Abstract
In this study, we evaluated C-kit immunohistochemical expression and C-kit and platelet derived growth factor receptor A (PDGFRA) gene mutations in triple negative breast cancer. 171 cases were analyzed by immunohistochemical staining for the expression of C-kit and 45 cases, including 10 C-kit negative cases and 35 C-kit positive cases, were performed for C-kit gene mutations in exons 9, 11, 13 and 17 and PDGFRA gene mutations in exons 12 and 18. C-kit expression was detected in 42.1% of triple negative breast cancers. Only 1 activating mutation was detected in exon 11 of C-kit gene in 1 case. No activating mutations were found in the other 44 cases. C-kit expression is a frequent finding in triple negative breast cancers; 1 activating mutation which was also found in gastrointestinal stromal tumors was detected; a few cases might benefit from imatinib.
Keywords: C-KIT, PDGFRA, mutation, triple negative breast cancer, immunohistochemical expression
Introduction
Breast cancer is one of the most common malignant diseases in women, representing 22% of all female cancers world widely [1]. Triple negative breast cancers, characterized by lack of expression of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER-2), constitutes a distinct subtype of breast carcinoma. Despite advances of treatment in breast cancer, triple negative breast cancer is likely to have a relatively aggressive clinical behavior and poor outcome, and no proven molecular targeted therapies are available for the treatment of triple negative breast cancer up to now [2,3].
C-kit and platelet derived growth factor receptor A (PDGFRA) are members of the type III tyrosine kinase gene family. Gastrointestinal stromal tumors (GISTs), which are the most common mesenchymal tumors of the gastrointestinal tract, are resistant to radiation and conventional chemotherapy. Most of GISTs express C-kit protein and harbor activating mutations in exons 9, 11, 13 and 17 of C-kit gene or exons 12, 18 of PDGFRA gene [4,5]. In view of the importance of C-kit and PDGFRA in GISTs, tyrosine kinase inhibitors, such as imatinib mesylate, have been successfully used to treat GISTs. Up to 90% of GISTs showing C-kit expression or mutation could benefit from imatinib mesylate [6,7].
Previous studies show that overexpression of C-kit is detected in part of triple negative breast cancer. It is hoped that triple negative breast cancers might benefit from imatinib mesylate [8-11]. To our knowledge, there are no reports on the presence of C-kit and PDGFRA mutations in triple negative breast cancers. The aim of the current study was to evaluate the expression of C-kit protein and the mutations of C-kit gene and PDGFRA gene in triple negative breast cancers in the hopes of identifying whether the patients could benefit from C-kit tyrosine kinase inhibitor such as imatinib.
Materials and methods
Cases selection
The patients diagnosed as primary breast cancer admitted to Nanjing Jinling Hospital between 2002 and 2010 were reviewed by 2 professional pathologists. All the cases recognized as triple negative breast cancer were reviewed to identify the histological subtypes of these tumors.
Immunohistochemistry
Paraffin sections were cut at 4-μm thickness, deparaffinized with xylene, and rehydrated. The slides were then subjected to pressure cooker antigen retrieval, using citrate buffer at pH 6.0 for 5 minutes. After the slides were stained with the primary antibodies against C-kit (1:1000, DAKO Corporation, Carpentaria, CA), immunodetection was performed with the MaxVision Kit (Maixin Biol, Fu Zhou, China) with 3,3’-diaminobenzidine chromogen as substrate. The slides were then counterstained with Harris hematoxylin. For C-kit staining status, if more than 1% of tumor cells were cytoplasm and/or membrane staining, these cases were defined as positive for C-kit.
C-kit gene mutation analysis
Genomic DNA was extracted and purified from paraffin wax embedded blocks with a commercially available kit (E.Z.N.A. Tissue DNA Kit; Omega Bio-Tek, Doraville, GA) according to the protocols of manufacturers. Polymerase chain reactions (PCR) were designed and DNA was subjected to PCR amplification of exons 9, 11, 13 and 17 of the C-kit gene and exons 12 and 18 of PDGFRA gene. Primer sequences are shown in Table 1. Direct sequencing of all the PCR products were performed using ABI BigDye Terminator v3.1 Cycling Kits (AB Applied Biosystems, Carlsbad, CA) and an ABI 3130xl sequencer (AB Applied Biosystems) following the manufacturer’s protocols. Sequencing was performed twice for each sample to avoid the possibility of PCR fidelity artefacts and carried out in both directions. Every case with observed mutation would identify the mutational status of its normal tissue.
