Abstract
We have compared mutation analysis by Amplification Refractory Mutation System (ARMS) and epidermal growth factor receptor (EGFR) mutant-specific antibodies for their ability to detect two common activating EGFR mutations in a cohort of 115 advanced non-small cell lung cancer (NSCLC), including cytology material, core biopsy, and bronchoscopic biopsies. Assessment of EGFR mutation status was performed by using antibodies and ARMS assay specific to the two major forms of mutant EGFR, exon 19 deletion E746-A750 (c.2235_2249del15 or c.2236_2250del15, p. Glu746_Ala750 del) and exon 21 L858R point mutation (c.2573T>G, p.Leu858Arg). In this study the optimal buffer for antigen retrieval was sodium citrate (pH 6.0). Q score was used to evaluate the specific mutant EGFR proteins expression. Validation using clinical material showed deletions in exon 19 were detected in 19.1% and L858R mutation in 20% of all cases by ARMS assay. A cutoff value of score 1 was used as positive by IHC. No wild type cases were immuno-reactive. The antibodies performed well in cytology, core biopsies and bronchoscopic biopsies. There were only one false positive case using L858R IHC (sensitivity 100%, specificity 98.5%, positive predictive value 96%, negative predictive value 100%). All 23 E746-A750 exon 19 deletions identified by mutation analysis were positive by IHC. The sensitivity of exon 19 IHC for E746-A750 was 100%, specificity 100%, positive predictive value 100% and negative predictive value 100%. The result of the IHC stains was finely correlated with mutations status determined by ARMS assay. Although inferior to molecular genetic analysis of the EGFR gene, IHC is highly specific and sensitive for the targeted EGFR mutations. The antibodies are likely to be of clinical value in cases especially where limited tumor material is available, or in situations where molecular genetic analysis is not readily available.
Keywords: NSCLC, cytology, EGFR mutation, immunohistochemistry
Introduction
Identification of tumors harboring sensitizing epidermal growth factor receptor (EGFR) mutations is important in selecting patients likely to respond to EGFR-tyrosine kinase inhibitor (TKI) therapy. Somatic mutations within the tyrosine kinase (TK) domain of the epidermal growth factor receptor (EGFR) gene are found in approximately 40% of lung adenocarcinomas and a small number of squamous cell cancer in Asian populations [1,2]. This discovery caused a wave of enthusiasm in the therapy of such an aggressive tumor. Study of the mutational state of EGFR became a matter of urgent necessity in patients with advanced non-small cell lung cancer. Activating mutations in EGFR occur in exons 18-21 in non-small cell lung cancer with in-frame deletions in exon 19 (most frequently E746-A750) and the L858R missense mutation in exon 21 (Leu858Arg) being the commonest, accounting for approximately 80-90% of cases in most published studies [3,4].
There is still no standardized test approved and the current diversity of methods for conducting this test is creating serious logistical problems worldwide. Direct sequencing of PCR products has been the most commonly used methodology for this purpose. The main drawbacks of this method are its low sensitivity (20-50%) and the significant risk of contamination involved in handling post-PCR products [5]. There is a continuing clinical demand and application for technology that can detect very low levels of mutant EGFR DNA amongst a high wild type (WT) background before anti-EGFR therapy decision. Recent advances in molecular techniques have enabled the development of more sensitive methods for detecting mutations with real-time quantitative PCR, using specific probes or amplified refractory mutation system (ARMS) technology.
Most recently, the development of EGFR mutant-specific antibodies for immunohistochemistry (IHC) has presented a new method for consideration [6,7]. Many molecular tests are available for EGFR mutation detection, but they are less widely available and generally have longer turnaround times than immunohistochemistry (IHC). Molecular tests are also relatively expensive and require larger amounts of tumor tissue than IHC. Although several independent groups have investigated the sensitivity and specificity of these antibodies in the detection of EGFR mutations in non-small cell lung cancer (NSCLC). Most of them confirmed a high degree of specificity, but the reported sensitivities were quite variable ranging from 24% to 100% [6,8]. This inconsistency may be related to differences in methodology and interpretation, as well as population specific differences in gene mutations and differences in the level of protein expression. None of them systematically study or compare IHC and ARMS in biopsy and cytology specimens, which suggests that further study is needed before EGFR mutation-specific IHC can be implemented as a clinical tool.
