Abstract
Replication restart primosome is a complex dynamic system that is essential for bacterial survival. This system uses various proteins to reinitiate chromosomal DNA replication to maintain genetic integrity after DNA damage. The replication restart primosome in Escherichia coli is composed of PriA helicase, PriB, PriC, DnaT, DnaC, DnaB helicase, and DnaG primase. The assembly of the protein complexes within the forked DNA responsible for reloading the replicative DnaB helicase anywhere on the chromosome for genome duplication requires the coordination of transient biomolecular interactions. Over the last decade, investigations on the structure and mechanism of these nucleoproteins have provided considerable insight into primosome assembly. In this review, we summarize and discuss our current knowledge and recent advances on the DNA-binding mode of the primosomal proteins PriA, PriB, and DnaT.
1. Introduction
Genome integrity should be maintained from generation to generation to ensure proper cell function and survival [1–3]. In bacteria, some exogenous and endogenous sources of DNA damage can inactivate a large proportion of replication forks [4, 5]. When DNA is damaged, the replication machinery, originally initiated at oriC, can be arrested and disassembled anywhere along the DNA, leading to replication failure [5, 6]. To reload DnaB helicase for oriC-independent DNA replication, collapsed DNA replication forks must be reactivated by the replication restart primosome [7, 8]. Primosome is the protein complex responsible for the conversion of single-stranded circular DNA to the replicative-form DNA in the replication cycle of ϕX174 phage [9, 10]. After DNA repair, the replication restart primosome [11–13], a formidable enzymatic machine, can translocate along the single-stranded DNA-binding protein (SSB), unwind the duplex DNA, and prime the Okazaki fragments required for the progression of replication forks [14]. In Escherichia coli, the replication restart primosome is composed of PriA helicase, PriB, PriC, DnaB helicase, DnaC, DnaT, and DnaG primase [3]. To date, two DnaB helicase-recruiting pathways are known: PriA-PriB-DnaT-DnaC-dependent and PriC-DnaC-dependent systems; the former system uses fork structures without gaps in the leading strand, whereas the latter system preferentially uses fork structures with large gaps (>5 nucleotides) in the leading strand [3]. As shown in Figure 1, PriA can bind directly and assemble a primosome on the template without gaps in the leading strand, and PriC initiates the assembly of a primosome on a fork containing gaps in the leading strand.
Figure 1.
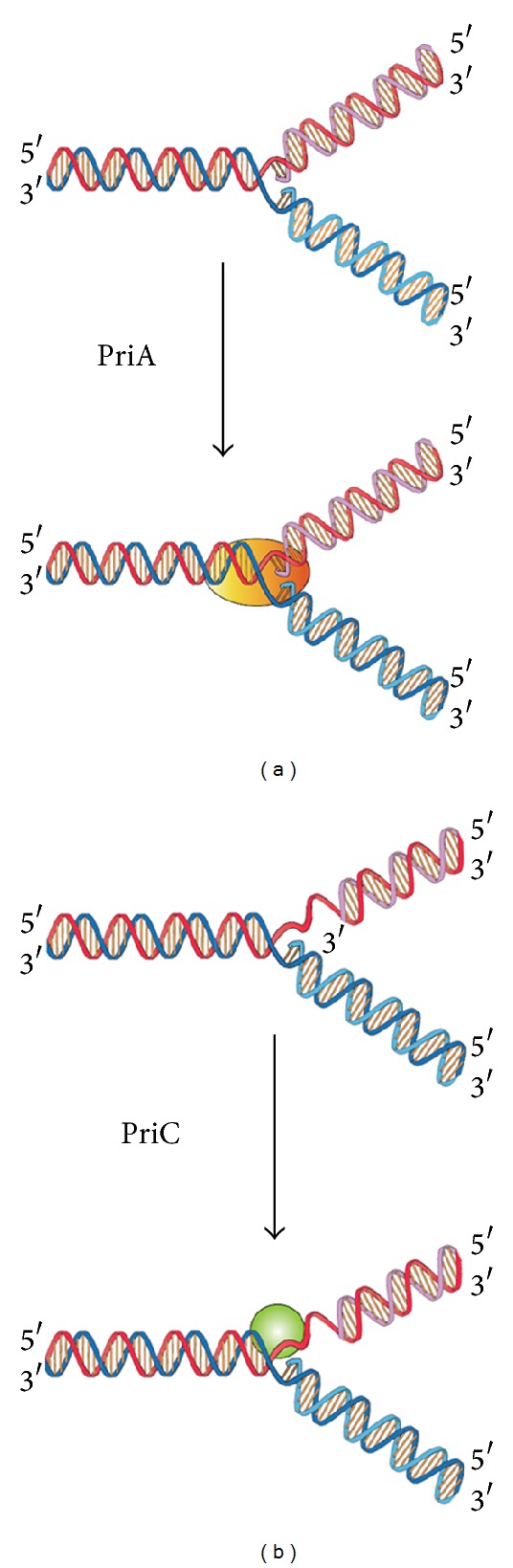
Two DnaB helicase-recruiting pathways for DNA replication restart at the stalled replication fork in vitro. The PriA-directed pathway (i.e., PriA-PriB-DnaT-DnaC-dependent reaction) preferentially uses fork structures without gaps in the leading strand, whereas the PriC-directed pathway (i.e., PriC-DnaC-dependent system) preferentially uses fork structures containing large gaps (>5 nucleotides) in the leading strand.
A hand-off mechanism for PriA-directed primosome assembly [15] has been proposed (Figure 2), whereby (i) PriA recognizes and binds to a replication fork; (ii) PriB joins PriA to form a PriA-PriB-DNA ternary complex; (iii) DnaT participates in this nucleocomplex to form a triprotein complex, in which PriB is released from ssDNA due to recruitment of DnaT; (iv) the PriA-PriB-DnaT-DNA quaternary complex loads the DnaB/C complex; (v) DnaB is loaded on the lagging strand template. Genetic analyses suggest that these primosomal proteins are essential replication proteins for bacterial cell growth [12, 16–21]. These proteins are required for reinitiating chromosomal DNA replication in bacteria; thus, blocking their activities would be detrimental to bacterial survival [22, 23]. Several primosomal proteins, such as PriA, PriB, PriC, and DnaT, are not found in humans; thus, these proteins may be potential targets in developing antibiotics for the six antibiotic-resistant pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter sp.) [24, 25]. The recently discovered inhibitor CGS 15943 targets Neisseria gonorrhoeae PriA helicase with an IC50 of 114 ± 24 μM [26].
Figure 2.
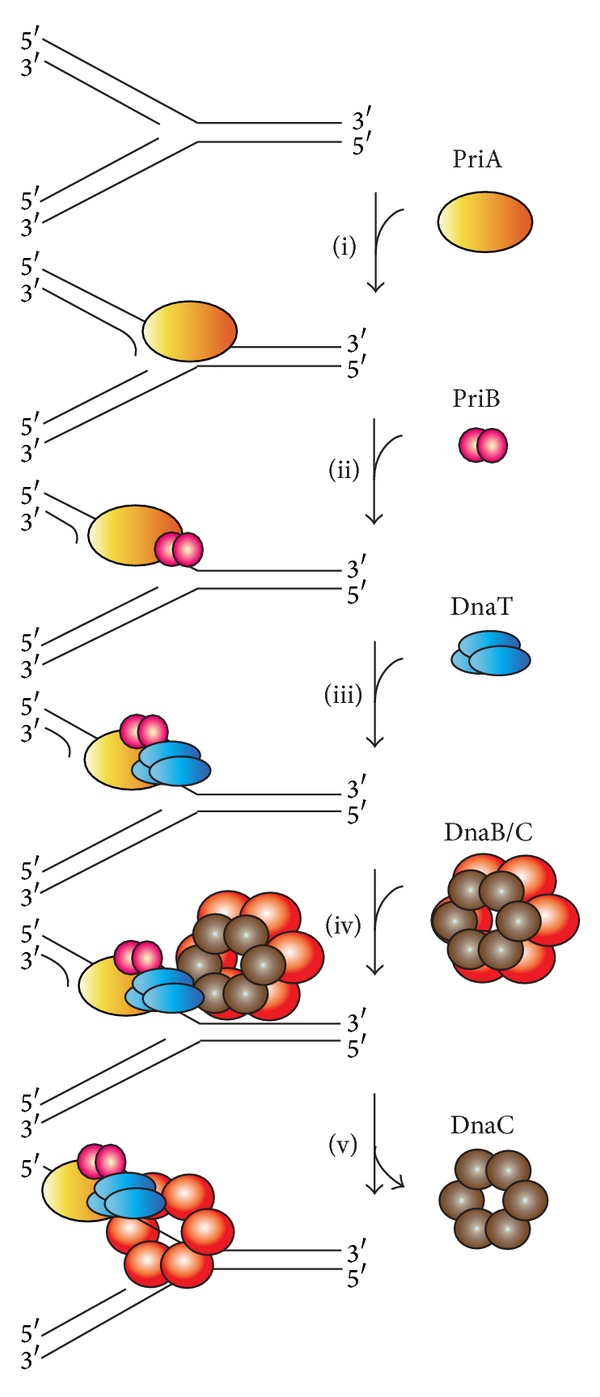
A hand-off mechanism for the replication restart primosome assembly. The proposed assembly mechanism is as follows. (i) PriA recognizes and binds to a replication fork, (ii) PriB joins PriA to form a PriA-PriB-DNA ternary complex, (iii) DnaT participates in this nucleocomplex to form a triprotein complex, in which PriB is released from ssDNA due to recruitment of DnaT, (iv) the PriA-PriB-DnaT-DNA quaternary complex loads the DnaB/C complex, and (v) DnaB is loaded on the lagging strand template.