Table 1.
Polymerase Chain Reaction Primer Sequences and Annealing Temperatures
| Gene/Exon | Primer sequence (5’→3’) | Annealing temperatures |
|---|---|---|
| C-kit | ||
| 9F | ATTTATTTTCCTAGAGTAAGCCAGGG | 58°C |
| 9R | ATCATGACTGATATGGTAGACAGAGC | |
| 11F | CCAGAGTGCTCTAATGACTG | 58°C |
| 11R | ACCCAAAAAGGTGACATGGA | |
| 13F | ATCAGTTTGCCAGTTGTGCT | 58°C |
| 13R | TTTATAATCTAGCATTGCC | |
| 17F | TGTATTCACAGAGACTTGGC | 58°C |
| 17R | GGATTTACATTATGAAAGTCA | |
| PDGFRA | ||
| 12F | TCCAGTCACTGTGCTGCTTC | 58°C |
| 12R | GCAAGGGAAAAGGGAGTCTT | |
| 18F | ACCATGGATCAGCCAGTCTT | 58°C |
| 18R | TGAAGGAGGATGAGCCTGAC |
Results
171 patients in this research, including 2 male patient, with an overall mean age of 45 years (range 24 to 80 years), were classified as triple negative breast cancers account for 16.8% of all breast cancers. For histological subtypes (Table 2), these cases included invasive ductal carcinoma (150/171, 87.7%), metaplastic carcinoma (6/171, 3.5%), medullary carcinoma (9/171, 5.3%), apocrine carcinoma (1/171, 0.6%), invasive micropapillary carcinoma (1/171, 0.6%), invasive lobular carcinoma (2/171, 1.2%) and lipid-rich carcinoma (2/171, 1.2%). 72 cases were positive for C-kit (Figure 1), included invasive ductal carcinoma (62/72, 86.1%), metaplastic carcinoma (5/72, 6.9%), medullary carcinoma (3/72, 4.2%), apocrine carcinoma (1/72, 1.4%) and invasive micropapillary carcinoma (1/72, 1.4%). 99 cases were negative for C-kit. A subset of 10 C-kit negative cases and 35 C-kit positive cases were selected for DNA extraction and gene mutation analysis. Sequencing of exons 9, 11, 13 and 17 of C-kit gene and exons 12 and 18 of PDGFRA gene only revealed 1829 T > C alteration in exon 11 of C-kit gene in 1 case (Figure 2), with a predicted missense substitution of valine by alanine. The patient with mutational activation of C-kit gene was a 42-year-old Chinese female, diagnosed as invasive ductal carcinoma in histological grade III. No activating mutations of exons 9, 11, 13, and 17 in C-kit gene and exons 12 and 18 in PDGFRA gene were found in normal tissue of this case and the other 44 cases.
Table 2.
The Results of C-kit Immunohistochemical expression in triple negative breast cancer
| Total No. | C-kit expression | ||
|---|---|---|---|
|
| |||
| Positive No. (%) | Negative No. (%) | ||
| All Samples | 171 | 72 (42.1) | 99 (57.9) |
| Histological subtypes | |||
| Ductal Carcinoma | 150 | 62 (41.3) | 88 (58.7) |
| Metaplastic Carcinoma | 6 | 5 (83.3) | 1 (16.7) |
| Medullary Carcinoma | 9 | 3 (33.3) | 6 (66.7) |
| Other Subtypes* | 6 | 2 (33.3) | 4 (66.7) |
Other subtypes include apocrine carcinoma (n = 1), invasive micropapillary carcinoma (n = 1), invasive lobular carcinoma (n = 2) and lipid-rich carcinoma (n = 2).