In the study reported here we optimized the methodology and interpretive aspects of IHC for detection of EGFR mutations, and evaluated the success of this effort by comparison with ARMS technology. This study investigated the staining protocol, staining pattern, scoring methods, and cut off value to determine the diagnostic power of EGFR mutation-specific IHC in advanced Chinese NSCLC patients.
Methods
Patient samples
Samples for study were selected according to the following criteria: Advanced non small cell lung carcinoma, cytology or biopsy material available, and no pre-TKI therapy. A total of 115 cases were collected retrospectively from the Department of Pathology, Southeast University Zhongda Hospital during November 2011 to April 2014. All specimens were dissected and immersed in 10% neutral buffered formalin, then fixed overnight. Tissue specimens and cytology cell blocks were processed by standard automatic tissue processor and embedded in paraffin block routinely. Informed consent for the use of these specimens for medical studies was obtained.
Immunohistochemistry
115 tissue blocks were cut into 4-μm-thick whole sections. EGFR mutation specific antibodies were Rabbit XPW mAbs obtained from Cell Signaling Technology (Danvers, MA), 6B6 specific for the E746-A750 del mutation, and 43B2 for the L858R mutation. The antibodies were diluted 1:150 with antigen retrieval buffer. The antigen retrieval buffers tested were sodium citrate (pH 6.0), EDTA (pH 7.8) and EDTA (pH 8.8), and sodium citrate (pH 6.0) was chosen for the highest sensitivity with the lowest background. Cytokeratin 7 IHC was used as a quality control for tissue and protocol. Conditions and dilutions for the antibodies were optimized in the laboratory according to manufacturer’s instructions.
IHC scoring
The expression of mutant EGFR proteins was scored according to the quickscore [9,10] (Q score) based on estimating the percentage (P) of staining tumor cells (0-100%) and the intensity (I) of staining (0, complete absence of staining; 1, faint cytoplasmic staining; 2, moderate and incomplete membranous staining; 3, strong membranous staining). the slides were scored by multiplying the percentage of positive tumor cells by the intensity (Q=P×I; maximum=300). A positive result was Q score≥1. Both the intensity and percentage of stained cells were assessed at low magnification (objective magnification ×10). The distribution of staining, membrane or cytoplasm, was assessed at high magnification (objective magnification ×40). Four experienced pathologists (Lihua Zhang, Xueqing Wang, Ruiping Li and Xiang Fan) reviewed all of the slides independently, and then replicated the analysis 4 to 6 weeks later. Discrepant cases were reviewed In a multi-head microscope and reported as a consensus.
ARMS assay
The DNA extraction was performed with QIAampTM DNA FFPE Tissue kit and automated on the One-drop 1000 according to the manufacturer’s instructions. The patients’ DNA was tested by using AD×EGFR Mutations Detection Kit (Amoy Diagnostics, Xiamen, China), which has received State Food and Drug Administration (SFDA)’s approval for clinical usage in mainland China. The kit used the principle of Amplified Refractory Mutation System (ARMS) and covered the 19E746_A750 deletions or exon 21 L858R point mutations. The assay was carried out according to the manufacturer’s protocol with the ABI Step-one Plus real-time PCR system. A positive or negative result could be reached if it met the criterion that was defined by the manufacturer’s instruction.
Ethical approval
All experiments above have been performed with the approval of Southeast University Zhongda Hospital Ethics Committee.
Results
Regarding the pre-analytical phase in the study of EGFR mutations, it is important to note that microdissection was performed on 65 of the 115 tumors analyzed (45%). After DNA extraction, at least 80 ng/ml of DNA were obtained from 75% of the samples. Of the samples that yielded a lower DNA concentration, 100% were cytology material.
EGFR mutations were more common in females and in never smokers. Although the relationship was not statistically significant in our data, EGFR mutations occurred also more frequently in ACs with acinar, papillary and leptic patterns compared to other histological types (Table 1). Interestingly, it is important to note that our EGFR detection rates for the cytology material and small biopsies (bronchoscopic and CNBs) were similar to the rates for surgical specimens in our laboratory (data not shown): 45 out of 115 (39%) and 19 out of 46 (41%), respectively.
