Abstract
Accumulated evidence has revealed the presence of Notch receptor polymorphisms in non-tumorous diseases; however, few studies have investigated the association of Notch polymorphisms with breast cancer risk. A total of 100 invasive ductal carcinoma (IDC) and 50 ductal carcinoma in situ (DCIS) patients and 100 usual ductal hyperplasia (UDH) controls were genotyped for the following Notch receptor single nucleotide polymorphisms (SNPs) using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry: Notch1, rs3124591; Notch2, rs11249433; Notch3, rs3815188, and rs1043994; and Notch4, rs367398, and rs520692. Immunohistochemistry was used to determine the effect of Notch polymorphisms on corresponding Notch protein expression in successfully genotyped patients. The frequency of rs3124591 TC genotype was significantly higher in IDC (24.7%, 20/81) and DCIS (30%, 12/40) patients than in UDH controls (8%, 8/97) (P = 0.002 and P = 0.011, respectively). However, the distribution of other SNP genotypes was not significantly different between IDC and DCIS patients and UDH controls. The frequency of TC genotype was significantly higher in poorly differentiated tumors than in well-differentiated and moderately differentiated tumors (P = 0.022). Importantly, a positive correlation between the rs3124591 TC genotype and high Notch1 protein expression was observed in DCIS patients (P = 0.043) but not in IDC patients. This is the first study to suggest an increased risk of IDC and DCIS of the breast for the Notch1 rs3124591 variant. Furthermore, given the inconsistent associations between the rs3124591 variant and Notch1 expression in IDC and DCIS, this variant may affect breast cancer risk through mechanisms in the latter stage other than alterations in Notch1 protein expression.
Keywords: Notch1, invasive ductal carcinoma, single nucleotide polymorphisms, association study
Introduction
Breast carcinoma is a major cause of cancer-specific mortality worldwide and efforts aimed at the prevention and early detection of this disease remain important. Breast cancer remains the most common cancer in women in China with age-standardized rates by world population of 21.6 per 100,000 [1]. Over the past few decades, new techniques and methods for breast cancer diagnosis and treatment have been developed, leading to increased survival rates in breast cancer patients. However, many breast cancers still cannot be detected at an early stage, further indicating that more efforts should be exerted toward cancer preventive measures.
The Notch gene family encodes a group of evolutionally conserved transmembrane receptors (Notch1-4), which are expressed on the cell surface as heterodimers. Notch is thought to play a crucial role in a number of normal cellular processes such as epithelial cell polarity/adhesion, proliferation, and apoptosis. Notch signaling is activated by the binding of Notch ligand to epidermal growth factor-like repeats on an adjacent Notch receptor [2,3]. Increasing evidence supports a role for aberrant Notch signaling in breast cancer. Notch receptors have been reported to have opposing functions in normal and cancerous breast tissues. High Notch1 expression has been shown to promote tumor formation in normal breast tissues, whereas Notch2 has been found to play a tumor suppressive role in human breast cancer [4]. Recent reports have demonstrated that overexpression of Notch3IC (containing the constitutively active intracellular domain of Notch3) in transgenic mice leads to developmental disorders of the mammary gland [5]. Notch3 was also found to be required for the proliferation of human epidermal growth factor receptor-2 (HER-2)-negative breast cancer cell lines [5,6]. Moreover, aberrant activation of Notch1 and Notch4 in mammary epithelium caused luminal cell hyperplasia and tumorgenesis by decreasing apoptosis and increasing mitotic rate [7]. An association of Notch1 and Notch4 with poor prognostic factors in triple-negative breast cancer cases has also been suggested [7]. Although Notch signaling is known to regulate multiple cellular processes that affect cancer development, only a few mechanisms have been identified that are capable of explaining the pleiotropic responses to aberrant Notch activation. Thus, the underlying mechanisms by which Notch proteins contribute to the cancer phenotype remain largely elusive.
To date, many authors have investigated the role of Notch gene polymorphisms in disease pathogenesis [8,9]. Previous reports have demonstrated the presence of Notch polymorphisms in non-tumorous diseases, such as Notch1 rs3124591 in bicuspid aortic valve [10], Notch3 rs3815188 and rs1043994 in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) [11], and Notch4 rs367398 and rs520692 in schizophrenia [12]. Recent single nucleotide polymorphism (SNP) and mutation association studies have revealed the functional significance of Notch genetic alterations and their association with breast cancer risk [13,14]. Novel non-synonymous somatic Notch1 mutations have been observed in breast cancers, implicating the role of Notch1 mutations in breast tumorgenesis [15]. Additionally, several lines of evidence have identified an association between the Notch2 rs11249433 genetic variant and breast cancer risk [13,14,16]. Nevertheless, few studies have addressed the association of polymorphisms in other Notch loci with breast cancer susceptibility. Hence, the present study was designed to evaluate the association of Notch SNPs with breast cancer susceptibility with the aim of identifying patients likely to benefit from early prophylaxis and treatment.