Apocrine carcinoma (n = 1) and invasive micropapillary carcinoma (n = 1) was positive for C-kit while the other four cases were negative for C-kit.
Figure 1.
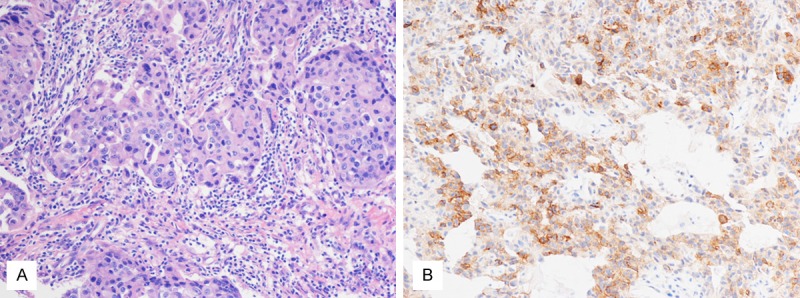
A. The histological feature of triple negative breast cancers (× 200); B. Membranous and stromal expression of C-kit in a triple negative breast cancer (× 200).
Figure 2.
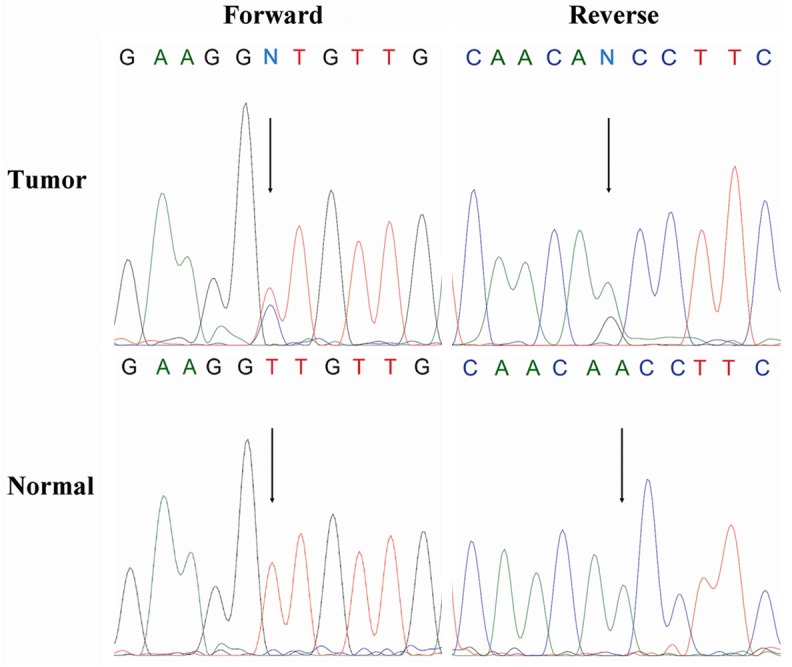
Forward/reverse sequencing chromatograms for mutation identified in exon 11 of C-kit gene in tumor and matched normal samples. The nucleotide change is 1829 T > C that would lead to substitution of alanine for valine.
Discussion
Triple negative breast cancers, associated with high grade, poor prognosis and young patient age, are negative for ER, PR and Her-2 and can not be treated with the current molecular targeted therapy of breast cancer, such as antiestrogen hormonal therapies or trastuzumab [2,3]. C-kit, in consideration of overexpression in triple negative breast cancer, is one of the hot therapeutic targets in the present researches.
Several studies have investigated the expression of C-kit protein in breast tumors. Simon et al. reported that only 2.1% of ductal carcinomas and 0.5% of lobular carcinomas expressed C-kit while 19.1% of medullary carcinomas expressed C-kit [12]. It has been frequently reported that C-kit is expressed in 11-22.7% of basal-like breast cancers as a subtype which lack expression of ER, PR and Her-2 [8-10]. The results of our current study showed that 41.7% of the triple negative breast cancers were positive for C-kit. The discrepancy might be due to the use of different antibodies, staining protocols, and scoring method.