Table 1.
Clinicopathological characteristics in lung cancer patients
| Total (n=115) | Biopsy | Cytology | |
|---|---|---|---|
| Age | |||
| ≤60 | 45 | 38 | 7 |
| >60 | 70 | 56 | 14 |
| Gender | |||
| M | 71 | 56 | 15 |
| F | 44 | 38 | 6 |
| Histology | |||
| Adenocarcinoma with acinar patterns | 49 | 49 | - |
| Adenocarcinoma with papillary and leptic patterns | 8 | 8 | - |
| Adenocarcinoma with solid patterns | 56 | 56 | - |
| Squamous cell carcinoma | 2 | 2 | - |
| TNM stage | |||
| III | 48 | 48 | 0 |
| IV | 67 | 46 | 21 |
IHC results
The staining distribution included cytoplasm only or cytoplasm together with membrane (Figure 1). Normal tissue adjacent to carcinoma was negative. In our study, 95% of cases the tumor cells were stained in some areas and completely negative in other areas. Only in 15% of the biopsy cases were either negative or positive in 100% of the tumor cells, and the intensity score was ranging from 2 to 3. Overall, the staining pattern showed characteristics of heterogeneity more than homogeneity. Based on the scoring systems, for L858R-specific IHC the percentage of positive cases was 20% (23/115); for E746_A750 del-specific IHC it was 20% (23/115) (Table 2).
Figure 1.
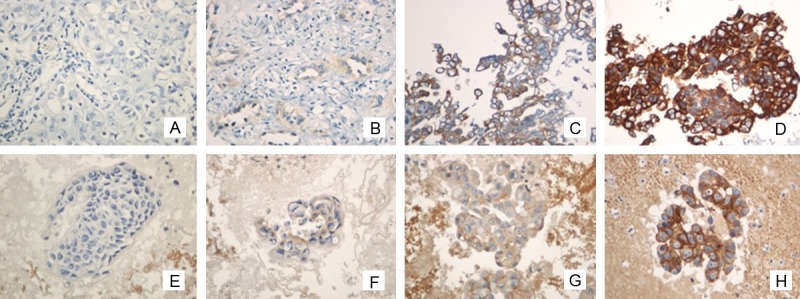
Representative immunostaining of both histology and cytology preparations in lung cancer patients. EGFR mutation specifi c antibodies are clearly stained in cancer cells and EGFR mutation was detected in both samples by DNA-based assay (×400, in all figures). Immunohistochemistry using anti- EGFR mutation showed that the tumor cells were Scored 0 in histology (A), scored 1 in histology (B), scored 2 in histology (C), scored 3 in histology (D), Scored 0 in cytology (E), Scored 1 in cytology (F), Scored 2 in cytology (G), and Scored 3 in cytology (H).
Table 2.
Summary of immunostaining results and mutation status based on ARMS assay
| ARMS assay | |||||
|---|---|---|---|---|---|
|
|
|||||
| Immunohistochemistry | mutation+ | mutation- | |||
|
|
|||||
| No | E746_A750 del | L858R | |||
| Positive (score≥1) | 46 | 45 (97.8%) | 1 (2.2%) | ||
| biopsy | 37 | 18 | 19 | 0 | |
| cytology | 9 | 5 | 3 | 1 | |
| Negative (score<1) | 69 | 0 (0%) | 69 (100%) | ||
| biopsy | 62 | 0 | 0 | 62 | |
| cytology | 7 | 0 | 0 | 7 | |
| Total | 115 | 45 (39.1) | 70 (60.9%) | ||
Concordance analysis of IHC and ARMS assay
The correlation between the expression of mutation-specific proteins and EGFR-mutational status is presented in Table 2. Of the 23 cases with E746_A750 del in exon19, an IHC score great than or equal to 1 was observed in all 23 cases. Of the 22 cases with L858R mutation in exon 21, an IHC score be equal or greater than 1 was observed in 23 (Table 2). Sensitivity, specificity, PPV, and NPV were calculated according to each IHC score (Table 3). When both sensitivity and specificity were taken into account, an IHC Score of 1 was defined as the cutoff point. For the detection of E746_A750 del by IHC, the sensitivity was 100%, the specificity was 100%, the PPV was 100%, and the NPV was 100%. For the detection of L858R in exon 21 by IHC, the sensitivity was 100%, the specificity was 98.5%, the PPV was 96%, and the NPV was 100%.