Over the past 10 years, considerable progress has been made in the structural mechanisms of the replication restart primosome assembly. The structural information is a prerequisite for formulating any model of the assembly mechanism of the primosome (Table 1). In the following sections, we summarize and discuss our current knowledge and recent advances on the DNA-binding mode of the primosomal proteins PriA, PriB, and DnaT.
Table 1.
List of the structures of the primosomal proteins available in Protein Data Bank.
| PDB ID | X-ray | NMR | Length | |
|---|---|---|---|---|
|
PriA |
2D7E | The N-terminal domain of Escherichia coli PriA | 105 | |
| 2DwN | The N-terminal domain of Escherichia coli PriA bound to AG | 105 | ||
| 2D7G | The N-terminal domain of Escherichia coli PriA bound to AA | 105 | ||
| 2D7H | The N-terminal domain of Escherichia coli PriA bound to CCC | 105 | ||
| 2Dwl | The N-terminal domain of Escherichia coli PriA bound to AC | 105 | ||
| 2Dwm | The N-terminal domain of Escherichia coli PriA bound to AT | 105 | ||
| 4NL4 | Klebsiella pneumoniae PriA bound to ADP | 731 | ||
| 4NL8 | Klebsiella pneumoniae PriA bound to SSB C-terminal tail peptide | 731 | ||
|
| ||||
| PriB | 2CCZ | Escherichia coli PriB bound to ssDNA (15 mer) | 104 | |
| 1V1Q | Escherichia coli PriB | 104 | ||
| 1WOC | Escherichia coli PriB | 100 | ||
| 1TXY | Escherichia coli PriB | 100 | ||
| 2PNH | Escherichia coli PriB E39A | 100 | ||
| 4APV | Klebsiella pneumoniae PriB | 102 | ||
| 3K8A | Neisseria gonorrhoeae PriB | 100 | ||
| 4FDB | Ralstonia solanacearum PriB | 99 | ||
| 3EN2 | Ralstonia solanacearum PriB | 95 | ||
| 3FHW | Bordetella parapertussis PriB | 102 | ||
| 3KLW | Bordetella pertussis PriB | 98 | ||
| 4GS3 | The N-terminal domain of Thermoanaerobacter tengcongensis PriB | 104 | ||
|
| ||||
| DnaT | None | |||
|
| ||||
|
DnaB |
4ESV | Geobacillus stearothermophilus DnaB bound to DNA (14 mer) | 441 | |
| 2R6E | Geobacillus stearothermophilus DnaB | 441 | ||
| 2R6D | Geobacillus stearothermophilus DnaB | 441 | ||
| 2R6A | Geobacillus stearothermophilus DnaB bound to DnaG | 441 | ||
| 2R6C | Geobacillus stearothermophilus DnaB bound to DnaG | 441 | ||
| 4M4W | Geobacillus stearothermophilus DnaB bound to DnaG and DnaI | 454 | ||
| 2R5U | The N-terminal domain of Mycobacterium tuberculosis DnaB | 167 | ||
| 2Q6T | Thermus aquaticus DnaB | 440 | ||
| 3GXV | The N-terminal domain of Helicobacter pylori DnaB | 121 | ||
| 4A1F | The C-terminal domain of Helicobacter pylori DnaB | 323 | ||
| 4NMN | Aquifex aeolicus DnaB bound to ADP | 434 | ||
| 2VYF | Geobacillus kaustophilus DnaC | 441 | ||
| 2VYE | Geobacillus kaustophilus DnaC bound to ssDNA (9 mer) | 441 | ||
| 1B79 | The N-terminal domain of Escherichia coli DnaB | 128 | ||
| 1JWE | The N-terminal domain of Escherichia coli DnaB | 114 | ||
|
| ||||
| DnaC | 3EC2 | Aquifex aeolicus DnaC 42–221 | The N-terminal domain of Bacillus subtilis DnaI | 180 |
| 3ECC | Aquifex aeolicus DnaC bound to ADP | 185 | ||
| 2W58 | Geobacillus kaustophilus DnaI | 199 | ||
| 4M4W | Geobacillus stearothermophilus DnaB bound to DnaG and DnaI | 278 | ||
| 2QGZ | Streptococcus pyogenes DnaI | 308 | ||
| 2K7R | 106 | |||
|
| ||||
| DnaG | 3B39 | Escherichia coli DnaG 109–427 bound to ssDNA (15 mer) | 321 | |
| 1DD9 | Escherichia coli DnaG 115–428 | 338 | ||
| 1DDE | Escherichia coli DnaG 115–428 | 338 | ||
| 1T3W | The C-terminal domain of Escherichia coli DnaG | 148 | ||
| 2HAJ | Escherichia coli DnaG 447–581 | 135 | ||
| 4E2K | Staphylococcus aureus DnaG 108–428 | 321 | ||
| 4EDG | Staphylococcus aureus DnaG 108–428 bound to ATP | 321 | ||
| 4EDK | Staphylococcus aureus DnaG 108–428 bound to GTP | 319 | ||
| 4EDT | Staphylococcus aureus DnaG 108–428 bound to ppGpp | 321 | ||
| 4EDV | Staphylococcus aureus DnaG 108–428 bound to ppGpp | 321 | ||
| 4EE1 | Staphylococcus aureus DnaG 108–428 bound to CTP | 321 | ||
| 4EDR | Staphylococcus aureus DnaG 108–428 bound to UTP | 321 | ||
| 2LZN |
Staphylococcus aureus DnaG 462-605 |
143 | ||
| 1Z8S | Bacillus stearothermophilus DnaG 452–597 | 146 | ||
| 4EHS | The C-terminal domain of Helicobacter pylori DnaG 438–559 | 122 | ||
| 4M4W | Geobacillus stearothermophilus DnaB bound to DnaI and DnaG | 143 | ||
| 2R6A | Geobacillus stearothermophilus DnaB bound to DnaG | 143 | ||
| 2R6C | Geobacillus stearothermophilus DnaB bound to DnaG | 143 | ||
| 2AU3 | Aquifex aeolicus DnaG 1–403 | 403 | ||
|
| ||||
| PriC | 2RT6 | The N-terminal domain of Escherichia coli PriC | 98 | |
Length and amino acid residues.
2. Structural Insights into the DNA-Binding Mode
2.1. PriA Helicase
PriA functions as a scaffold that recruits other primosomal proteins. It was originally discovered as an essential factor for the conversion of single-stranded circular DNA to the replicative-form DNA of ϕX174 single-stranded phage in vitro [27]. The priA mutant of E. coli exhibits complex phenotypes that include reduced viability, chronic induction of SOS response, rich media sensitivity, decreased homologous recombination, sensitivity to UV irradiation, defective double-stranded break repair, and both induced and constitutive stable DNA replication [6, 12, 28–30]. The native PriA is a monomer with a molecular mass of ~82 kDa. The tertiary structure of the monomer contains two functional domains, namely, the helicase domain (HD), which encompasses ~540 amino acid residues from the C-terminus, and the DNA-binding domain, which comprises ~181 amino acid residues from the N-terminus [31–33]. PriA is a DEXH-type helicase that unwinds DNA with a 3′ to 5′ polarity [34]. Fuelled by the binding and hydrolysis of ATP, PriA moves along the nucleic acid filaments with other primosomal proteins and separates double-stranded DNA into their complementary single strands [35]. PriA preferentially binds to a D-loop-like structure by recognizing a bend at the three-way branched DNA structures and duplex DNA with a protruding 3′ single strand [32, 36, 37]. PriA interacts with SSB [38], PriB [15, 39, 40], and DnaT [15]. PriA can unwind the nascent lagging strand DNA to create a suitable binding site to help PriC load the DnaB helicase onto stalled replication forks where a gap exists in the nascent leading strand [41, 42]. The crystal structures of the N-terminal 105 amino acid residue segment of E. coli PriA (EcPriA) in complex with different deoxydinucleotides show a feasible interaction model for the base-non-selective recognition of the 3′-terminus of DNA between the nucleobase and the DNA-binding sites of EcPriA [43].