From the above discussion, it would appear that C-kit overexpression was detected in about 40% of triple negative breast cancers. The overexpression of C-kit in triple negative breast cancer implied that these cases might potentially be treated with tyrosine kinase receptor inhibitors imatinib mesylate. However, only 1 activating mutation in exon 11 of C-kit gene was found while no activating mutations were found in the other 44 cases, similar to a small sample in that no C-kit mutations were found in exons 2, 8, 9, 11, 13 and 17 in the set of 10 cases showing intense C-kit staining [12]. The results indicated that the overexpression of C-kit protein in triple negative breast cancer lacked relation with activating mutations of C-kit and PDGFRA. Except for triple negative breast cancer, C-kit expression was found frequently in primary pulmonary adenoid cystic carcinoma with absence of C-kit activating mutations [13] and C-kit expression was a frequent finding in phyllodes tumors while only 1 silent germline point mutation was detected [14]. Indeed, with the exceptions of the GISTs, there is a lack of correlation between C-kit activating mutations and C-kit expression in multiple types of tumors [15].
Imatinib, the first developed molecular targeted agent for chronic myelogenous leukemia (CML) and GISTs, inhibits not only Bcr-Abl kinases but also C-kit and platelet derived growth factor receptor (PDGFR) [16,17]. Previous studies have shown that the response to the treatment of imatinib have a closer relationship with the mutational activation of C-kit gene but not with the expression of C-kit protein [18-20]. If this is true, the 44 cases without activating mutations of C-kit gene or PDGFRA gene might not benefit from imatinib. This is in agreement with the research in that imatinib could enhance the malignant behavior of human breast carcinoma cells in vitro [21]. A clinical trial also indicated that imatinib, as a single agent, had no clinical activity in metastatic breast cancers [22]. It can be seen that imatinib mesylate might not provide any benefit for most of the triple negative breast cancer patients. However, 1 missense mutation in exon 11 of C-kit gene, which is also found in GISTs, was detected in our series [4]. This is a 42-year-old Chinese female patient diagnosed as invasive ductal carcinoma in grade III, with no special clinicopathological features. Because this patient harbored an activating mutation and might achieve a response to treatment with imatinib, it suggested that the patients showing C-kit stromal and/or membranous staining in immunohistochemistry would better do gene mutation analysis for the hope of identifying the cases that might benefit from imatinib although this amount might be very small. Actually, we have also recognized that our relatively small sample for gene mutation analysis of C-kit gene may have influenced our results. Therefore, further investigations, on a larger and more heterogeneous population, should be carried out to validate and extend our results to ensure what type of breast cancer might harbor activating mutation of C-kit gene.
Dasatinib, a novel orally active small molecular targeting agent inhibiting of both the src and abl kinases in the treatment of imatinib-resistant Philadelphia Chromosome-positive leukemias [23], has captured attention. Finn et al. showed that dasatinib selectively inhibited growth of basal-type/triple-negative breast cancer cell lines growing in vitro [24]. Lombardo et al. showed that dasatinib not only inhibited the src and abl kinases but also inhibited C-kit [25]. Dasatinib may be a potential molecular targeted drug for C-kit positive triple negative breast cancers. The effect of Dasatinib in triple negative breast cancers needs further research.
In conclusion, we have shown that 42.1% of triple negative breast cancers express C-kit protein; 1 activating mutation was detected in our series; the expression of C-kit in triple negative breast cancer lacked correlation with activating mutations in C-kit and PDGFRA gene; and that a few patients may achieve a response to C-kit tyrosine kinase inhibitor such as imatinib. Further research is required to investigate the potential effect of dasatinib on triple negative breast cancers expressing C-kit and PDGFRA in clinical trials.