Table 3.
Diagnostic power of mutation-specifi c antibodies comparing with ARMS
| Mutation-specific antibodies | EGFR mutations | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|
| Anti-EGFR E746_A750 del | E746_A750 del | ||||
| score≥1 as positive | 100% | 100% | 100% | 100% | |
| score≥2 as positive | 41% | 100% | 100% | 77% | |
| score≥3 as positive | 53% | 91% | 75% | 79% | |
| Anti-EGFR L858R | L858R | ||||
| score≥1 as positive | 100% | 98.5% | 96% | 100% | |
| score≥2 as positive | 44% | 97% | 88% | 79% | |
| score≥3 as positive | 69% | 74% | 55% | 83% |
Discussion
Lung cancer presents a major public health problem and is responsible for the largest number of cancer related deaths world-wide for both men and women, causing approximately 1.2 million deaths per year. Approximately 70% of lung cancers are diagnosed in advanced stages where small biopsies and cytological specimens are the only source of material for both diagnosis and mutation testing [11,12]. The discovery that activating mutations in the epidermal growth factor receptor (EGFR) are associated with response to treatment with the small molecule tyrosine kinase inhibitors (TKI) has revolutionized the field of thoracic oncology. EGFR mutation status is the best predictor of response to tyrosine kinase inhibitors (TKIS) in primary lung NSCLC, varying by gender, ethnicity and smoking status. Approximately 40% of Asian patients with NSCLC of the lung harbor EGFR mutations [5]. The two most common EGFR mutations associated with NSCLC are in-frame deletions in exon 19 (the most common E746-A750 15-bp deletion) and the point mutation replacing leucine with arginine at codon 858 in exon 21 (L858R). These two mutations are responsible for 90% of the EGFR mutations in lung NSCLC patients [2,4].
Recently two monoclonal antibodies, specific to the two most common forms of mutated EGFR protein have become commercially available [6]. Early studies have shown promising results regarding the use of these antibodies in screening patients for TKI therapy [13]. In our current study we report our experience with these new antibodies in identifying mutant EGFR protein in cytology and small biopsy specimens. The results of mutation detection using immunohistochemistry were correlated to results obtained by amplification refractory mutation system (ARMS) assay which is known of high sensitivity and specificity.
Direct DNA sequencing and PCR based assays have been used as methods for detection of EGFR mutations in tumor tissue. Mutation testing in the clinical setting has become standard of care. Nonetheless, limitations of the use of molecular testing at the financial level include high costs of tests and reagents in China [14]. As a result, many patients that could benefit from targeted therapy with EGFR-TKI inhibitors are not tested. Most patients with pulmonary carcinoma are diagnosed with advanced disease, and are not candidates for surgical based therapy. In many cases, cytology, small biopsy material or samples from metastatic site may be the only tissue available for diagnostic, prognostic and predictive testing. Some of these limited samples, however, fail molecular testing mostly due to scant cellularity and the low quality of DNA [15]. Taking these considerations, a wider use and accessibility of immunohistochemistry for mutation specific antibodies could offer an alternative or adjunct testing to direct sequencing and other molecular methods.
Yu et al. generated monoclonal antibodies specific to exon 19 E746-A750 deletions and exon 21 L858R mutation and reported a sensitivity of the immunohistochemical assays of 92% in 340 cases of NSCL cancer specimens compared 99% for DNA sequencing. Recently, several studies examined the presence of EGFR mutations in NSCLC by IHC using the same two antibodies and the reported sensitivity ranged from 24% to 100% and specificity ranged from 77% to 100% [6,8,13]. The new mutant-specific antibody clones (SP111 and SP125) used by An Na Seo et al did not proved to harbor the high accuracy compared to the old ones (6B6 and 43B2) [8]. The molecular methods they use varied from direct DNA sequencing to PNA-LNA PCR clamp assay, and none of these studies systematically compared the ICH with ARMS for their ability to detect the two specific EGFR mutations in small biopsy and cytology samples in advanced lung cancer patients.