Materials and methods
Study population and DNA extraction
The study was approved by the Human Ethics Committee of Shihezi University. We consecutively enrolled 100 invasive ductal carcinoma (IDC; aged 33-80 years), 50 ductal carcinoma in situ (DCIS; aged 35-50 years), and 100 usual ductal hyperplasia (UDH; aged 20-65 years) female patients who were admitted to the Department of Pathology, Shihezi University School of Medicine. A term of free informed consent was signed by all sample donors prior to sample collection. Patients diagnosed with IDC did not receive radiotherapy or chemotherapy before surgery. Clinical and pathological information was obtained from medical records and pathology reports. The final diagnosis was confirmed by two pathologists according to the clinical and histological criteria of the Pathology and Genetics Tumours of the Breast and Female Genital Organs (National Comprehensive Cancer Network, 2003). Estrogen receptor (ER), progesterone receptor (PR), and HER2 status were abstracted from the medical records of patients. All surgical specimens were embedded in paraffin, sectioned into 5-µm-thick slices, and subjected to conventional hematoxylin and eosin staining.
Genomic DNA was isolated from paraffin-embedded tissues using the phenol-chloroform method [17], dissolved in sterile double-distilled water, and stored at -80°C for 12-24 h to preserve DNA integrity.
SNP site selection and primer design
In the present study, six Notch SNPs (rs3124591, rs11249433, rs3815188, rs1043994, rs367398, and rs520692) were selected from the National Center for Biotechnology Information SNP database. Association studies for these polymorphisms have been reported previously [10-13]. Primers for polymerase chain reaction (PCR) were designed using Sequenom MassARRAY Assay Design 3.0 (Sequenom, San Diego, CA, USA).
PCR base-specific cleavage and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS)
The PCR reactions were carried out in a 5 μL reaction system containing 25 mM MgCl2, 25 μM deoxyribonucleotides (dNTPs), 0.1 pmol/μL of each primer, 0.5 U Hot Star Taq DNA polymerase (Qiagen, Chatsworth, CA, USA), and buffer supplied with the enzyme (final concentration 1 ×). The reaction mix was preactivated for 4 min at 94°C. The reactions were carried out for 45 cycles of 94°C for 20 s, 56°C for 30 s, and 72°C for 1 min, followed by elongation at 72°C for 3 min. dNTPs for amplification were dephosphorylated by adding 1.53 μL H2O, 0.17 μL shrimp alkaline phosphatase (SAP) buffer (10 ×), and 0.51 U (0.3 μL) SAP. The reaction was incubated at 37°C for 40 min, 85°C for 5 min, and 12°C for 30 min. The single base extension reaction was performed in a 9 μL reaction volume containing 0.619 μL H2O, 0.2 μL iPLEX Buffer Plus (10 ×), 0.2 μL iPLEX Termination mix, 0.94 μL iPLEX Extend Primer mix, 0.041 μL iPLEX enzyme (iPLEX, Sequenom), 5 μL PCR reaction product, and 2 μL SAP treatment product. The extension cycling conditions were as follows: 94°C for 30 s, followed by 94°C for 5 s, 52°C for 5 s, 80°C for 1 min, and 72°C for 3 min. The cleavage products were desalted with 6 mg of cationic resin mixture (Sequenom), sealed, and incubated for 10 min. Desalted cleavage products were assessed by spotting onto a 384-element matrix arrayed silicon SpectroCHIP (Sequenom). Mass spectra were collected using a MassARRAY™ system mass spectrometer (Sequenom), and TYPER4.0 was used to analyze the data.
Immunohistochemistry
To study the expression of Notch1 protein, we used FFPE tissue samples collected from successfully genotyped patients with IDC and DCIS. Several serial sections from different regions of the tumors were stained using rabbit anti-human Notch1 antibody (SIGMA#N4788) for determining protein expression. Staining and evaluation was performed as described earlier [18,19]. For comparison, all sections were processed in parallel.
