Abstract
Background: MiR-24/378 is thought to be onco-miRNAs for their ability of enhancing tumor growth. The objective of this study was to evaluate the potential predictive value of miR-24/378 expression in formalin-fixed paraffin-embedded tissues of breast cancer patients. Methods: The expression of miR-24/378 was examined in 101 breast cancer patients and 40 controls using real-time quantitative PCR. All statistical analyses were performed using SPSS16.0. Results: We found that miR-24 and miR-378 were significantly up-regulated in breast cancer patients compared with controls (all P < 0.01). The expression levels of the two miRNAs were highly correlated with each other in breast cancer patients, with r = 0.778 between miR-24 and miR-378. Moreover, the two miRNAs exhibited great capability of discriminating between cancer patients and controls by ROC analysis. MiR-24 and miR-378 showed 0.79 and 0.807 AUC values respectively. Conclusions: Over-expression of miR-24 and miR-378 in FFPE tissue of breast cancer patients might conduct as an ideal source for biomarker discovery and validation in breast cancer patients.
Keywords: Biomarkers, breast cancer, formalin-fixed paraffin-embedded, miR-24, miR-378
Introduction
MiRNAs are small (about 21 nucleotides long) non-coding RNA and post-transcriptionally regulate gene expression by silencing different target mRNAs [1,2]. MiRNAs are important post-transcriptional regulators of gene expression in plants, metazoans, and mammals [3]. By negatively regulating the stability or translational efficiency of target mRNAs, miRNAs play important roles in diverse regulatory pathways, including tumor growth [4], cancer development[5,6], tumor invasion and metastasis [7], control of development [8], apoptosis [9,10], cell proliferation [11], cell differentiation [12], cellular self-renewal [13], division [14], protein secretion [15] and viral infection [16].
Our previous studies have shown that miR-24 promotes tumor invasion and metastasis [7], miR-93 enhances tumor metastasis and growth [17,18], and miR-378 promotes tumor growth and cellular self-renewal [4,13]. MiR-24, miR-93 and miR-378 are all considered to be onco-microRNAs for their role in enhancing tumor growth. Aberrant expression of miRNA genes contributes to the formation and progression of cancer [6]. Recent studies have indicated that miR-24 is highly expressed in the serum of breast cancer patients [19], non small cell lung cancer [20] and oral squamous cell carcinoma [21]. Qian et al demonstrated that over-expression of miR-378 is frequently observed in AML [22]. MiR-378 was also found up-regulated in the serum of renal cell carcinoma patients [23].
Fresh and frozen tissue samples are the appropriate source of DNA, RNA and protein for clinic and research. However, due to the limitation and inefficiency of prospectively collecting fresh and frozen samples, scientists are starting to look for alternative stocks of archived samples with potential utility for disease analysis such as formalin-fixed paraffin-embedded (FFPE) tissues [24]. RNA appears to be degraded in FFPE tissue [25]. Interestingly, miRNAs have been demonstrated to be stable when preserved in FFPE tissue at room temperature because of their shorter lengths. The expression of miRNAs in various cancers using FFPE tissues has been successfully analyzed [26]. A high correlation between miRNA expression levels derived from FFPE and matched fresh-frozen tissues have been revealed [24]. FFPE tissues are ideal for miRNA study and immunohistochemistry (IHC) which carries abundant pathologicaldata. FFPE tissues might conduct as an ideal source for miRNA expression study, biomarker discovery and validation [27].
Emerging evidence has suggested that miR-24/378 is highly expressed in different cancers and could conduct as potential biomarkers for cancer detection. However, little is known about the expression level of miR-24/378 in FFPEtissue of breast cancer patients and the correlations between miR-24/378 and breast cancer. In this study, we investigated expression of miR-24/378 simultaneously in FFPE tissue of breast cancer patients as well as the correlationsbetween them and breast cancer. Moreover, we further analyzed potential predictive value of miR-24/378 as novel potential biomarkers for breast cancer patients.