Ellison et al. reported ARMS routinely being able to detect at least 1% mutant in a back-ground of normal DNA [16] Our immunostaining results for the EGFR mutation-specific antibodies showed a sensitivity of 100%, specificity of 99%, positive predictive value of 100% and negative predictive value of 99%. The ARMS assays we used did not cover the several exon 19 deletions other than the most common E746_A750 del, which is the target of the exon 19 del antibodies, which explains the high sensitivity and specificity results. Our data confirmed that IHC assay is very reliable in detecting its specific mutation targets. Hopefully, improved antibodies that could detect other less frequent deletions and mutations could be available to strengthen the use of immunohistochemistry as a tool for EGFR mutations screening.
IHC is known to sometimes suffer from high inter-laboratory variability in assay performance, and high inter-observer variability in assay interpretation. These drawbacks may explain the variability in results of the studies described before. There is still much work to be done before IHC can be considered an adequate substitute for direct analysis of mutations in the EGFR gene in NSCLC. Antigen retrieval is the key step in IHC affecting the background. We found that slides treated by sodium citrate (pH 6.0) showed the best histological pictures with strongly specific staining and minimal background. Scoring system also plays a critical role in obtaining a reliable IHC result. In our study, we found the positivity cut off of score 1 improve sensitivity and did not result in false positive interpretations. This may due to our fine antigen retrieval and tissue processing.
In conclusion, we endorse an algorithm proposed for screening cases for EGFR mutations where cases positive on IHC would be reported as positive for specific mutations, whereas negative cases would still be subject to standard molecular testing for the other none-classic mutants. Another important observation is the fact that our results indicate the use of these antibodies on cytology and small biopsy samples is as efficient as ARMS. These two mutation specific antibodies can be used in a clinical setting. Their use may have a role in cases where the diagnostic material is unsatisfactory for molecular use such as in scant cell block slide or liquid-based cytology.
Acknowledgements
The authors wish to thank Xiaoli Zhu for clinical, laboratory and logistic support. This work was supported by grants from the natural science foundation of Jiangsu Province (BK2012750), and the Natural Science Foundation of China (81101856).
Disclosure of conflict of interest
None.
References
- 1.Cheng L, Alexander RE, Maclennan GT, Cummings OW, Montironi R, Lopez-Beltran A, Cramer HM, Davidson DD, Zhang S. Molecular pathology of lung cancer: key to personalized medicine. Mod Pathol. 2012;25:347–369. doi: 10.1038/modpathol.2011.215. [DOI] [PubMed] [Google Scholar]
- 2.Bonanno L, Schiavon M, Nardo G, Bertorelle R, Bonaldi L, Galligioni A, Indraccolo S, Pasello G, Rea F, Favaretto A. Prognostic and predictive implications of EGFR mutations, EGFR copy number and KRAS mutations in advanced stage lung adenocarcinoma. Anticancer Res. 2010;30:5121–5128. [PubMed] [Google Scholar]
- 3.Kawahara A, Yamamoto C, Nakashima K, Azuma K, Hattori S, Kashihara M, Aizawa H, Basaki Y, Kuwano M, Kage M, Mitsudomi T, Ono M. Molecular diagnosis of activating EGFR mutations in non-small cell lung cancer using mutation-specific antibodies for immunohistochemical analysis. Clin Cancer Res. 2010;16:3163–3170. doi: 10.1158/1078-0432.CCR-09-3239. [DOI] [PubMed] [Google Scholar]