Statistical analysis
Each SNP was tested for deviation from the Hardy-Weinberg equilibrium (HWE) using chi-squared (χ2) or Fisher’s exact tests depending on allele and genotype frequency (Online Encyclopedia for Genetic Epidemiology studies, USA: http://www.oege.org/software/hwe-mr-calc.shtml). The associations of each genotype or combinations of genotypes with clinicopathological features of IDC patients were compared using the χ2 test for categorical variables. The association between rs3124591 genotypes and Notch1 expression was also analyzed using the χ2 test. All association analyses were performed using the Statistical Package for the Social Sciences version 17.0 (SPSS, Chicago, IL, USA). All P-values were two-sided, and P-values < 0.05 were considered statistically significant.
Results
SNP detection
SNP alleles were determined using a MALDI-TOF MS-based approach followed by statistical analysis. MALDI-TOF MS has been a powerful and reliable tool for the determination of a limited number of genetic variants in tissue sample sets. The Notch gene polymorphisms were prudently detected in the study population. The rate of successful genotyping for the Notch gene polymorphisms in IDC, DCIS, and UDH patients ranged from 80% to 97%. Genotype distribution and allele frequencies for the six successfully genotyped Notch SNPs in all IDC, DCIS, and control specimens are shown in Table 1. The genotype distribution of all polymorphisms followed HWE in the UDH controls (all P > 0.05).
Table 1.
Genotype and allele frequencies of Notch polymorphisms in patients with IDC and DCIS and in controls
| Notch polymorphisms | Genotypes | No. of IDC/DCIS/UDH | IDC vs. UDH | DCIS vs. UDH | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| P | OR (95% CI) | P | OR (95% CI) | |||
| Notch1 (rs3124591) | CC | 0/0/0 | _ | _ | _ | _ |
| TC | 20/12/8 | 0.002 | 3.708 (1.534, 8.966) | 0.011 | 3.482 (1.283, 9.449) | |
| TT | 61/28/89 | 1 | 1 | |||
| CC + TC | 11/12/8 | 0.002 | 3.708 (1.534, 8.966) | 0.011 | 3.482 (1.283, 9.449) | |
| Allele | C | 20/12/8 | 0.566 | 0.798 (0.369, 1.727) | 0.002 | 4.103 (1.608, 10.469) |
| T | 142/68/186 | 1 | 1 | |||
| Notch2 (rs11249433) | CC | 0/0/0 | _ | _ | _ | _ |
| TC | 9/5/8 | 0.817 | 1.125 (1.414, 3.054) | 0.933 | 1.265 (0.390, 4.104) | |
| TT | 85/42/85 | 1 | 1 | |||
| CC + TC | 9/5/8 | 0.817 | 1.125 (1.414, 3.054) | 0.933 | 1.265 (0.390, 4.104) | |
| Allele | C | 9/5/8 | 0.821 | 1.119 (0.422, 2.965) | 0.935 | 1.250 (0.397, 3.932) |
| T | 179/89/178 | 1 | 1 | |||
| Notch3 (rs3815188) | AA | 15/6/14 | 0.803 | 0.879 (0.382, 2.104) | 0.524 | 0.701 (0.235, 2.093) |
| GA | 33/17/42 | 0.196 | 0.658 (0.348, 1.242) | 0.296 | 0.662 (0.306, 1.436) | |
| GG | 43/22/36 | 1 | 1 | |||
| AA + GA | 48/23/56 | 0.267 | 0.718 (0.399, 1.291) | 0.278 | 0.672 (0.327, 1.379) | |
| Allele | A | 63/29/70 | 0.495 | 0.862 (0.563, 1.321) | 0.315 | 0.762 (0.447, 1.297) |
| G | 119/61/14 | 1 | 1 | |||
| Notch3 (rs1043994) | AA | 3/1/1 | 0.582 | 3.227 (0.328, 31.80) | 0.532 | 2.219 (0.135, 36.60) |
| GA | 20/12/18 | 0.627 | 1.195 (0.582, 2.455) | 0.36 | 1.479 (0.638, 3.431) | |