Figure 3(a) shows that the alignment consensus of 150 sequenced PriA homologs by ConSurf [44] reveals the degree of variability at each position along the primary sequence. The highly variable amino acid residues are colored teal, whereas the highly conserved are colored burgundy. A consensus sequence was established by determining the most commonly found amino acid residue at each position relative to the primary sequence of K. pneumoniae PriA (KpPriA). The amino acid sequences of KpPriA and EcPriA share 88% identity [45]. The N-terminal 19–219 amino acid residues in PriA are not highly conserved. The crystal structure of KpPriA has been recently determined [45]. KpPriA has six subdomains (Figure 3(b)), namely, a 3′ DNA-binding domain (3′BD; orange), a winged-helix domain (WH; green), two lobes of the helicase core (colored hot pink and blue, resp.), a Cys-rich region (CRR; dark blue), and a C-terminal domain (CTD; red). The 3′BD and WH comprise the N-terminal DNA-binding domain (DBD), and the other four subdomains (two lobes of the helicase core, CRR, and CTD) comprise the HD. Asp17, located in the 3′BD of EcPriA, is crucial for the 3′ base-non-selective recognition of DNA [43], and Arg697, located in the CTD of KpPriA, is crucial for the C-terminal tail of SSB (SSB-Ct) binding and induction of structural changes in the SSB-DNA complex [45]; both are significantly invariable. Many biochemical and genetic studies have been performed on the DNA-binding mode of PriA [7, 8], but the structural basis for the full length PriA-DNA complex is still lacking.
Figure 3.
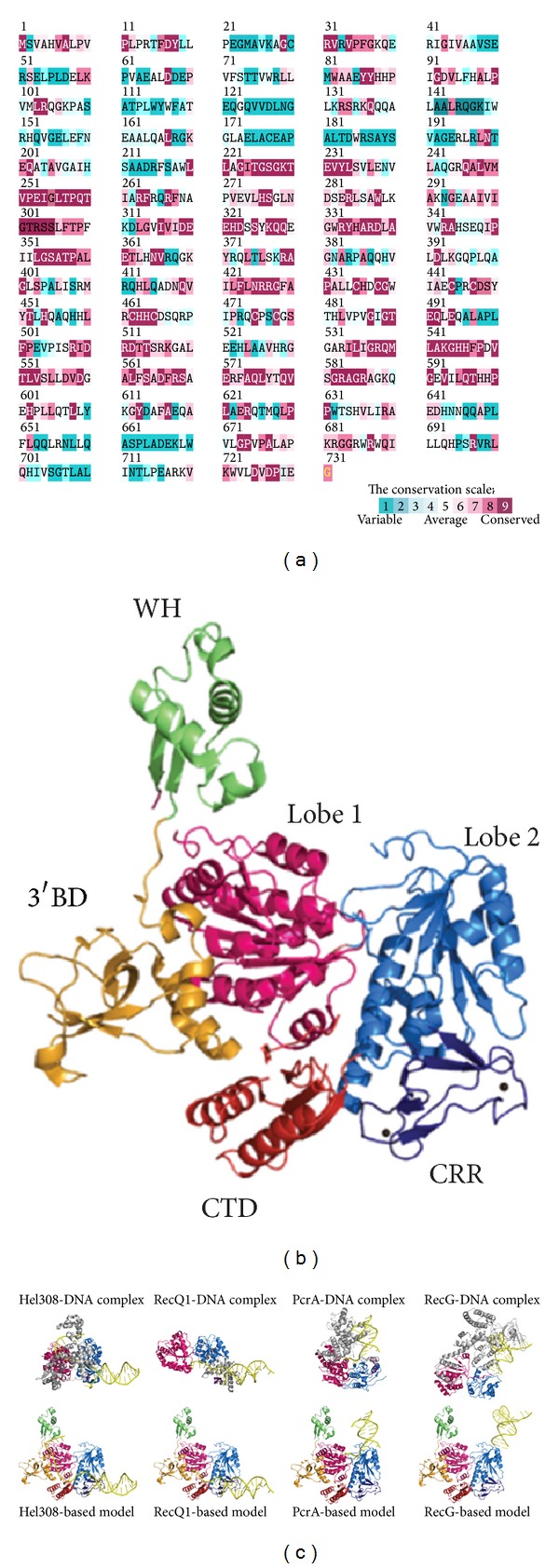
(a) Amino acid sequence alignment of KpPriA. An alignment consensus of 150 sequenced PriA homologs by the program ConSurf reveals the degree of variability at each position along the primary sequence. Highly variable amino acids are colored teal, whereas those highly conserved are colored burgundy. A consensus sequence was established by determining the most commonly found amino acid residue at each position relative to the primary sequence of KpPriA. The N-terminal 19–219 amino acid residues in PriA are not highly conserved. Asp17, located in the 3′BD of EcPriA, is crucial for the 3′ base-non-selective recognition of DNA, and Arg697, located in the CTD of KpPriA, is crucial for the SSB-Ct binding and induction of structural changes in the SSB-DNA complex; both are significantly invariable. (b) Crystal structure of KpPriA. KpPriA has six subdomains (Protein Data Bank entry: 4NL4), namely, a 3′ DNA-binding domain (3′BD; orange), a winged-helix domain (WH; green), two lobes of the helicase core (colored hot pink and blue, resp.), a Cys-rich region (CRR; dark blue), and a C-terminal domain (CTD; red). 3′BD and WH comprise the N-terminal DNA-binding domain (DBD), and the other four subdomains (two lobes of the helicase core, CRR, and CTD) comprise the helicase domain (HD). (c) Putative DNA-binding mode of KpPriA. The DNA-binding models of KpPriA are directly constructed by manually superimposing the KpPriA with DNA-bound crystal structure of Hel308 (Protein Data Bank entry: 2P6R), RecQ1 (Protein Data Bank entry: 2WWY), PcrA (Protein Data Bank entry: 3PJR), and RecG (Protein Data Bank entry: 1GM5). Considering the known ssDNA-binding site at DBD and the putative wedge element in KpPriA located at CRR, KpPriA may use the Hel308-based model to bind DNA. The β-hairpin, an important motif for DNA strand separation by helicase, is colored in magenta.
To elucidate the structural mechanism of DNA binding and unwinding by PriA, Bhattacharyya et al. [45] compared the structure of the full length KpPriA with those of other DNA helicases of superfamily II, namely, RecQ1 (Protein Data Bank entry: 2WWY) [46, 47] and Hel308 (Protein Data Bank entry: 2P6R) [48]. The structures of these helicases have been solved in complex with substrate DNA. RecQ1 and Hel308 bind to single-stranded DNA tailed duplex and unwind via the DNA unwinding wedge element, a prominent β-hairpin for strand separation [47, 49]. PriA also shares sequence similarity with other helicases, such as PcrA (Protein Data Bank entry: 3PJR) [50], a DNA helicase of superfamily I, and RecG (Protein Data Bank entry: 1GM5) [51], a DNA helicase of superfamily II. The structures of these helicases bound to DNA, along with KpPriA, are shown in Figure 3(c) for comparison. According to the crystal structures of the helicase-DNA complex, the two lobes of the helicase core of KpPriA (colored hot pink and blue, resp.) are aligned and manually superimpose the location of the dsDNA from the complex structure with KpPriA structure. These modeled structures of KpPriA show that the DNA-binding modes and thus the DNA-unwinding modes are different. Considering the known ssDNA-binding site at DBD and the putative wedge element in KpPriA located at CRR, KpPriA may use the Hel308-based model to bind DNA. The DNA-binding mode, fork DNA recognition site(s), and the helicase translocation using either the inchworm stepping or Brownian motor mechanism [52] must be further confirmed by additional biophysical and structural studies.
