett C, Sugarbaker D, Mayer R. Phase III trial of trimodality therapy with cisplatin, fluorouracil, radiotherapy, and surgery compared with surgery alone for esophageal cancer: CALGB 9781. J Clin Oncol. 2008;26:1086–1092. doi: 10.1200/JCO.2007.12.9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 5.Hoshino M, Fukui H, Ono Y, Sekikawa A, Ichikawa K, Tomita S, Imai Y, Imura J, Hiraishi H, Fujimori T. Nuclear expression of phosphorylated EGFR is associated with poor prognosis of patients with esophageal squamous cell carcinoma. Pathobiology. 2007;74:15–21. doi: 10.1159/000101047. [DOI] [PubMed] [Google Scholar]
- 6.Mann SS, Hammarback JA. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J Biol Chem. 1994;269:11492–11497. [PubMed] [Google Scholar]
- 7.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem. 2003;278:51841–51850. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- 9.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanzawa T, Kondo Y, Ito H, Kondo S, Germano I. Induction of autophagic cell death in malignant glioma cells by arsenic trioxide. Cancer Res. 2003;63:2103–2108. [PubMed] [Google Scholar]
- 12.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 13.Miracco C, Cosci E, Oliveri G, Luzi P, Pacenti L, Monciatti I, Mannucci S, De Nisi MC, Toscano M, Malagnino V, Falzarano SM, Pirtoli L, Tosi P. Protein and mRNA expression of autophagy gene Beclin 1 in human brain tumours. Int J Oncol. 2007;30:429–436. [PubMed] [Google Scholar]
- 14.Ahn CH, Jeong EG, Lee JW, Kim MS, Kim SH, Kim SS, Yoo NJ, Lee SH. Expression of beclin-1, an autophagy-related protein, in gastric and colorectal cancers. APMIS. 2007;115:1344–1349. doi: 10.1111/j.1600-0463.2007.00858.x. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Lu Y, Lu C, Zhang L. Beclin-1 expression is a predictor of clinical outcome in patients with esophageal squamous cell carcinoma and correlated to hypoxia-inducible factor (HIF)-1alpha expression. Pathol Oncol Res. 2009;15:487–493. doi: 10.1007/s12253-008-9143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato K, Ogura T, Kishimoto A, Minegishi Y, Nakajima N, Miyazaki M, Esumi H. Critical roles of AMP-activated protein kinase in constitutive tolerance of cancer cells to nutrient deprivation and tumor formation. Oncogene. 2002;21:6082–6090. doi: 10.1038/sj.onc.1205737. [DOI] [PubMed] [Google Scholar]
- 17.Pouyssegur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–443. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- 18.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuervo AM. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Qadir MA, Kwok B, Dragowska WH, To KH, Le D, Bally MB, Gorski SM. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat. 2008;112:389–403. doi: 10.1007/s10549-007-9873-4. [DOI] [PubMed] [Google Scholar]
- 21.Samaddar JS, Gaddy VT, Duplantier J, Thandavan SP, Shah M, Smith MJ, Browning D, Rawson J, Smith SB, Barrett JT, Schoenlein PV. A role for macroautophagy in protection against 4-hydroxytamoxifen-induced cell death and the development of antiestrogen resistance. Mol Cancer Ther. 2008;7:2977–2987. doi: 10.1158/1535-7163.MCT-08-0447. [DOI] [PubMed] [Google Scholar]
- 22.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 23.Yoshioka A, Miyata H, Doki Y, Yamasaki M, Sohma I, Gotoh K, Takiguchi S, Fujiwara Y, Uchiyama Y, Monden M. LC3, an autophagosome marker, is highly expressed in gastrointestinal cancers. Int J Oncol. 2008;33:461–468. [PubMed] [Google Scholar]
- 24.Rice TW, Blackstone EH, Rusch VW. 7th edition of the AJCC Cancer Staging Manual: esophagus and esophagogastric junction. Ann Surg Oncol. 2010;17:1721–1724. doi: 10.1245/s10434-010-1024-1. [DOI] [PubMed] [Google Scholar]
- 25.Rhodes A, Jasani B, Barnes DM, Bobrow LG, Miller KD. Reliability of immunohistochemical demonstration of oestrogen receptors in routine practice: interlaboratory variance in the sensitivity of detection and evaluation of scoring systems. J Clin Pathol. 2000;53:125–130. doi: 10.1136/jcp.53.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta. 2009;1793:664–673. doi: 10.1016/j.bbamcr.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 27.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DiPaola RS, Dvorzhinski D, Thalasila A, Garikapaty V, Doram D, May M, Bray K, Mathew R, Beaudoin B, Karp C, Stein M, Foran DJ, White E. Therapeutic starvation and autophagy in prostate cancer: a new paradigm for targeting metabolism in cancer therapy. Prostate. 2008;68:1743–1752. doi: 10.1002/pros.20837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han C, Sun B, Wang W, Cai W, Lou D, Sun Y, Zhao X. Overexpression of microtubule-associated protein-1 light chain 3 is associated with melanoma metastasis and vasculogenic mimicry. Tohoku J Exp Med. 2011;223:243–251. doi: 10.1620/tjem.223.243. [DOI] [PubMed] [Google Scholar]
- 30.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 31.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]