| GG | 66/32/71 | 1 | 1 | |||
| AA + GA | 23/13/19 | 0.455 | 1.302 (0.651, 2.606) | 0.316 | 1.518 (0.669, 3.446) | |
| Allele | A | 26/14/20 | 0.323 | 1.368 (0.733, 2.553) | 0.299 | 1.474 (0.706, 3.075) |
| G | 152/76/160 | 1 | 1 | |||
| Notch4 (rs367398) | AA | 6/6/5 | 0.847 | 1.131 (0.324, 3.944) | 0.318 | 2.352 (0.653, 8.467) |
| GA | 38/17/36 | 0.986 | 0.995 (0.546, 1.812) | 0.84 | 0.926 (0.437, 1.962) | |
| GG | 52/25/49 | 1 | 1 | |||
| AA + GA | 44/23/41 | 0.97 | 1.011 (0.568, 1.801) | 0.791 | 1.100 (0.545, 2.219) | |
| Allele | A | 50/29/47 | 0.962 | 1.011 (0.637, 1.606) | 0.32 | 1.322 (0.762, 2.294) |
| G | 142/63/135 | 1 | 1 | |||
| Notch4 (rs520692) | GG | 0/0/2 | 0.234 | 1.030 (0.989, 1.074) | 0.543 | 1.030 (0.989, 1.074) |
| AG | 21/11/25 | 0.444 | 0.770 (0.394, 1.504) | 0.606 | 0.807 (0.356, 1.826) | |
| AA | 72/36/66 | 1 | 1 | |||
| GG + AG | 21/11/27 | 0.315 | 0.713 (0.368, 1.381) | 0.479 | 0.747 (0.332, 1.679) | |
| Allele | G | 21/11/29 | 0.224 | 0.689 (0.377, 1.259) | 0.38 | 0.717 (0.341, 1.509) |
| A | 165/83/157 | 1 | 1 | |||
Genotype and allele frequencies of Notch gene polymorphisms
As shown in Table 1, the distribution of the rs3124591 TC genotype was significantly different between IDC and DCIS patients and UDH controls. Using the TT genotype as a reference, the TC genotype was significantly associated with an increased risk of IDC and DCIS (P = 0.002, odds ratio [OR] = 3.708; P = 0.011, OR = 3.482). Interestingly, the mutant homozygous genotype CC was absent in our study population. In addition, the C allele of rs3124591 was significantly associated with high risk of DCIS (P = 0.002, OR = 4.104) but not IDC (P = 0.566, OR = 0.798). However, significant associations of other SNP genotypes and alleles with IDC or DCIS risk were not observed. The genotype and allele frequencies of rs11249433, rs3815188, rs1043994, rs367398, and rs520692 were similar between IDC and DCIS patients and UDH controls (P > 0.05).
Association between the rs3124591 polymorphism and Notch1 protein expression in IDC and DCIS patients
To examine the correlation between the rs3124591 polymorphism and Notch1 protein expression in IDC and DCIS patients, immunohistochemistry was performed on formalin-fixed, paraffin-embedded tissue sections from 81 IDC patients and 40 DCIS patients. We found that Notch1 protein expression was significantly higher in IDC patients compared with DCIS patients (72.8% [69/81] vs. 42.5% [17/40]; P < 0.001; Figure 1). Expectedly, as shown in Table 2, Notch1 protein expression was significantly higher in DCIS patients with the TC genotype (P = 0.043). Although Notch1 protein expression was higher in IDC patients with the TC genotype, this association did not reach significance (P = 0.159).
Figure 1.
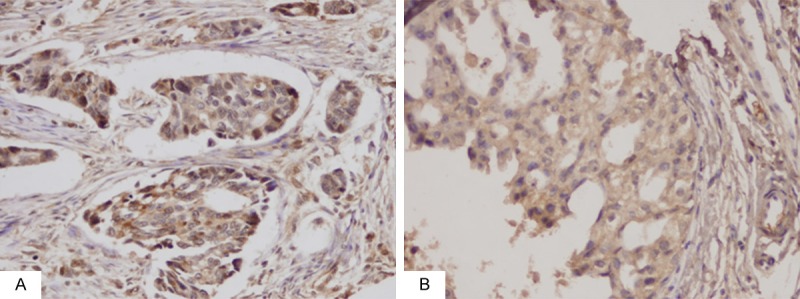
Significantly higher expression of Notch1 protein in IDC was observed than that in DCIS. A. Expression of Notch1 protein in IDC (original magnification, 200 ×). B. Expression of Notch1 protein in DCIS (original magnification, 200 ×).
Table 2.