2.2. PriB Protein
PriB is a basic accessory protein in PriA-directed DNA replication restart primosome [11, 13]. It was originally discovered as an essential factor for the conversion of single-stranded circular DNA to the replicative-form DNA of ϕX174 single-stranded phage in vitro. In contrast to the ϕX174 model, del(priB)302 mutant has almost wild-type phenotypes [53], suggesting that PriB is not absolutely required for bacterial DNA replication. PriB was formerly known as the “n protein” because it can be inactivated by treatment with N-ethylmaleimide [54]. In a PriA-PriB-DnaT-dependent reaction, PriB is the second protein to be assembled in the protein-DNA complex. It stabilizes the binding of PriA to DNA hairpin [35, 55] and then stimulates PriA helicase activity [40, 56]. The PriA stimulation by PriB correlates with the ability of PriB to form a stable PriA-PriB-DNA complex [40]. PriB also facilitates the association of DnaT with PriA [57]. More than one PriA-PriB complex is possibly involved in the initiation of primosome formation, and the effect of PriB on the PriA-DNA association is dependent on the DNA structure [58]. PriB interacts with PriA [15, 39], DnaT [15, 59, 60], SSB [54, 61], and itself [61, 62] and does not interact with DnaA, DnaB, DnaC, or DnaG [61]. The mechanisms of DnaC-DnaB complex loading by PriA-PriB-DnaT complex at the forks and then DnaB-DnaG complex formation remain unclear.
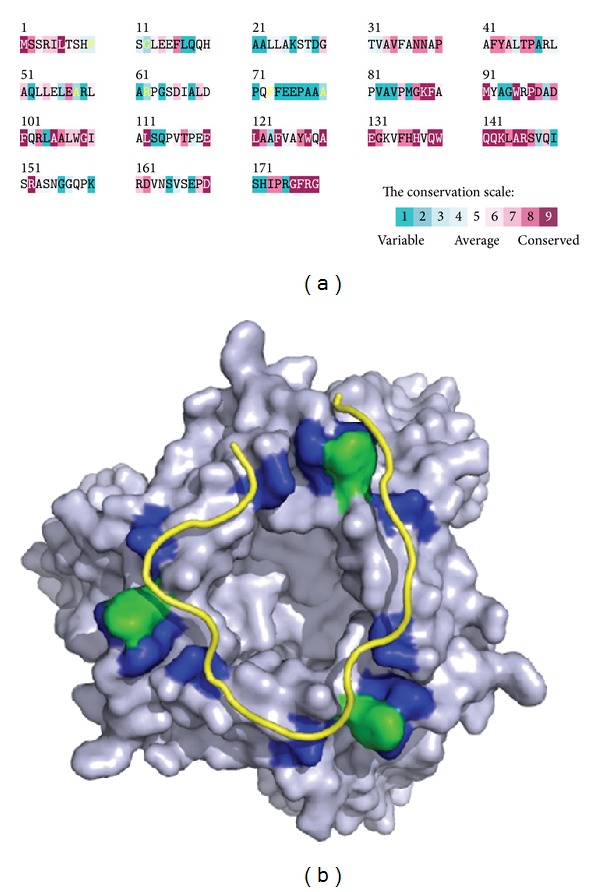
PriB is a homodimer with polypeptide chains of 104 amino acid residues [63–65] (Figure 4(a)). Each PriB monomer has an oligonucleotide/oligosaccharide-binding (OB) fold structure [66–69] with three flexible β-hairpin loops: L12 (residues 20–24), L23 (residues 37–44), and L45 (residues 81–88) (Figure 4(b)). PriB can bind to ssDNA [15, 39, 40, 54, 56, 62–65, 70–73], ssRNA [65], double-stranded DNA [56, 70], and circular ϕX viral DNA [73]. Although PriB is a dimer, it has only one DNA-binding site [73], which is located in loop L45 centrally within the dimer, and this site occupies a total site size of 12 ± 1 nucleotides [72]. The N-terminal 1–49 amino acid residue region of PriB is crucial for dimerization, whereas the C-terminal 50–104 amino acid residue region is crucial for ssDNA binding [71]. PriB shares structural similarity with the N-terminal DNA-binding domain of the E. coli SSB (EcSSB) [63–65, 74, 75]. Sequence comparisons and operon organization analyses also show that PriB evolves from the duplication of the SSB gene [76], but they differ in their ssDNA-binding properties and strategies [70, 73]. For example, EcSSB possesses three conserved aromatic residues (Trp40, Trp54, and Phe60) in the L45 loop of the OB fold. These residues serve important functions in ssDNA binding. Two of these residues (Trp40 and Phe60 in EcSSB) are replaced with nonconserved amino acids in the PriB family. In contrast to the EcSSB-DNA complex, the L23 loop from each subunit of PriB makes a close contact with the β-barrel core. The longer and extended L23 loops in EcSSB greatly increase the interactions between EcSSB and ssDNA [73, 75].
Figure 4.
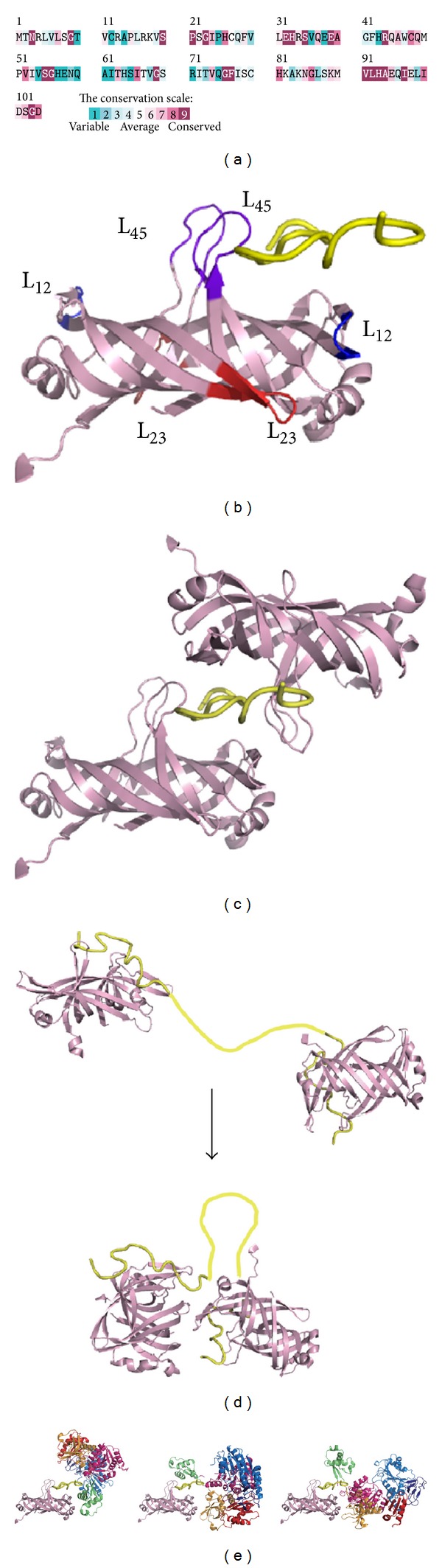
(a) Amino acid sequence alignment of EcPriB. An alignment consensus of 111 sequenced PriB homologs by the program ConSurf reveals the degree of variability at each position along the primary sequence. In general, the overall amino acid sequences among PriB proteins are not highly conserved, including many residues found important for ssDNA binding by EcPriB, such as Phe42, Trp47, Lys82, Lys84, and Lys89. (b) EcPriB is a homodimer with polypeptide chains of 104 amino acid residues. Each PriB monomer has an OB-fold structure with three flexible β-hairpin loops: L12 (residues 20–24; colored in blue), L23 (residues 37–44; colored in red), and L45 (residues 81–88; colored in purple blue). The ssDNA in the complex is shown in gold. (c) Crystal structure of EcPriB in complex with DNA. The complex structure of EcPriB (Protein Data Bank entry: 2CCZ) shows that a single dT15 ssDNA periodically interacts with two OB folds from two symmetrically related EcPriB dimers in the crystal and that the DNA is sandwiched by PriB dimers via their L45 loops. (d) Possible working model of interaction between two PriB proteins on ssDNA. PriB proteins cooperatively bind to ssDNA in two steps: two PriB proteins independently interact with ssDNA and then interact with each other through His64 on the ssDNA. The ssDNA in the complex is shown in gold. The region in ssDNA that does not directly interact with PriB, proposed in this two-step binding model, is colored in yellow. (e) Proposed models for PriA-DNA-PriB structure. These models are based on these observations: (1) two PriB dimers are complexed with a single dT15; (2) PriA has a highly electropositive ssDNA-binding region in DBD, and the basic DBD in PriA may be involved in complex with PriB; (3) DBD of PriA alone in solution forms a dimer and not a monomer as the full-length PriA.