Materials and methods
Patients and samples
FFPE tissue samples from 101 breast cancer patients in the Affiliated People’s Hospital of Jiangsu University was collected in this study. Additionally, FFPE tissue samples collected from 40 cancer free donors were used as controls. All samples were obtained according to the guidelines of The Affiliated People’s Hospital’s protocol including patient consent and specimen collection. The diagnosis and classification of breast cancer patients were based on the Tumor-Node-Metastasis (TNM) system of American Joint Committee on Cancer (AJCC) [28]. All cases were diagnosed with histologically and clinically confirmed stages I, II and III breast cancer.
RNA extraction and reverse transcription
Total RNA was extracted from 101 FFPE tissue of breast cancer patients and 40 controls using the RecoverAll™ Total Nucleic Acid Isolation Kit (Ambion, catalog no: AM 1975) and reverse transcribed to cDNA using miScript Reverse Transcription Kit (Qiagen, catalog no. 218061).
Real-time quantitative PCR
Real-time quantitative PCR (RQ-PCR) was performed according to the manufacturer’s instructions using miScript SYBR green PCR kit (Qiagen, catalog no. 218073) with the manufacturer-provided miScript Universal primer and miRNA-Specific forward primer: TGGCTGAGTTCAGGAGGAACA (miR-24), CTCCTGACTCCAGGTCCTGTGT (miR-378). RQ-PCR amplification was described in our previous study [22].
Statistical analyses
All statistical analyses were performed using spss16.0. The comparison of miRNA expression levels between patients and controls was performed using the Mann-Whitney test. MiR-24/378 expression levels were presented as median of copies which were normalized to U6 expression. Spearman correlation coefficient was used to analyze the correlation of miR-24 and miR-378 expression. The χ2 test was used to analyze the association of miR-24 and miR-378 expression with clinicopathological characteristics. Receiver operator characteristic (ROC) curve analysis was applied to assess the diagnostic significance of miRNAs. AUC (area under a curve) was the area under the ROC curve, representing the accuracy of using certain miRNA expression to discriminate patients from controls. Cutoff was the selected value of certain miRNA, which could differentiate patients from controls with the most optimized sensitivity and specificity. The correlations between miRNAs and breast cancer were further determined using univariable logistic regression as well as multivariable logistic regression. All values shown are two-sided, and a P-value <0.05 was considered statistically significant.
Results
Expression and correlation of miR24/378 in FFPE tissues of breast cancer patients
Both miR-24 and miR-378 exhibited higher expression levels in breast cancer patients compared with controls (all P<0.01) (Table 1) (Figure 1A, 1B). The median of miR-24 level was 3.262 and 0.275 in breast cancer patients and in controls respectively. The median of miR-378 level was 1.002 in breast cancer patients and 0.096 in controls.
Table 1.
Expression level of miR24/378 and their diagnostic significance in breast cancer patients
| Median in cancer patients | Median in control | P value | AUC | Cutoffs | Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|---|---|---|
| miR-24 | 3.262 | 0.275 | <0.01 | 0.79 | 0.2066 | 62.5 | 90 |
| miR-378 | 1.002 | 0.096 | <0.01 | 0.807 | 0.0427 | 68 | 85 |
Figure 1.
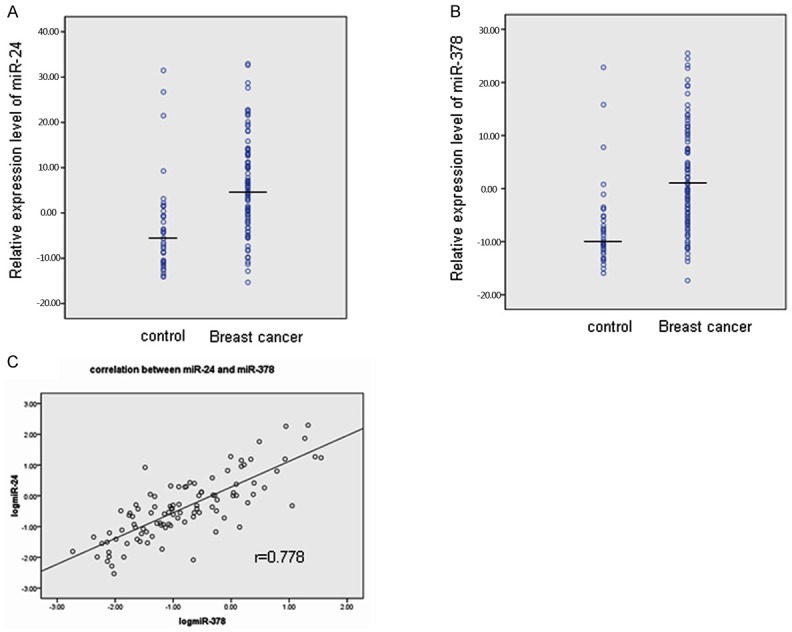
Relative expression level of miR-24 (A) and miR-378 (B) in 101 breast cancer patients and 40 controls. MiRNA expression levels are presented after log10 transformation, medium of each miRNA in both control and breast cancer group was indicated as marked. Correlation between miR-24 and miR-378, the spearman correlation indicated high association between the two miRNAs, with r = 0.778 between miR-24 and miR-378 as shown in the scatter plot (C).