Correlations between rs3124591 genotype and Notch1 expression in IDC and DCIS
| IDC | |||
| Notch1 expression | Genotype | P | |
|
|
|||
| TT (n) | TC (n) | ||
| High | 42 | 17 | 0.159 |
| Low | 19 | 3 | |
| DCIS | |||
| Notch1 expression | Genotype | P | |
|
|
|||
| TT (n) | TC (n) | ||
| High | 9 | 8 | 0.043 |
| Low | 19 | 4 | |
Associations of the rs3124591 TC genotype and Notch1 protein expression with clinicopathological features of IDC
In light of the significant association of the rs3124591 TC genotype with IDC development, we determined the association of this genotype with clinicopathological features of IDC. The details of this analysis are shown in Table 3. We found that the frequency of the TC genotype was significantly higher in poorly differentiated tumors than in well-differentiated and moderately differentiated tumors. However, the distribution of this SNP genotype was not affected by age, lymph node metastasis, or ER, PR, or HER-2 status. Additionally, the expression of Notch1 protein was found to be significantly higher in IDC patients with lymph node metastasis than in those without lymph node metastasis. In contrast, significant associations of Notch1 protein expression with age, tumor differentiation, or ER, PR, or HER-2 status were not observed.
Table 3.
Association of rs3124591 genotype and Notch1 protein expression with clinicopathologic features in patients with IDC
| Clinicopathological parameters | Notch1 genotype | P | Notch1 expression | P | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| TT (n) | TC (n) | High (n) | Low (n) | |||
| Age at diagnosis | ||||||
| < 51 years | 30 | 14 | 0.105 | 29 | 15 | 0.126 |
| > 51 years | 31 | 6 | 30 | 7 | ||
| Lymph node status | ||||||
| Positive | 31 | 11 | 0.745 | 35 | 7 | 0.028 |
| Negative | 30 | 9 | 24 | 15 | ||
| Tumor differentiation | ||||||
| Well & moderate | 53 | 12 | 0.022 | 47 | 18 | 1.000 |
| Poor | 8 | 8 | 12 | 4 | ||
| ER status | ||||||
| ER (-/+) | 28 | 13 | 0.138 | 28 | 13 | 0.352 |
| ER (2+/3+) | 33 | 7 | 31 | 9 | ||
| PgR status | ||||||
| PgR (-/+) | 33 | 13 | 0.393 | 35 | 11 | 0.451 |
| PgR (2+/3+) | 28 | 7 | 24 | 11 | ||
| HER-2 status | ||||||
| HER-2 (-/+) | 23 | 11 | 0.174 | 27 | 7 | 0.258 |
| HER-2 (2+/3+) | 38 | 9 | 32 | 15 | ||
Discussion
Identification of shared genetic determinants for early prophylaxis and treatment of breast cancer is the emerging premise underlying the results of accumulative association studies. In the present study, we evaluated for the first time the associations of the more-widely studied Notch SNPs (Notch1, rs3124591; Notch2, rs11249433; Notch3, rs1043994, rs3815188; Notch4, rs520692 and rs367398) with breast cancer risk to better characterize associations across the Notch gene in a Chinese population. We found that the rs3124591 variant was associated with an increased risk of IDC and DCIS and poorly differentiated IDC. Moreover, we found that the mutant heterozygote TC genotype of rs3124591 was positively associated with high Notch1 protein expression in DCIS but not in IDC, suggesting a possible mechanism for this variant in the early stage of Notch1-associated IDC. However, further studies with larger sample sizes are needed to confirm the role and mechanism of the Notch1 SNP rs3124591 in IDC development.
In recent years, significant research efforts have focused on identifying genetic variants of the Notch gene that are associated with breast cancer [13,16,20,21]. Several lines of evidence have demonstrated the presence of Notch gene polymorphisms in various non-tumorous diseases. For instance, the Notch3 rs3815188 polymorphism has been observed in patients with CADASIL in two reports [11,22], and the Notch4 rs367398 and rs520692 polymorphisms are frequently found in patients with psychopathic disorders [12,23]. However, few studies have reported the association between Notch gene polymorphisms and breast cancer risk with the exception of the Notch2 SNP rs11249433. In this study, we examined the common variants in Notch genes in UDH, DCIS, and IDC patients, including the Notch1 SNP rs3124591 (exon 34 of 9q34), Notch2 SNP rs11249433 (pericentromeric region of 1p11.2), Notch3 SNPs rs3815188 and rs1043994 (exon 3 and 4 of 19p13.1, respectively), and Notch4 SNPs rs520692 and rs367398 (exon 5 and promoter region of 6p21.3, respectively). To the best of our knowledge, this is the first study to investigate the presence of the rs3124591 genotype in IDC and DCIS patients in a Chinese population. Although our study group was small, we showed that the frequency of the rs3124591 TC genotype was significantly higher in IDC and DCIS patients than in UDH controls. This finding suggests that the rs3124591 TC genotype is associated with an increased risk of IDC, which is supported by the association of this variant genotype with poorly differentiated IDCs.