Figure 4(a) shows the alignment consensus of 111 sequenced PriB homologs by ConSurf [44]. The alignment indicates that the overall amino acid sequences among PriB proteins are not highly conserved; only 21 amino acid residues are significantly conserved: Asn3, Gly9, Ser20, Pro21, Gly23, Glu32, His33, Ser35, Glu39, Arg44, Ser55, Gly56, Gly69, Gly76, Phe77, Val91, Leu92, His93, Ala94, Ile97, and Gly103. Many residues important for ssDNA binding by E. coli PriB (EcPriB), such as Phe42 [64], Trp47 [64, 73], Lys82 [64, 73], Lys84 [73], and Lys89 [73], are not conserved. PriB may be a nonessential facilitating factor in DNA replication restart [53], and many prokaryotic genomes do not contain a recognizable homolog of priB [39]. Hence, we speculate that these residues among PriB proteins for binding ssDNA do not need to be precisely conserved.
We previously described the crystal structure of EcPriB in complex with ssDNA dT15 (Protein Data Bank entry: 2CCZ) [73]. A single dT15 ssDNA periodically interacts with two OB folds from two symmetrically related EcPriB dimers in the crystal, sandwiched by PriB dimers via their L45 loops (Figure 4(c)). Although the precise function of more than one PriB self-assembled on DNA to form a high-density nucleoprotein complex is still unclear, PriB binds DNA with strong cooperativity [70, 72, 73] in two steps (Figure 4(d)): two PriB proteins independently interact with ssDNA in primary binding mode, and then the proteins interact with each other through His64 on the ssDNA [77]. Whether the resultant ssDNA bound by more than one PriB forms a unique structure suitable for further assembly process for the primosome is not clearly known. The complex structure [73] and the thermodynamic analysis [72] indicate that the PriB dimer behaves like a protein with half-site reactivity, where only one monomer of the PriB dimer can engage in interactions with the DNA and the partner protein. The importance of the binding site on PriB for ssDNA to overlap the binding sites for PriA and DnaT needs to be investigated [15]. Each preprimosome may contain two PriB dimers [60]; whether or not this cocrystal structure, in which two PriB dimers are complexed with a single dT15 ssDNA, is an artificial or an actual binding mode for ssDNA by PriB also remains unclear. PriA may have a function similar to a monomer of the symmetrical PriB dimer in the crystal to stabilize the partially disordered ssDNA because the cooperation between PriB and PriA may be necessary to form a stable PriA-DNA-PriB complex. That is, the PriB-ssDNA-PriB complex (Figure 4(c)) may mimic the structure of the PriA-ssDNA-PriB complex (Figure 4(e)). We proposed three binding ways by use of the crystal structures of PriA and PriB. EcPriA has a highly electropositive ssDNA-binding region (amino acid residues 1–198) containing 8 Lys and 14 Arg residues in DBD; thus, the basic DBD in EcPriA may be involved in complex with EcPriB [73]. The DBD of EcPriA alone in solution forms a dimer and not a monomer as EcPriA [31], suggesting that another unknown stabilization factor is needed. The DBD of PriA and one of the monomers of PriB may bind to ssDNA cooperatively to decrease the dissociation rate of PriA from the DNA during helix unwinding [73]. The crystal structure of PriA in complex with PriB and DNA is necessary to elucidate the assembly mechanism of the replication restart primosome.
More than a mere ssDNA-binding protein, PriB can bind both ssDNA and dsDNA with comparable affinity [70]. SSB can also bind dsDNA but with far less affinity than ssDNA [78]. According to the crystal structures of some dimeric proteins complexed with dsDNA found in the Protein Data Bank, PriB binds dsDNA in three possible ways (Figure 5). First, PriB may bind to dsDNA via the replication terminator protein- (RTP-) binding mode (Protein Data Bank entry: 1F4K) [79]. RTP, a dimeric WH protein [80, 81], uses two recognition helices to bind the major grooves of dsDNA. The PriB dimer also has two helices but does not contain any aromatic or positively charged residues as RTP. Thus, PriB binds to dsDNA via the RTP-binding mode that can be ruled out. Second, PriB may bind to dsDNA via the HU-binding mode (Protein Data Bank entry: 1P51) [82, 83]. HU is a dimeric nucleoid-associated protein that mainly uses two β sheets to bind dsDNA. Third, PriB may bind dsDNA in a manner similar to binding ssDNA. The structure-based mutational analysis indicates that the residues in PriB crucial for ssDNA binding are also crucial for dsDNA binding [70]. These residues responsible for ssDNA and dsDNA binding are almost overlapped; thus, PriB may use a similar approach to bind to the phosphate backbone of ssDNA and dsDNA through several positively charged residues. This phenomenon may be the reason for the comparable binding affinities of PriB to ssDNA and dsDNA. We speculate that, during evolution [76], the conserved aromatic and other residues in the L45 loop of the OB fold in SSB are changed into nonconserved and positively charged residues in PriB to more precisely fit the requirement for assembly of the replication restart primosome at the stalled DNA forks.
Figure 5.
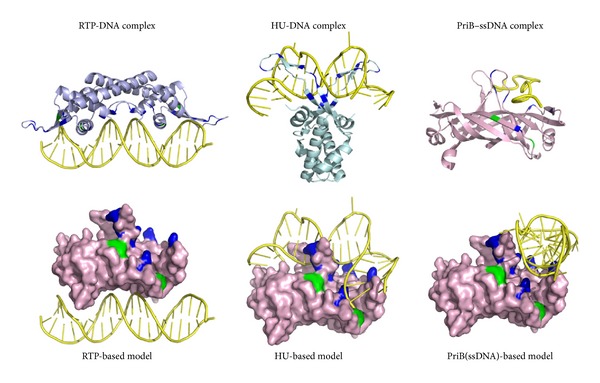
Putative dsDNA-binding mode of PriB. The DNA-binding models of PriB are directly constructed by manually superimposing the PriB dimer with DNA-bound crystal structure of RTP (Protein Data Bank entry: 1F4K), HU (Protein Data Bank entry: 1P51), and B-form dsDNA. The hydrophobic (green) and basic residues (blue) of RTP, Lys14, Arg16, Lys51, Arg59, Lys71, Lys74, Lys76, Lys77, Lys81, Lys91, Tyr58, and Tyr88, located on the dsDNA-binding surface, are indicated. The basic residues Arg53, Arg55, Lys56, Arg58, Arg61, Lys64, Lys68, and Arg75 of HU located on the dsDNA-binding surface are also indicated. Considering the known dsDNA-binding sites in PriB, PriB may use the HU-based model to bind dsDNA. Alternatively, PriB may use a similar approach to bind ssDNA and dsDNA because the residues responsible for ssDNA and dsDNA binding are almost overlapped.
2.3. DnaT Protein
DnaT is an essential protein in the assembly of the PriA-directed DNA replication restart primosome [6, 11–13, 15, 55, 57]. It provides a specific recognition site for loading the replicative DnaB helicase during the promosome assembly [15, 42]. DnaT, formerly known as the “protein i” [84–86], was originally discovered as a critical factor for the conversion of single-stranded circular DNA to the replicative-form DNA of ϕX174 single-stranded phage [9] and pBR322 plasmid replication, but not for R1 plasmid replication [87]. Genetic analysis for E. coli DnaT suggests that a replication protein is essential for bacterial cell growth because the colony size, cell morphology, inability to properly partition nucleoids, UV sensitivity, and basal SOS expression of the dnaT822 mutant are similar to those of priA2::kan mutants [18]. DnaT is required for E. coli growth at elevated pressure [88] and for the lytic cycle of Mu growth [89]. DnaT is a homotrimer of ~22 kDa subunits [86, 90], but it also exists in solution as a monomer-trimer equilibrium system [91]. In a PriA-PriB-DnaT-dependent reaction, DnaT is the third protein to be assembled in the protein-DNA complex (Figure 2). The association of DnaT with PriA is facilitated by PriB [57]. Although the function of DnaT in the recruitment of DnaB helicase has been proposed, the fundamental function of DnaT for the replication restart primosome assembly is not widely known.