Disclosure of conflict of interest
None.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 2.Lara-Medina F, Perez-Sanchez V, Saavedra-Perez D, Blake-Cerda M, Arce C, Motola-Kuba D, Villarreal-Garza C, Gonzalez-Angulo AM, Bargallo E, Aguilar JL, Mohar A, Arrieta O. Triple-negative breast cancer in Hispanic patients: high prevalence, poor prognosis, and association with menopausal status, body mass index, and parity. Cancer. 2011;117:3658–3669. doi: 10.1002/cncr.25961. [DOI] [PubMed] [Google Scholar]
- 3.Abulkhair O, Moghraby JS, Badri M, Alkushi A. Clinicopathologic features and prognosis of triple-negative breast cancer in patients 40 years of age and younger in Saudi Arabia. Hematol Oncol Stem Cell Ther. 2012;5:101–106. doi: 10.5144/1658-3876.2012.101. [DOI] [PubMed] [Google Scholar]
- 4.Battochio A, Mohammed S, Winthrop D, Lefresne S, Mulder K, Chu Q, O’Hara C, Lai R. Detection of c-KIT and PDGFRA gene mutations in gastrointestinal stromal tumors: comparison of DHPLC and DNA sequencing methods using a single population-based cohort. Am J Clin Pathol. 2010;133:149–155. doi: 10.1309/AJCP1FNW7RGZFTYU. [DOI] [PubMed] [Google Scholar]
- 5.Zong L, Chen P. Prognostic value of KIT/PDGFRA mutations in gastrointestinal stromal tumors: a meta-analysis. World J Surg Oncol. 2014;12:71. doi: 10.1186/1477-7819-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonescu CR. Targeted therapies in gastrointestinal stromal tumors. Semin Diagn Pathol. 2008;25:295–303. doi: 10.1053/j.semdp.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Serrano C, George S. Recent advances in the treatment of gastrointestinal stromal tumors. Ther Adv Med Oncol. 2014;6:115–127. doi: 10.1177/1758834014522491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim MJ, Ro JY, Ahn SH, Kim HH, Kim SB, Gong G. Clinicopathologic significance of the basal-like subtype of breast cancer: a comparison with hormone receptor and Her2/neu-overexpressing phenotypes. Hum Pathol. 2006;37:1217–1226. doi: 10.1016/j.humpath.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Lerma E, Peiro G, Ramon T, Fernandez S, Martinez D, Pons C, Munoz F, Sabate JM, Alonso C, Ojeda B, Prat J, Barnadas A. Immunohistochemical heterogeneity of breast carcinomas negative for estrogen receptors, progesterone receptors and Her2/neu (basal-like breast carcinomas) Mod Pathol. 2007;20:1200–1207. doi: 10.1038/modpathol.3800961. [DOI] [PubMed] [Google Scholar]
- 10.Nalwoga H, Arnes JB, Wabinga H, Akslen LA. Expression of EGFR and c-kit is associated with the basal-like phenotype in breast carcinomas of African women. APMIS. 2008;116:515–525. doi: 10.1111/j.1600-0463.2008.01024.x. [DOI] [PubMed] [Google Scholar]
- 11.Kashiwagi S, Yashiro M, Takashima T, Aomatsu N, Kawajiri H, Ogawa Y, Onoda N, Ishikawa T, Wakasa K, Hirakawa K. c-Kit expression as a prognostic molecular marker in patients with basal-like breast cancer. Br J Surg. 2013;100:490–496. doi: 10.1002/bjs.9021. [DOI] [PubMed] [Google Scholar]
- 12.Simon R, Panussis S, Maurer R, Spichtin H, Glatz K, Tapia C, Mirlacher M, Rufle A, Torhorst J, Sauter G. KIT (CD117)-positive breast cancers are infrequent and lack KIT gene mutations. Clin Cancer Res. 2004;10:178–183. doi: 10.1158/1078-0432.ccr-0597-3. [DOI] [PubMed] [Google Scholar]
- 13.Aubry MC, Heinrich MC, Molina J, Lewis JE, Yang P, Cassivi SD, Corless CL. Primary adenoid cystic carcinoma of the lung: absence of KIT mutations. Cancer. 2007;110:2507–2510. doi: 10.1002/cncr.23075. [DOI] [PubMed] [Google Scholar]