Furthermore, Spearman correlation analysis revealed high association between the two miRNAs, with r = 0.778 between miR-24 and miR-378 as indicated in the scatter plot (Figure 1C).
A panel of highly specific and sensitive miRNAs for determination of breast cancer
ROC analysis was then performed to evaluate the capability of using the miRNAs to discriminate between 101 patients and 40 controls. Optimum cutoff values were determined. As indicated in Table 1 and Figure 2, miR-24 showed 0.79 AUC with 62.5% sensitivity and 90% specificity in discriminating breast cancer patients from controls (Figure 2A); while miR-378 showed 0.807 AUC with 68% sensitivity and 85% specificity (Figure 2B). The findings imply that the selected miRNAs could hold promise as biomarkers for breast cancer. As shown in Figure 2C, the combination of the two miRNAs produced 0.711 AUC, which was not significantly difference from that of each individualmiRNA.
Figure 2.
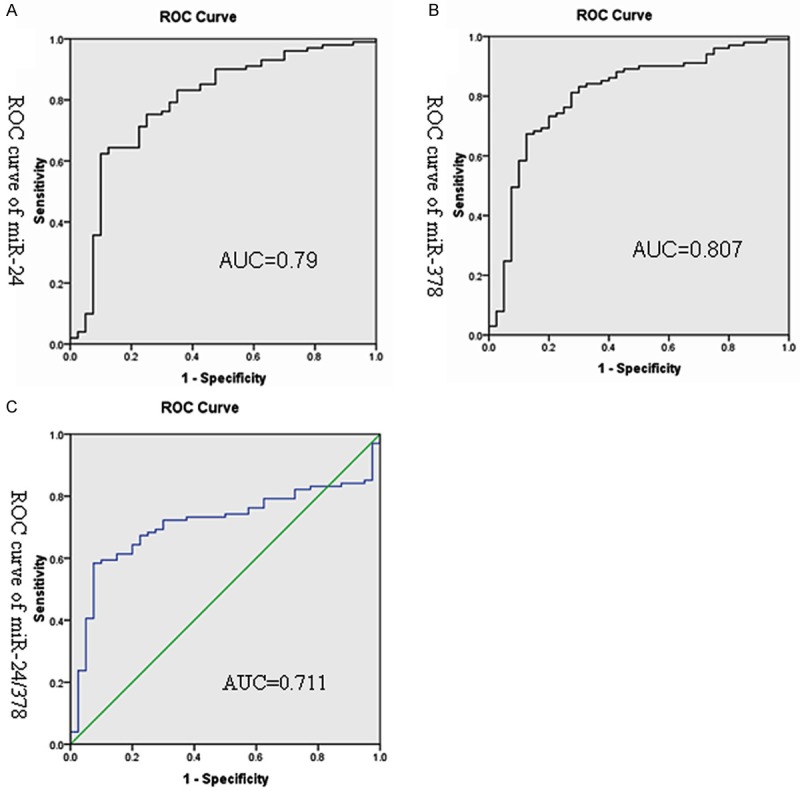
ROC curve analysis of the two miRNAs in 101 breast cancer patients and 40 controls. The area under the ROC curve (AUC) indicates the accuracy for differentiating breast cancer patients from controls in terms of sensitivity and specificity. ROC curve of miR-24 (A), miR-24 showed 0.79 AUC with 62.5% sensitivity and 85% specificity. ROC curve of miR-378 (B), miR-378 showed 0.807 AUC with 68% sensitivity and 90% specificity. Combination of the two miRNAs produced 0.711 AUC (C).