We have recently identified and characterized that DnaT is a ssDNA-binding protein [90]. Based on the alignment consensus of 29 sequenced DnaT homologs by ConSurf [44], we found that the amino acid residues in the C-terminal region of K. pneumoniae DnaT (KpDnaT) are highly conserved (Figure 6(a)) and that KpDnaT contains 10 Arg, 5 Lys, and 18 aromatic amino acid residues (11 Phe, 4 Trp, and 3 Tyr). We attempted to assess whether or not KpDnaT, especially at the C-terminal region, has ssDNA-binding activity because the aromatic stacking and electropositive interactions serve important functions in ssDNA binding by proteins [73, 75, 92–94]. KpDnaT can form distinct complexes with ssDNA of different lengths, and the size of the binding site is 26 ± 2 nucleotides for a trimeric KpDnaT [90]. Although DnaT is not an OB-fold protein predicted from sequence analysis and structure modeling, the activity for ssDNA binding by DnaT, assayed in the same manner, is even higher than that of PriB, an OB-fold protein [90]. The two-domain structure for DnaT is characterized by the involvement of the N-terminal domain (amino acid residues 1–83) in PriB binding and the C-terminal domain (amino acid residues 84–179) in ssDNA binding [59].
Figure 6.
(a) Amino acid sequence alignment of KpDnaT. An alignment consensus of 29 sequenced DnaT homologs by the program ConSurf reveals the degree of variability at each position along the primary sequence. In general, the amino acid residues in the C-terminal region of KpDnaT are highly conserved. (b) Modeled structure of KpDnaT. The structure of KpDnaT is modeled by the bioinformatic program (PS)2 and then manually built using threefold symmetry with a 25 mer ssDNA (gold). The highly conserved hydrophobic (green) and basic residues (blue) of KpDnaT, His136, His137, Trp140, Lys143, Arg146, and Arg151 located on the potential ssDNA-binding surface are indicated.
To date, little is known about the ssDNA-binding mode of non-OB-fold proteins, particularly trimeric proteins. No protein with amino acid sequence similar to DnaT is found in the structure databank. Thus, homology modeling for the DnaT structure by several homology-based programs is not successful, including the use of SWISS-MODEL (http://swissmodel.expasy.org/) [95]. To obtain an indepth understanding of the structure-function relationship of KpDnaT, its 3D structure has been modeled by the bioinformatic program (PS)2 [96, 97]. (PS)2 (http://140.113.239.111/~ps2v2/docs.php) is an automatic homology modeling server that combines both sequence and secondary structure information to detect the homologous proteins with remote similarity and target-template alignment [96, 97]. The modeled structure of KpDnaT, manually built using threefold symmetry with a hit of alpha-aminotransferase from Pyrococcus horikoshii (Protein Data Bank entry: 1GD9) suggested from (PS)2, is a ring-shaped trimer (Figure 6(b)) [59, 90]. Based on the structural model of KpDnaT, we suggested that the positively charged (blue) and aromatic residues (green) located in the C-terminus of DnaT are involved in ssDNA binding: H136, H137, W140, K143, R146, and R151 (Figure 6(b)). These residues in DnaT are significantly conserved among the 29 sequenced DnaT proteins (Figure 6(a)). F73 and F74 are also potential binding sites for ssDNA, but they are not completely conserved in DnaT family. The ring-like structure of KpDnaT is slightly similar to that of the hexameric (comprised of three dimers) DnaC helicase from Geobacillus kaustophilus, a DnaB-like helicase [92]. DnaT may bind to DnaB with a stoichiometry of 1 : 2, one DnaT monomer to a DnaB dimer. However, the DnaT structure is only a modeled structure, and these speculations, including the putative DNA and DnaB-binding modes of DnaT, must be further confirmed by additional biophysical studies.
3. Perspectives
Most DNA helicases of superfamily I and superfamily II are almost nonhexameric and have poor dsDNA unwinding activities when acting alone in vitro [98]. Some helicases might function as ssDNA translocases rather than helicases, and self-assembly and/or interactions with accessory proteins are required to activate helicase activity [98]. Several monomeric ssDNA translocases of superfamily I can potentially displace proteins that are bound to ssDNA by translocating along the ssDNA and be activated by self-assembly, removal of an autoinhibitory domain, or direct interactions with an accessory protein(s) [38, 40, 99–101]. For PriA, the self-assembly and removal of an autoinhibitory domain for higher helicase activity have not been reported. However, poor helicase activity for PriA, which can be significantly stimulated by PriB [40] and SSB [38], is found. Based on the structure of KpPriA bound to an SSB C-terminal peptide (Trp-Met-Asp-Phe-Asp-Asp-Asp-Ile-Pro-Phe) and the study of a single-molecule FRET (smFRET), Bhattacharyya et al. [45] proposed a pushing mechanism, which is similar to that for the RecA recombinase [102], for PriA-mediated replication restart. For SSB-bound DNA replication forks, PriA translocase activity may push SSB along the lagging-strand template to expose additional ssDNA for PriB and DnaT binding and that will ultimately serve as a binding site for DnaB [45]. This model provides structural insight into the molecular mechanism for initiating replication restart primosome assembly. The interaction of PriB with PriA is weak, and the stimulation of PriA by PriB via an interaction with ssDNA is not DNA structure-specific [40]. Thus, the targeting of stalled forks and recombination intermediates during replication restart likely correlates with PriA alone. More structural studies for these primosomal proteins are still necessary to elucidate the interaction between PriB and DnaT, as well as the release from the replication restart system. Several studies have raised new interesting questions as to whether or not PriA, PriB, and DnaT are always synchronically expressed for physiological needs and whether or not PriB and DnaT have additional functions for other systems. PriA and DnaT are required for E. coli growth at elevated pressure [88]; however, why PriB is not necessary to be synchronically expressed has yet to be determined. Many prokaryotic genomes do not contain a recognizable homolog of priB and dnaT (e.g., P. aeruginosa; Table 2). Thus, further operon and gene regulation analyses for PriB and DnaT expression, not limited to replication restart, should be also investigated in combination with the biochemical and structural investigations.
Table 2.
Examples for some different PriA-directed primosome systems.
| PriA size (amino acid residues) |
Partner proteins found in NCBI | |
|---|---|---|
| Escherichia coli and Klebsiella pneumoniae | 731 | PriB, PriC, DnaT, DnaC, DnaB helicase, and DnaG |
| Staphylococcus aureus | 802 | DnaD, DnaB, DnaI, DnaC helicase, and DnaG |
| Pseudomonas aeruginosa | 739 | Only DnaB helicase and DnaG are found |
PriA is conserved in bacteria, but its primosomal partners are not.