- 14.Bose P, Dunn ST, Yang J, Allen R, El-Khoury C, Tfayli A. c-Kit expression and mutations in phyllodes tumors of the breast. Anticancer Res. 2010;30:4731–4736. [PubMed] [Google Scholar]
- 15.Miettinen M, Lasota J. KIT (CD117): a review on expression in normal and neoplastic tissues, and mutations and their clinicopathologic correlation. Appl Immunohistochem Mol Morphol. 2005;13:205–220. doi: 10.1097/01.pai.0000173054.83414.22. [DOI] [PubMed] [Google Scholar]
- 16.Heinrich MC, Griffith DJ, Druker BJ, Wait CL, Ott KA, Zigler AJ. Inhibition of c-kit receptor tyrosine kinase activity by STI 571, a selective tyrosine kinase inhibitor. Blood. 2000;96:925–932. [PubMed] [Google Scholar]
- 17.Wang WL, Healy ME, Sattler M, Verma S, Lin J, Maulik G, Stiles CD, Griffin JD, Johnson BE, Salgia R. Growth inhibition and modulation of kinase pathways of small cell lung cancer cell lines by the novel tyrosine kinase inhibitor STI 571. Oncogene. 2000;19:3521–3528. doi: 10.1038/sj.onc.1203698. [DOI] [PubMed] [Google Scholar]
- 18.Lux ML, Rubin BP, Biase TL, Chen CJ, Maclure T, Demetri G, Xiao S, Singer S, Fletcher CD, Fletcher JA. KIT extracellular and kinase domain mutations in gastrointestinal stromal tumors. Am J Pathol. 2000;156:791–795. doi: 10.1016/S0002-9440(10)64946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dy GK, Miller AA, Mandrekar SJ, Aubry MC, Langdon RM Jr, Morton RF, Schild SE, Jett JR, Adjei AA. A phase II trial of imatinib (ST1571) in patients with c-kit expressing relapsed small-cell lung cancer: a CALGB and NCCTG study. Ann Oncol. 2005;16:1811–1816. doi: 10.1093/annonc/mdi365. [DOI] [PubMed] [Google Scholar]
- 20.Lasota J. Not all c-kit mutations can be corrected by imatinib. Lab Invest. 2007;87:317. [PubMed] [Google Scholar]
- 21.Rappa G, Anzanello F, Lorico A. Imatinib mesylate enhances the malignant behavior of human breast carcinoma cells. Cancer Chemother Pharmacol. 2011;67:919–926. doi: 10.1007/s00280-010-1394-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cristofanilli M, Morandi P, Krishnamurthy S, Reuben JM, Lee BN, Francis D, Booser DJ, Green MC, Arun BK, Pusztai L, Lopez A, Islam R, Valero V, Hortobagyi GN. Imatinib mesylate (Gleevec) in advanced breast cancer-expressing C-Kit or PDGFR-beta: clinical activity and biological correlations. Ann Oncol. 2008;19:1713–1719. doi: 10.1093/annonc/mdn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, Paquette R, Cortes J, O’Brien S, Nicaise C, Bleickardt E, Blackwood-Chirchir MA, Iyer V, Chen TT, Huang F, Decillis AP, Sawyers CL. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 24.Finn RS, Dering J, Ginther C, Wilson CA, Glaspy P, Tchekmedyian N, Slamon DJ. Dasatinib, an orally active small molecule inhibitor of both the src and abl kinases, selectively inhibits growth of basal-type/“triple-negative” breast cancer cell lines growing in vitro. Breast Cancer Res Treat. 2007;105:319–326. doi: 10.1007/s10549-006-9463-x. [DOI] [PubMed] [Google Scholar]
- 25.Lombardo LJ, Lee FY, Chen P, Norris D, Barrish JC, Behnia K, Castaneda S, Cornelius LA, Das J, Doweyko AM, Fairchild C, Hunt JT, Inigo I, Johnston K, Kamath A, Kan D, Klei H, Marathe P, Pang S, Peterson R, Pitt S, Schieven GL, Schmidt RJ, Tokarski J, Wen ML, Wityak J, Borzilleri RM. Discovery of N-(2-chloro-6-methyl-phenyl)-2-(6-(4-(2-hydroxyethyl)-piperazin-1-yl)-2-methylpyrimidin-4-ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J Med Chem. 2004;47:6658–6661. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]