MiR-24 and miR-378 levels were also found significantly correlated with breast cancer by univariable logistic regression (P < 0.01, Table 2). However, in multivariate regression analysis, which included both of the two miRNAs, the correlation between miRNAs and breast cancer was greatly attenuated as show in Table 3.
Table 2.
Univariable Logistic regression model of miR24/378
| Regression coefficient (β) | SE | Wald. χ2 | P value | Exp (β) | |
|---|---|---|---|---|---|
| miR-24 | 2.437 | 0.501 | 22.982 | <0.01 | 11.839 |
| miR-378 | 2.283 | 0.489 | 21.824 | <0.01 | 9.802 |
Table 3.
Multivariate logistical model of miR24/378
| Regression coefficient (β) | SE | Wald. χ2 | P value | Exp (β) | |
|---|---|---|---|---|---|
| MiR-24 | 1.151 | 0.962 | 1.431 | 0.232 | 3.162 |
| MiR-378 | 1.127 | 0.606 | 3.459 | 0.063 | 3.086 |
Correlation of miR-24 and miR-378 expression with clinicopathological characteristics of breast cancer
To further evaluate whether high-expression of miR-24 and miR-378 was related to the clinical progression of breast cancer, we analyzed the association of the two miRNAs with the clinicopathological status of breast cancer patients. High expression of miR-24/378 was not related to most of the clinicopathological characteristics of patients with breast cancer (Table 4).
Table 4.
Clinicopathological characteristics and expression of miR-24 and miR-378 in breast cancer
| Characteristics | Cases | miR24 expression level | miR378 expression level | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Low | high | P | Low | high | P | ||
| Age (years) | |||||||
| ≤40 | 19 | 10 | 9 | 0.085 | 8 | 11 | 0.627 |
| 40-55 | 34 | 18 | 16 | 19 | 15 | ||
| >55 | 48 | 23 | 25 | 24 | 24 | ||
| Histological grade | |||||||
| I | 47 | 20 | 27 | 0.093 | 22 | 25 | 0.686 |
| II | 28 | 19 | 9 | 16 | 12 | ||
| III | 26 | 12 | 14 | 13 | 13 | ||
| N stage | |||||||
| 0 | 60 | 27 | 23 | 0.786 | 27 | 33 | 0.461 |
| 1-3 | 23 | 14 | 9 | 13 | 10 | ||
| 4-9 | 10 | 7 | 3 | 7 | 3 | ||
| >10 | 8 | 5 | 3 | 4 | 4 | ||
| ER status | |||||||
| negative | 41 | 24 | 17 | 0.181 | 20 | 21 | 0.776 |
| positive | 60 | 27 | 33 | 31 | 29 | ||
| PR status | |||||||
| negative | 44 | 25 | 19 | 0.264 | 24 | 20 | 0.474 |
| positive | 57 | 26 | 31 | 27 | 300.807 | ||
Discussion
Aberrant expression patterns of miRNAs have been reported in various cancers and dysregulation of miRNAs are revealed to highly correlate with the progression and prognosis of different cancers [19-23]. Breast canceris thoughtto be a heterogeneous neoplasm, which involves various profile alterations in both expression of miRNA and mRNA [27]. Abundant studies about the expression level of various miRNAs and their functions in breast cancer have been reported [27,29]. Mar-Aguilar F et al demonstrated up-regulation of miR-21, miR-191 and down-regulation of miR-125b in breast cancer patients [30]. Over-expression of serum miR-155 was found in breast cancer patients [31]. Recent studies revealed that miRNAs might be ideal potential candidates as therapeutic targets and novel biomarkers [27].
In this study, we detected the expression of miR-24 and miR-378 in FFPE tissue of breast cancers using RQ-PCR. We found that miR-24 and miR-378 showed higher expression levels in breast cancer patients compared with controls.