Acknowledgments
The authors would like to thank three anonymous reviewers and the editor for their comments. This research was supported by a grant from the National Science Council, Taiwan (NSC 102-2320-B-040-019 to C.Y. Huang).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Smeenk G, van Attikum H. The chromatin response to DNA breaks: leaving a mark on genome integrity. Annual Review of Biochemistry. 2013;82:55–80. doi: 10.1146/annurev-biochem-061809-174504. [DOI] [PubMed] [Google Scholar]
- 2.Panier S, Durocher D. Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nature Reviews Molecular Cell Biology. 2013;14:661–672. doi: 10.1038/nrm3659. [DOI] [PubMed] [Google Scholar]
- 3.Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nature Reviews Molecular Cell Biology. 2006;7(12):932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- 4.Maher RL, Branagan AM, Morrical SW. Coordination of DNA replication and recombination activities in the maintenance of genome stability. Journal of Cellular Biochemistry. 2011;112(10):2672–2682. doi: 10.1002/jcb.23211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nature Reviews Molecular Cell Biology. 2002;3(11):859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 6.Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404(6773):37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 7.Masai H, Tanaka T, Kohda D. Stalled replication forks: making ends meet for recognition and stabilization. BioEssays. 2010;32(8):687–697. doi: 10.1002/bies.200900196. [DOI] [PubMed] [Google Scholar]
- 8.Gabbai CB, Marians KJ. Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival. DNA Repair. 2010;9(3):202–209. doi: 10.1016/j.dnarep.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marians KJ. Prokaryotic DNA replication. Annual Review of Biochemistry. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 10.Schekman R, Weiner A, Kornberg A. Multienzyme systems of DNA replication: proteins required for chromosome replication are resolved with the aid of a simple viral DNA template. Science. 1974;186(4168):987–993. doi: 10.1126/science.186.4168.987. [DOI] [PubMed] [Google Scholar]
- 11.Sandler SJ, Marians KJ. Role of PriA in replication fork reactivation in Escherichia coli. Journal of Bacteriology. 2000;182(1):9–13. doi: 10.1128/jb.182.1.9-13.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandler SJ. Multiple genetic pathways for restarting DNA replication forks in Escherichia coli K-12. Genetics. 2000;155(2):487–497. doi: 10.1093/genetics/155.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marians KJ. PriA-directed replication fork restart in Escherichia coli . Trends in Biochemical Sciences. 2000;25(4):185–189. doi: 10.1016/s0968-0004(00)01565-6. [DOI] [PubMed] [Google Scholar]
- 14.Patel SS, Pandey M, Nandakumar D. Dynamic coupling between the motors of DNA replication: hexameric helicase, DNA polymerase, and primase. Current Opinion in Chemical Biology. 2011;15(5):595–605. doi: 10.1016/j.cbpa.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopper M, Boonsombat R, Sandler SJ, Keck JL. A hand-off mechanism for primosome assembly in replication restart. Molecular Cell. 2007;26(6):781–793. doi: 10.1016/j.molcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boonsombat R, Yeh S, Milne A, Sandler SJ. A novel dnaC mutation that suppresses priB rep mutant phenotypes in Escherichia coli K-12. Molecular Microbiology. 2006;60(4):973–983. doi: 10.1111/j.1365-2958.2006.05147.x. [DOI] [PubMed] [Google Scholar]
- 17.Sandler SJ. Requirements for replication restart proteins during constitutive stable DNA replication in Escherichia coli K-12. Genetics. 2005;169(4):1799–1806. doi: 10.1534/genetics.104.036962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCool JD, Ford CC, Sandler SJ. A dnaT mutant with phenotypes similar to those of a priA2::kan mutant in Escherichia coli K-12. Genetics. 2004;167(2):569–578. doi: 10.1534/genetics.103.025296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinds T, Sandler SJ. Allele specific synthetic lethality between priC and dnaAts alleles at the permissive temperature of 30°C in E. coli K-12. BMC Microbiology. 2004;4, article 47 doi: 10.1186/1471-2180-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandler SJ, McCool JD, Do TT, Johansen RU. PriA mutations that affect PriA-PriC function during replication restart. Molecular Microbiology. 2001;41(3):697–704. doi: 10.1046/j.1365-2958.2001.02547.x. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Xu L, Sandler SJ, Marians KJ. Replication fork assembly at recombination intermediates is required for bacterial growth. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(7):3552–3555. doi: 10.1073/pnas.96.7.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marceau AH, Bernstein DA, Walsh BW, Shapiro W, Simmons LA, Keck JL. Protein interactions in genome maintenance as novel antibacterial targets. PLoS ONE. 2013;8(3) doi: 10.1371/journal.pone.0058765.e58765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu D, Bernstein DA, Satyshur KA, Keck JL. Small-molecule tools for dissecting the roles of SSB/protein interactions in genome maintenance. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(2):633–638. doi: 10.1073/pnas.0909191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bush K. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae . Current Opinion in Microbiology. 2010;13(5):558–564. doi: 10.1016/j.mib.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clinical Infectious Diseases. 2009;48(1):1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 26.Sunchu B, Berg L, Ward HE, Lopper ME. Identification of a small molecule PriA helicase inhibitor. Biochemistry. 2012;51(51):10137–10146. doi: 10.1021/bi301100w. [DOI] [PubMed] [Google Scholar]
- 27.Arai K, Kornberg A. Unique primed start of phage phi X174 DNA replication and mobility of the primosome in a direction opposite chain synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(1):69–73. doi: 10.1073/pnas.78.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masai H, Asai T, Kubota Y, Arai K, Kogoma T. Escherichia coli PriA protein is essential for inducible and constitutive stable DNA replication. EMBO Journal. 1994;13(22):5338–5345. doi: 10.1002/j.1460-2075.1994.tb06868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nurse P, Zavitz KH, Marians KJ. Inactivation of the Escherichia coli PriA DNA replication protein induces the SOS response. Journal of Bacteriology. 1991;173(21):6686–6693. doi: 10.1128/jb.173.21.6686-6693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kornberg A. Replication deficiencies in priA mutants of Escherichia coli lacking the primosomal replication n' protein. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(8):3029–3032. doi: 10.1073/pnas.88.8.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szymanski MR, Bujalowski PJ, Jezewska MJ, Gmyrek AM, Bujalowski W. The N-terminal domain of the Escherichia coli pria helicase contains both the DNA- and nucleotide-binding sites. Energetics of domain-DNA interactions and allosteric effect of the nucleotide cofactors. Biochemistry. 2011;50(43):9167–9183. doi: 10.1021/bi201100k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen H, North SH, Nakai H. Properties of the PriA helicase domain and its role in binding PriA to specific DNA structures. Journal of Biological Chemistry. 2004;279(37):38503–38512. doi: 10.1074/jbc.M404769200. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka T, Mizukoshi T, Taniyama C, Kohda D, Arai K, Masai H. DNA binding of PriA protein requires cooperation of the N-terminal D-loop/arrested-fork binding and C-terminal helicase domains. Journal of Biological Chemistry. 2002;277(41):38062–38071. doi: 10.1074/jbc.M204397200. [DOI] [PubMed] [Google Scholar]
- 34.Ouzounis CA, Blencowe BJ. Bacterial DNA replication initiation factor priA is related to proteins belonging to the “DEAD-box” family. Nucleic Acids Research. 1991;19(24, article 6953) doi: 10.1093/nar/19.24.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng JY, Marians KJ. The ordered assembly of the phiX174-type primosome. I. Isolation and identification of intermediate protein-DNA complexes. Journal of Biological Chemistry. 1996;271(26):15642–15648. doi: 10.1074/jbc.271.26.15642. [DOI] [PubMed] [Google Scholar]
- 36.Nurse P, Liu J, Marians KJ. Two modes of PriA binding to DNA. Journal of Biological Chemistry. 1999;274(35):25026–25032. doi: 10.1074/jbc.274.35.25026. [DOI] [PubMed] [Google Scholar]
- 37.McGlynn P, Al-Deib AA, Liu J, Marians KJ, Lloyd RG. The DNA replication protein PriA and the recombination protein RecG bind D-loops. Journal of Molecular Biology. 1997;270(2):212–221. doi: 10.1006/jmbi.1997.1120. [DOI] [PubMed] [Google Scholar]
- 38.Cadman CJ, McGlynn P. PriA helicase and SSB interact physically and functionally. Nucleic Acids Research. 2004;32(21):6378–6387. doi: 10.1093/nar/gkh980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong J, George NP, Duckett KL, DeBeer MAP, Lopper ME. The crystal structure of Neisseria gonorrhoeae PriB reveals mechanistic differences among bacterial DNA replication restart pathways. Nucleic Acids Research. 2009;38(2):499–509. doi: 10.1093/nar/gkp1031.gkp1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cadman CJ, Lopper M, Moon PB, Keck JL, McGlynn P. PriB stimulates PriA helicase via an interaction with single-stranded DNA. The Journal of Biological Chemistry. 2005;280(48):39693–39700. doi: 10.1074/jbc.M508521200. [DOI] [PubMed] [Google Scholar]