- 4.Kawahara A, Azuma K, Sumi A, Taira T, Nakashima K, Aikawa E, Abe H, Yamaguchi T, Takamori S, Akiba J, Kage M. Identification of non-small-cell lung cancer with activating EGFR mutations in malignant effusion and cerebrospinal fluid: rapid and sensitive detection of exon 19 deletion E746-A750 and exon 21 L858R mutation by immunocytochemistry. Lung Cancer. 2011;74:35–40. doi: 10.1016/j.lungcan.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Azuma K, Okamoto I, Kawahara A, Taira T, Nakashima K, Hattori S, Kinoshita T, Takeda M, Nakagawa K, Takamori S, Kuwano M, Ono M, Kage M. Association of the expression of mutant epidermal growth factor receptor protein as determined with mutation-specific antibodies in non-small cell lung cancer with progression-free survival after gefitinib treatment. J Thorac Oncol. 2012;7:122–127. doi: 10.1097/JTO.0b013e31822eeba2. [DOI] [PubMed] [Google Scholar]
- 6.Yu J, Kane S, Wu J, Benedettini E, Li D, Reeves C, Innocenti G, Wetzel R, Crosby K, Becker A, Ferrante M, Cheung WC, Hong X, Chirieac LR, Sholl LM, Haack H, Smith BL, Polakiewicz RD, Tan Y, Gu TL, Loda M, Zhou X, Comb MJ. Mutation-specific antibodies for the detection of EGFR mutations in non-small-cell lung cancer. Clin Cancer Res. 2009;15:3023–3028. doi: 10.1158/1078-0432.CCR-08-2739. [DOI] [PubMed] [Google Scholar]
- 7.Brevet M, Arcila M, Ladanyi M. Assessment of EGFR mutation status in lung adenocarcinoma by immunohistochemistry using antibodies specific to the two major forms of mutant EGFR. J Mol Diagn. 2010;12:169–176. doi: 10.2353/jmoldx.2010.090140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seo AN, Park TI, Jin Y, Sun PL, Kim H, Chang H, Chung JH. Novel EGFR mutation-specific antibodies for lung adenocarcinoma: highly specific but not sensitive detection of an E746_A750 deletion in exon 19 and an L858R mutation in exon 21 by immunohistochemistry. Lung Cancer. 2014;83:316–323. doi: 10.1016/j.lungcan.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Wu SG, Chang YL, Lin JW, Wu CT, Chen HY, Tsai MF, Lee YC, Yu CJ, Shih JY. Including total EGFR staining in scoring improves EGFR mutations detection by mutation-specific antibodies and EGFR TKIs response prediction. PLoS One. 2013;6:e23303. doi: 10.1371/journal.pone.0023303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, Wang X, Xue L, Xu N, Ye X, Zeng H, Lu S, Huang J, Akesu S, Xu C, He D, Tan Y, Hong Q, Wang Q, Zhu G, Hou Y, Zhang X. The use of mutation-specific antibodies in predicting the effect of EGFR-TKIs in patients with non-small-cell lung cancer. J Cancer Res Clin Oncol. 2013;140:849–857. doi: 10.1007/s00432-014-1618-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 12.Pao W, Ladanyi M. Epidermal growth factor receptor mutation testing in lung cancer: searching for the ideal method. Clin Cancer Res. 2007;13:4954–4955. doi: 10.1158/1078-0432.CCR-07-1387. [DOI] [PubMed] [Google Scholar]
- 13.Xiong Y, Bai Y, Leong N, Laughlin TS, Rothberg PG, Xu H, Nong L, Zhao J, Dong Y, Li T. Immunohistochemical detection of mutations in the epidermal growth factor receptor gene in lung adenocarcinomas using mutation-specific antibodies. Diagn Pathol. 2013;8:27. doi: 10.1186/1746-1596-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan XS, Liu B, Yu B, Shi SS, Wang X, Zhang J, Wang JD, Lu ZF, Ma HH, Zhou XJ. [Immunohistochemistry using epidermal growth factor receptor mutation-specific antibodies of del E746-A750 and L858R in lung adenocarcinomas] . Zhonghua Bing Li Xue Za Zhi. 2013;42:173–177. doi: 10.3760/cma.j.issn.0529-5807.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Hasanovic A, Ang D, Moreira AL, Zakowski MF. Use of mutation specific antibodies to detect EGFR status in small biopsy and cytology specimens of lung adenocarcinoma. Lung Cancer. 2012;77:299–305. doi: 10.1016/j.lungcan.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Ellison G, Donald E, McWalter G, Knight L, Fletcher L, Sherwood J, Cantarini M, Orr M, Speake G. A comparison of ARMS and DNA sequencing for mutation analysis in clinical biopsy samples. J Exp Clin Cancer Res. 2010;29:132. doi: 10.1186/1756-9966-29-132. [DOI] [PMC free article] [PubMed] [Google Scholar]