Due to the lack of current evidence about diagnostic ability of miR-24/378 in breast cancer patients, we performed ROC curve analysis. MiR24 expression in FFPE tissues of breast cancer produced an AUC of 0.79 with sensitivity of 62.5% and specificity of 90% in the identification of breast cancer. These results showed that miR-24 had considerable diagnostic power to discriminate breast cancer patients from controls. The result of ROC curve analysis alsorevealed that miR-378 produced an AUC of 0.807 with 68% sensitivity and 85% specificity, exhibiting similar diagnostic ability. The combination of the two miRNAs produced 0.711 AUC, showing no significant difference from 0.79 and 0.807 AUC values of each individual miRNA indistinguishing cancer patients from controls, which might imply that one single miRNA is already sufficient enough for the identification, or which can be exp-lained by the highly correlation between the two miRNAs. The association between miR-24/378 levels and breast cancer was further confirmed by multivariate and univariable logistic regression. However, compared with univariable model, in the regressionmodel which included both of the two miRNAs, the correlation between the two miRNAs and breast cancer was attenuated. This difference could be probably the results of the multi colinearity of the two miRNAs, since the two miRNAs were highly correlated with each other by spearman correlation analysis, which makes the estimation of individual miRNA’s predictive ability inaccurate in that model.
Additionally, the correlation of miR-24 and miR-378 with clinicopathological characteristics of breast cancer were also analyzed. We found that high expression of miR-24/378 was not related to most of the clinicopathological characteristics in breast cancer patients. MiR-24 and miR-378 are considered to be onco-microRNAs [4,7,13,17,18]. Based on our observations in the analysis of miR-24/378 expression level and clinical characteristics, the lack of relation between miR-24/378 and other clinicopathological characteristics might suggest that the mechanism underlying the high production of the two miRNAs is different from that of other clinic factors in breast cancer, which makes them potential diagnostic and prognostic biomarkers that are independent from other known biomarkers.
FFPE tissues could be long-term stored and can permanently preserve the architecture of tissue. The biomarkers derived from FFPE tissues are linked to pathological and clinical databases and provide efficient biological insight. Recent studies revealed that FFPE tissue is a reliable source for detection of miRNA expression levels as compared with frozen tissues [32-36]. Taken together, our study suggest that over-expression of miR-24 and miR-378 in FFPE tissues of breast cancer patients might conduct as a ideal source for biomarker discovery and validation in breast cancer patients.
In conclusions, over-expression of miR-24 and miR-378 in TTPE tissue of breast cancer patients might conduct as an ideal source for biomarker discovery and validation in breast cancer patients.
Acknowledgements
This study was supported by National Natural Science foundation of China (81172592, 81270630).
Disclosure of conflict of interest
None.
References
- 1.Hutvagner G, Zamore PDA. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 2.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 3.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A. 2007;104:20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye G, Fu G, Cui S, Zhao S, Bernaudo S, Bai Y, Ding Y, Zhang Y, Yang BB, Peng C. MicroRNA 376c enhances ovarian cancer cell survival by targeting activin receptor-like kinase 7:implications for chemoresistance. J Cell Sci. 2001;124:359–368. doi: 10.1242/jcs.072223. [DOI] [PubMed] [Google Scholar]