We also evaluated the association of Notch2 (rs11249433), Notch3 (rs3815188 and rs1043994) and Notch4 (rs367398 and rs520692) polymorphisms with the risk of IDC development. The distribution of rs11249433 alleles and genotypes was not different between Chinese IDC and DCIS patients and UDH controls, which is consistent with previous studies showing a negative association between the SNP rs11249433 and breast cancer in the Asian population [14]. Other studies have also shown that the rs11249433 polymorphism is not associated with breast cancer risk [24-25]. Fu et al. [24] demonstrated that the rs11249433 polymorphism was not associated with overall breast cancer risk but rather with an increased risk of ER-positive breast tumors without TP53 mutations. This increased risk was associated with elevated Notch2 expression. Jiang et al. [25] also found no obvious association between rs11249433 polymorphism and breast cancer risk in Chinese women. Additionally, a meta-analysis of Notch polymorphisms and breast cancer risk revealed an increased risk of breast cancer exclusively for ER-positive tumors in Caucasians but not Asians with the rs11249433 polymorphism on 1p11 [14]. These studies suggest that the association of the rs11249433 polymorphism with IDC risk is influenced by ethnicity and breast cancer subgroups. In the present study, we failed to show any significant correlation of the rs11249433 polymorphism with the risk of ER-positive breast cancer. However, we were likely unable to obtain an accurate estimate of this association because of the small sample size in our study. We found that the allele and genotype frequencies of the Notch3 polymorphisms rs3815188 and rs1043994 and the Notch4 polymorphisms rs367398 and rs520692 in IDC and DCIS patients were similar to those in UDH controls, suggesting that these SNPs were not associated with breast cancer risk. To our knowledge, this study is the first to analyze the association of Notch3 and Notch4 SNPs with breast cancer risk. Our findings require further validation in well-designed studies with larger sample sizes.
In consideration of the significant association of the rs3124591 polymorphism with IDC risk, we hypothesized that this SNP variant may influence IDC development through the regulation of Notch1 protein expression. Accumulative evidences have revealed that Notch1 is mutated in a high proportion of mouse mammary tumor virus-induced tumors, these mutations lead to a high expression of truncated Notch1 proteins and poor survival [26,27]. Therefore, we examined the association between the rs3124591 polymorphism and Notch1 protein expression in 81 IDC and 40 DCIS patients and found that the expression of Notch1 protein was significantly higher in IDC patients than in DCIS patients. Moreover, a higher expression of Notch1 protein was observed in IDC patients with lymph node metastasis than in those without lymph node metastasis, which is in accordance with a previous study [21]. Importantly, high frequency of the TC genotype in DCIS patients was associated with a concomitant increase in Notch1 protein expression, which was not found in IDC patients. This suggests that the impact of the rs3124591 variant on Notch1 protein expression may mainly occur early in IDC development. Given the inconsistent associations between the rs3124591 variant and Notch1 expression in IDC and DCIS, this variant may affect IDC risk through mechanisms in the latter stage other than alterations in Notch1 protein expression. A previous report has indicated that hypomethylation of the Notch1 oncogene may affect Notch1 expression in IDC patients [18]. The exact mechanism accounting for the influence of the rs3124591 variant on IDC risk remains to be determined.
In conclusion, our study demonstrates for the first time the association of the Notch1 SNP rs3124591 with IDC and DCIS risk. In addition, a link between the rs3124591 variant and unregulated Notch1 protein expression was indicated in DCIS patients but not in IDC patients, suggesting the involvement of this variant in the early stage of Notch1-associated IDC development.
Disclosure of conflict of interest
None.