- 6.Negrini M, Ferracin M, Sabbioni S, Croce CM. MicroRNAs in human cancer: from research to therapy. J Cell Sci. 2007;120:1833–1840. doi: 10.1242/jcs.03450. [DOI] [PubMed] [Google Scholar]
- 7.Du WW, Fang L, Li M, Yang X, Liang Y, Peng C, Qian W, O’Malley YQ, Askeland RW, Sugg SL, Qian J, Lin J, Jiang Z, Yee AJ, Sefton M, Deng Z, Shan SW, Wang CH, Yang BB. MicroRNA miR-24 enhances tumor invasion and metastasis by targeting PTPN9 and PTPRF to promote EGF signaling. J Cell Sci. 2013;126:1440–53. doi: 10.1242/jcs.118299. [DOI] [PubMed] [Google Scholar]
- 8.Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, Zhang Y, Xuan JW, Yee SP, Siragam V, Yang BB. MicroRNA miR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol. 2009;11:1031–1038. doi: 10.1038/ncb1917. [DOI] [PubMed] [Google Scholar]
- 9.Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, Yang B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120:3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- 10.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 11.Shatseva T, Lee DY, Deng Z, Yang BB. MicroRNA miR-199a-3p regulates cell proliferation and survival by targeting caveolin-2. J Cell Sci. 2011;124:2826–2836. doi: 10.1242/jcs.077529. [DOI] [PubMed] [Google Scholar]
- 12.Baumjohann D, Kageyama R, Clingan JM, Morar MM, Patel S, de Kouchkovsky D, Bannard O, Bluestone JA, Matloubian M, Ansel KM, Jeker LT. The microRNA cluster miR-17-92 promotes TFH cell differentiation and represses subset-in appropriate gene expression. Nat Immunol. 2012;14:840–848. doi: 10.1038/ni.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng Z, Du WW, Fang L, Shan SW, Qian J, Lin J, Qian W, Ma J, Rutnam ZJ, Yang BB. The intermediate filament vimentin mediates microRNA miR-378 function in cellular self-renewal by regulating the expression of the Sox2 transcription factor. J Biol Chem. 2013;288:319–331. doi: 10.1074/jbc.M112.418830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 15.Mello CC, Czech MP. Micromanaging insulin secretion. Nat Med. 2004;10:1297–1298. doi: 10.1038/nm1204-1297. [DOI] [PubMed] [Google Scholar]
- 16.Chellappan P, Vanitharani R, Fauquet CM. MicroRNA-binding viral protein interferes with Arabidopsis development. Proc Natl Acad Sci U S A. 2005;102:10381–10386. doi: 10.1073/pnas.0504439102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang L, Du WW, Yang W, Rutnam ZJ, Peng C, Li H, O’Malley YQ, Askeland RW, Sugg S, Liu M, Mehta T, Deng Z, Yang BB. MiR-93 enhances angiogenesis and metastasis by targeting LATS2. Cell Cycle. 2012;11:4352–4365. doi: 10.4161/cc.22670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang L, Deng Z, Shatseva T, Yang J, Peng C, Du WW, Yee AJ, Ang LC, He C, Shan SW, Yang BB. MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-β8. Oncogene. 2011;30:806–821. doi: 10.1038/onc.2010.465. [DOI] [PubMed] [Google Scholar]
- 19.Wu Q, Wang C, Lu Z, Guo L, Ge Q. Analysis of serum genome-wide microRNAs for breast cancer detection. Clin Chim Acta. 2012;413:1058–1065. doi: 10.1016/j.cca.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 20.Franchina T, Amodeo V, Bronte G, Savio G, Ricciardi GR, Picciotto M, Russo A, Giordano A, Adamo V. Circulating miR-22, miR-24 and miR-34a as novel predictive biomarkers to pemetrexed-based chemotherapy in advanced non small cell lung cancer. J Cell Physiol. 2013;229:97–99. doi: 10.1002/jcp.24422. [DOI] [PubMed] [Google Scholar]
- 21.Lin SC, Liu CJ, Lin JA, Chiang WF, Hung PS, Chang KW. miR-24 up-regulation in oral carcinoma: positive association from clinical and in vitro analysis. Oral Oncol. 2010;46:204–208. doi: 10.1016/j.oraloncology.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Qian J, Lin J, Qian W, Ma JC, Qian SX, Li Y, Yang J, Li JY, Wang CZ, Chai HY, Chen XX, Deng ZQ. Overexpression of miR-378 is frequent and may affect treatment outcomes in patients with acute myeloid leukemia. Leuk Res. 2012;37:765–768. doi: 10.1016/j.leukres.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 23.Redova M, Poprach A, Nekvindova J, Iliev R, Radova L, Lakomy R, Svoboda M, Vyzula R, Slaby O. Circulating miR-378 and miR-451 in serum are Potential biomarkers for renal cell carcinoma. J Transl Med. 2012;10:55–63. doi: 10.1186/1479-5876-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goswami RS, Waldron L, Machado J, Cervigne NK, Xu W, Reis PP, Bailey DJ, Jurisica I, Crump MR, Kamel-Reid S. Optimization and analysis of a quantitative real-time PCR-based technique to determine microRNA expression in formalin-fixed paraffin-embedded samples. BMC Biotechnol. 2012;10:47–59. doi: 10.1186/1472-6750-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macabeo-Ong M, Ginzinger DG, Dekker N, McMillan A, Regezi JA, Wong DT, Jordan RC. Effect of duration of fixation on quantitative reverse Transcription polymerase chain reaction analyses. Mod Pathol. 2002;15:979–987. doi: 10.1097/01.MP.0000026054.62220.FC. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Lu YE, Chen YA, Lu W, Xie X. MicroRNA profile of paclitaxel-resistant serous ovarian carcinoma based on formalin-fixed paraffin-embedded samples. BMC Cancer. 2013;13:216–223. doi: 10.1186/1471-2407-13-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu SW, Chen L, Man YG. miRNA Biomarkers in Breast Cancer Detection and Management. J Cancer. 2011;2:116–122. doi: 10.7150/jca.2.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singletary SE, Allred C, Ashley P, Bassett LW, Berry D, Bland KI, Borgen PI, Clark GM, Edge SB, Hayes DF, Hughes LL, Hutter RV, Morrow M, Page DL, Recht A, Theriault RL, Thor A, Weaver DL, Wieand HS, Greene FL. Staging system for breast cancer: revisions for the 6th edition of the AJCC cancer staging manual. Surg Clin North Am. 2013;83:803–819. doi: 10.1016/S0039-6109(03)00034-3. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Yang PY, Sun T, Li D, Xu X, Rui Y, Li C, Chong M, Ibrahim T, Mercatali L, Amadori D, Lu X, Xie D, Li QJ, Wang XF. miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat Cell Biol. 2013;15:284–294. doi: 10.1038/ncb2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mar-Aguilar F, Luna-Aguirre CM, Moreno-Rocha JC, Araiza-Chávez J, Trevino V, Rodríguez-Padilla C, Reséndez-Pérez D. Differential expression of miR-21, miR-125b and miR-191 in breast cancer tissue. Asia Pac J Clin Oncol. 2013;9:53–59. doi: 10.1111/j.1743-7563.2012.01548.x. [DOI] [PubMed] [Google Scholar]
- 31.Sun Y, Wang M, Lin G, Sun S, Li X, Qi J, Li J. Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS One. 2012;7:e47003. doi: 10.1371/journal.pone.0047003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, Hayashi K, Ju J. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13:1668–1674. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoefig KP, Thorns C, Roehle A, Kaehler C, Wesche KO, Repsilber D, Branke B, Thiere M, Feller AC, Merz H. Unlocking pathology archives for microRNA-profiling. Anticancer Res. 2008;28:119–123. [PubMed] [Google Scholar]
- 34.Laios A, O’Toole S, Flavin R, Martin C, Kelly L, Ring M, Finn SP, Barrett C, Loda M, Gleeson N, D’Arcy T, McGuinness E, Sheils O, Sheppard B, O’Leary J. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer. 2008;7:35–49. doi: 10.1186/1476-4598-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng ZQ, Qian J, Liu JQ, Lin J, Shao R, Yin JY, Tang Q, Zhang M, He L. Expression level of mir-93 in formalin-fixed paraffin-embedded tissues of breast cancer patients genetic testing and molecular biomarkers. Genet Test Mol Biomarkers. 2014;18:366–70. doi: 10.1089/gtmb.2013.0440. [DOI] [PubMed] [Google Scholar]
- 36.Deng ZQ, Yin JY, Tang Q, Liu FG, Jun Qian, Lin J, Shao R, Zhang M, He L. Over-expression of miR-98 in FFPE Tissues might serve as a valuable source for biomarker discovery in Breast Cancer Patients. International journal of clinical and experimental pathology. Int J Clin Exp Pathol. 2014;7:1166–71. [PMC free article] [PubMed] [Google Scholar]