References
- 1.Wang YC, Wei LJ, Liu JT, Li SX, Wang QS. Comparison of Cancer Incidence between China and the USA. Cancer Biol Med. 2012;9:128–132. doi: 10.3969/j.issn.2095-3941.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 2006;107:2223–2233. doi: 10.1182/blood-2005-08-3329. [DOI] [PubMed] [Google Scholar]
- 3.Alfadhli S, Nanda A. Genetic evidence for the involvement of NOTCH4 in rheumatoid arthritis and alopecia areata. Immunol Lett. 2013;150:130–133. doi: 10.1016/j.imlet.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Parr C, Watkins G, Jiang WG. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancer. Int J Mol Med. 2004;14:779–786. doi: 10.3892/ijmm.14.5.779. [DOI] [PubMed] [Google Scholar]
- 5.Hu C, Dievart A, Lupien M, Calvo E, Tremblay G, Jolicoeur P. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol. 2006;168:973–990. doi: 10.2353/ajpath.2006.050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamaguchi N, Oyama T, Ito E, Satoh H, Azuma S, Hayashi M, Shimizu K, Honma R, Yanagisawa Y, Nishikawa A, Kawamura M, Imai J, Ohwada S, Tatsuta K, Inoue J, Semba K, Watanabe S. NOTCH3 signaling pathway plays crucial roles in the proliferation of ErbB2-negative human breast cancer cells. Cancer Res. 2008;68:1881–1888. doi: 10.1158/0008-5472.CAN-07-1597. [DOI] [PubMed] [Google Scholar]
- 7.Speiser J, Foreman K, Drinka E, Godellas C, Perez C, Salhadar A, Ersahin C, Rajan P. Notch-1 and Notch-4 biomarker expression in triple-negative breast cancer. Int J Surg Pathol. 2012;20:139–145. doi: 10.1177/1066896911427035. [DOI] [PubMed] [Google Scholar]
- 8.Rubino E, Fenoglio P, Gallone S, Govone F, Vacca A, De Martino P, Giobbe ML, Boschi S, Pinessi L, Gentile S, Rainero I. Genetic variants in the NOTCH4 gene influence the clinical features of migraine. J Headache Pain. 2013;14:28. doi: 10.1186/1129-2377-14-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menon S, Cox HC, Kuwahata M, Quinlan S, MacMillan JC, Haupt LM, Lea RA, Griffiths LR. Association of a Notch 3 gene polymorphism with migraine susceptibility. Cephalalgia. 2011;31:264–270. doi: 10.1177/0333102410381143. [DOI] [PubMed] [Google Scholar]
- 10.Mohamed SA, Aherrahrou Z, Liptau H, Erasmi AW, Hagemann C, Wrobel S, Borzym K, Schunkert H, Sievers HH, Erdmann J. Novel missense mutations (p. T596M and p.P1797H) in NOTCH1 in patients with bicuspid aortic valve. Biochem Biophys Res Commun. 2006;345:1460–1465. doi: 10.1016/j.bbrc.2006.05.046. [DOI] [PubMed] [Google Scholar]
- 11.Roy B, Maksemous N, Smith RA, Menon S, Davies G, Griffiths LR. Two novel mutations and a previously unreported intronic polymorphism in the NOTCH3 gene. Mutat Res. 2012;732:3–8. doi: 10.1016/j.mrfmmm.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Wei J, Yu YQ, Liu SZ, Shi JP, Liu LL, Ju GZ, Yang JZ, Zhang D, Xu Q, Shen Y, Hemmings GP. Is NOTCH4 associated with schizophrenia? Psychiatr Genet. 2004;14:43–46. doi: 10.1097/00041444-200403000-00007. [DOI] [PubMed] [Google Scholar]
- 13.Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, Hankinson SE, Hutchinson A, Wang Z, Yu K, Chatterjee N, Garcia-Closas M, Gonzalez-Bosquet J, Prokunina-Olsson L, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Diver R, Prentice R, Jackson R, Kooperberg C, Chlebowski R, Lissowska J, Peplonska B, Brinton LA, Sigurdson A, Doody M, Bhatti P, Alexander BH, Buring J, Lee IM, Vatten LJ, Hveem K, Kumle M, Hayes RB, Tucker M, Gerhard DS, Fraumeni JJ, Hoover RN, Chanock SJ, Hunter DJ. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu S, Cai J, Wang H, Zhang H, Yang W. Association between 1p11-rs11249433 Polymorphism and Breast Cancer Susceptibility: evidence from 15 Case-Control Studies. PLoS One. 2013;8:e72526. doi: 10.1371/journal.pone.0072526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao X, Wood LD, Lindman M, Jones S, Buckhaults P, Polyak K, Sukumar S, Carter H, Kim D, Karchin R, Sjoblom T. Somatic mutations in the Notch, NF-KB, PIK3CA, and Hedgehog pathways in human breast cancers. Genes Chromosomes Cancer. 2012;51:480–489. doi: 10.1002/gcc.21935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campa D, Kaaks R, Le Marchand L, Haiman CA, Travis RC, Berg CD, Buring JE, Chanock SJ, Diver WR, Dostal L, Fournier A, Hankinson SE, Henderson BE, Hoover RN, Isaacs C, Johansson M, Kolonel LN, Kraft P, Lee IM, McCarty CA, Overvad K, Panico S, Peeters PH, Riboli E, Sanchez MJ, Schumacher FR, Skeie G, Stram DO, Thun MJ, Trichopoulos D, Zhang S, Ziegler RG, Hunter DJ, Lindstrom S, Canzian F. Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. J Natl Cancer Inst. 2011;103:1252–1263. doi: 10.1093/jnci/djr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi SR, Datar R, Liu C, Wu L, Zhang Z, Cote RJ, Taylor CR. DNA extraction from archival formalin-fixed, paraffin-embedded tissues: heat-induced retrieval in alkaline solution. Histochem Cell Biol. 2004;122:211–218. doi: 10.1007/s00418-004-0693-x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang N, Sun ZZ, Li F, Cao YW, Zhao CX, Liang WH, Sun HP, Li HA, Fu XG. [Detection and clinical significance of Notch1 methylation in breast cancer and intraductal proliferative breast lesions] . Zhonghua Bing Li Xue Za Zhi. 2011;40:324–329. [PubMed] [Google Scholar]
- 19.Wu M, Wei W, Xiao X, Guo J, Xie X, Li L, Kong Y, Lv N, Jia W, Zhang Y, Xie X. Expression of SIRT1 is associated with lymph node metastasis and poor prognosis in both operable triple-negative and non-triple-negative breast cancer. Med Oncol. 2012;29:3240–3249. doi: 10.1007/s12032-012-0260-6. [DOI] [PubMed] [Google Scholar]
- 20.Yao K, Rizzo P, Rajan P, Albain K, Rychlik K, Shah S, Miele L. Notch-1 and notch-4 receptors as prognostic markers in breast cancer. Int J Surg Pathol. 2011;19:607–613. doi: 10.1177/1066896910362080. [DOI] [PubMed] [Google Scholar]
- 21.Ma D, Dong X, Zang S, Ma R, Zhao P, Guo D, Dai J, Chen F, Ye J, Ji C. Aberrant expression and clinical correlation of Notch signaling molecules in breast cancer of Chinese population. Asia Pac $lxfS1$ 2011;7:385–391. doi: 10.1111/j.1743-7563.2011.01433.x. [DOI] [PubMed] [Google Scholar]
- 22.Testi S, Malerba G, Ferrarini M, Ragno M, Pradotto L, Mauro A, Fabrizi GM. Mutational and haplotype map of NOTCH3 in a cohort of Italian patients with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) J Neurol Sci. 2012;319:37–41. doi: 10.1016/j.jns.2012.05.025. [DOI] [PubMed] [Google Scholar]
- 23.Shibata N, Ohnuma T, Higashi S, Higashi M, Usui C, Ohkubo T, Watanabe T, Kawashima R, Kitajima A, Ueki A, Nagao M, Arai H. Genetic association between Notch4 polymorphisms and Alzheimer’s disease in the Japanese population. J Gerontol A Biol Sci Med Sci. 2007;62:350–351. doi: 10.1093/gerona/62.4.350. [DOI] [PubMed] [Google Scholar]
- 24.Fu YP, Edvardsen H, Kaushiva A, Arhancet JP, Howe TM, Kohaar I, Porter-Gill P, Shah A, Landmark-Hoyvik H, Fossa SD, Ambs S, Naume B, Borresen-Dale AL, Kristensen VN, Prokunina-Olsson L. NOTCH2 in breast cancer: association of SNP rs11249433 with gene expression in ER-positive breast tumors without TP53 mutations. Mol Cancer. 2010;9:113. doi: 10.1186/1476-4598-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang Y, Shen H, Liu X, Dai J, Jin G, Qin Z, Chen J, Wang S, Wang X, Hu Z, Shen H. Genetic variants at 1p11.2 and breast cancer risk: a twostage study in Chinese women. PLoS One. 2011;6:e21563. doi: 10.1371/journal.pone.0021563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girard L, Hanna Z, Beaulieu N, Hoemann CD, Simard C, Kozak CA, Jolicoeur P. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 27.Guo S, Liu M, Gonzalez-Perez RR. Role of Notch and its oncogenic signaling crosstalk in breast cancer. Biochim Biophys Acta. 2011;1815:197–213. doi: 10.1016/j.bbcan.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]