- 41.Heller RC, Marians KJ. Unwinding of the nascent lagging strand by Rep and PriA enables the direct restart of stalled replication forks. Journal of Biological Chemistry. 2005;280(40):34143–34151. doi: 10.1074/jbc.M507224200. [DOI] [PubMed] [Google Scholar]
- 42.Heller RC, Marians KJ. The disposition of nascent strands at stalled replication forks dictates the pathway of replisome loading during restart. Molecular Cell. 2005;17(5):733–743. doi: 10.1016/j.molcel.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 43.Sasaki K, Ose T, Okamoto N, et al. Structural basis of the 3′-end recognition of a leading strand in stalled replication forks by PriA. EMBO Journal. 2007;26(10):2584–2593. doi: 10.1038/sj.emboj.7601697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landau M, Mayrose I, Rosenberg Y, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Research. 2005;33(2):W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhattacharyya B, George NP, Thurmes TM, et al. Structural mechanisms of PriA-mediated DNA replication restart. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(4):1373–1378. doi: 10.1073/pnas.1318001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vindigni A, Marino F, Gileadi O. Probing the structural basis of RecQ helicase function. Biophysical Chemistry. 2010;149(3):67–77. doi: 10.1016/j.bpc.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 47.Pike ACW, Shrestha B, Popuri V, et al. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(4):1039–1044. doi: 10.1073/pnas.0806908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Büttner K, Nehring S, Hopfner K. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nature Structural and Molecular Biology. 2007;14(7):647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 49.Lucic B, Zhang Y, King O, et al. A prominent β-hairpin structure in the winged-helix domain of RECQ1 is required for DNA unwinding and oligomer formation. Nucleic Acids Research. 2011;39(5):1703–1717. doi: 10.1093/nar/gkq1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97(1):75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 51.Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107(1):79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- 52.Patel SS, Donmez I. Mechanisms of helicases. Journal of Biological Chemistry. 2006;281(27):18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 53.Sandler SJ, Marians KJ, Zavitz KH, Coutu J, Parent MA, Clark AJ. dnaC mutations suppress defects in DNA replication- and recombination-associated functions in priB and priC double mutants in Escherichia coli K-12. Molecular Microbiology. 1999;34(1):91–101. doi: 10.1046/j.1365-2958.1999.01576.x. [DOI] [PubMed] [Google Scholar]
- 54.Low RL, Shlomai J, Kornberg A. Protein n, a primosomal DNA replication protein of Escherichia coli: purification and characterization. Journal of Biological Chemistry. 1982;257(11):6242–6250. [PubMed] [Google Scholar]
- 55.Allen GC, Jr., Kornberg A. Assembly of the primosome of DNA replication in Escherichia coli. Journal of Biological Chemistry. 1993;268(26):19204–19209. [PubMed] [Google Scholar]
- 56.Feng C, Sunchu B, Greenwood ME, Lopper ME. A bacterial PriB with weak single-stranded DNA binding activity can stimulate the DNA unwinding activity of its cognate PriA helicase. BMC Microbiology. 2011;11, article 189 doi: 10.1186/1471-2180-11-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, Nurse P, Marians KJ. The ordered assembly of the φX174-type primosome. III. PriB facilitates complex formation between PriA and DnaT. Journal of Biological Chemistry. 1996;271(26):15656–15661. doi: 10.1074/jbc.271.26.15656. [DOI] [PubMed] [Google Scholar]
- 58.Szymanski MR, Jezewska MJ, Bujalowski W. Binding of two PriA-PriB complexes to the primosome assembly site initiates primosome formation. Journal of Molecular Biology. 2011;411(1):123–142. doi: 10.1016/j.jmb.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 59.Huang YH, Huang CY. The N-terminal domain of DnaT, a primosomal DNA replication protein, is crucial for PriB binding and self-trimerization. Biochemical and Biophysical Research Communications. 2013;442:147–152. doi: 10.1016/j.bbrc.2013.11.069. [DOI] [PubMed] [Google Scholar]
- 60.Ng JY, Marians KJ. The ordered assembly of the φX174-type primosome. II. Preservation of primosome composition from assembly through replication. The Journal of Biological Chemistry. 1996;271(26):15649–15655. doi: 10.1074/jbc.271.26.15649. [DOI] [PubMed] [Google Scholar]
- 61.Huang YH, Lin MJ, Huang CY. Yeast two-hybrid analysis of PriB-interacting proteins in replication restart primosome: a proposed PriB-SSB interaction model. The Protein Journal. 2013;32(6):477–483. doi: 10.1007/s10930-013-9509-y. [DOI] [PubMed] [Google Scholar]
- 62.Huang YH, Lin HH, Huang CY. A single residue determines the cooperative binding property of a primosomal DNA replication protein, PriB, to single-stranded DNA. Bioscience, Biotechnology and Biochemistry. 2012;76(6):1110–1115. doi: 10.1271/bbb.110938. [DOI] [PubMed] [Google Scholar]
- 63.Shioi S, Ose T, Maenaka K, et al. Crystal structure of a biologically functional form of PriB from Escherichia coli reveals a potential single-stranded DNA-binding site. Biochemical and Biophysical Research Communications. 2005;326(4):766–776. doi: 10.1016/j.bbrc.2004.11.104. [DOI] [PubMed] [Google Scholar]
- 64.Lopper M, Holton JM, Keck JL. Crystal structure of PriB, a component of the Escherichia coli replication restart primosome. Structure. 2004;12(11):1967–1975. doi: 10.1016/j.str.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 65.Liu JH, Chang TW, Huang CY, et al. Crystal structure of PriB, a primosomal DNA replication protein of Escherichia coli . The Journal of Biological Chemistry. 2004;279(48):50465–50471. doi: 10.1074/jbc.M406773200. [DOI] [PubMed] [Google Scholar]
- 66.Horvath MP. Structural anatomy of telomere OB proteins. Critical Reviews in Biochemistry and Molecular Biology. 2011;46(5):409–435. doi: 10.3109/10409238.2011.609295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flynn RL, Zou L. Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians. Critical Reviews in Biochemistry and Molecular Biology. 2010;45(4):266–275. doi: 10.3109/10409238.2010.488216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kerr ID, Wadsworth RIM, Cubeddu L, Blankenfeldt W, Naismith JH, White MF. Insights into ssDNA recognition by the OB fold from a structural and thermodynamic study of SulfolobusSSB protein. The EMBO Journal. 2003;22(11):2561–2570. doi: 10.1093/emboj/cdg272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. The EMBO Journal. 1993;12(3):861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang YH, Lo YH, Huang W, Huang CY. Crystal structure and DNA-binding mode of Klebsiella pneumoniae primosomal PriB protein. Genes to Cells. 2012;17(10):837–849. doi: 10.1111/gtc.12001. [DOI] [PubMed] [Google Scholar]
- 71.Hsieh HC, Huang CY. Identification of a novel protein, PriB, in Klebsiella pneumoniae . Biochemical and Biophysical Research Communications. 2011;404(1):546–551. doi: 10.1016/j.bbrc.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 72.Szymanski MR, Jezewska MJ, Bujalowski W. Interactions of the Escherichia coli Primosomal PriB Protein with the Single-stranded DNA. Stoichiometries, Intrinsic Affinities, Cooperativities, and Base Specificities. Journal of Molecular Biology. 2010;398(1):8–25. doi: 10.1016/j.jmb.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang CY, Hsu CH, Sun YJ, Wu HN, Hsiao CD. Complexed crystal structure of replication restart primosome protein PriB reveals a novel single-stranded DNA-binding mode. Nucleic Acids Research. 2006;34(14):3878–3886. doi: 10.1093/nar/gkl536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chan KW, Lee YJ, Wang CH, Huang H, Sun YJ. Single-stranded DNA-binding protein complex from Helicobacter pylori suggests an ssDNA-binding surface. Journal of Molecular Biology. 2009;388(3):508–519. doi: 10.1016/j.jmb.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 75.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nature Structural Biology. 2000;7(8):648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 76.Ponomarev VA, Makarova KS, Aravind L, Koonin EV. Gene duplication with displacement and rearrangement: origin of the bacterial replication protein PriB from the single-stranded DNA-binding protein Ssb. Journal of Molecular Microbiology and Biotechnology. 2003;5(4):225–229. doi: 10.1159/000071074. [DOI] [PubMed] [Google Scholar]
- 77.Fujiyama S, Abe Y, Takenawa T, et al. Involvement of histidine in complex formation of PriB and single-stranded DNA. Biochimica et Biophysica Acta. 2014;184:299–307. doi: 10.1016/j.bbapap.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 78.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiological Reviews. 1990;54(4):342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wilce JA, Vivian JP, Hastings AF, et al. Structure of the RTP-DNA complex and the mechanism of polar replication fork arrest. Nature Structural Biology. 2001;8(3):206–210. doi: 10.1038/84934. [DOI] [PubMed] [Google Scholar]
- 80.Tsai KL, Sun YJ, Huang CY, Yang JY, Hung MC, Hsiao CD. Crystal structure of the human FOXO3a-DBD/DNA complex suggests the effects of post-translational modification. Nucleic Acids Research. 2007;35(20):6984–6994. doi: 10.1093/nar/gkm703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsai KL, Huang CY, Chang CH, Sun YJ, Chuang WJ, Hsiao CD. Crystal structure of the human FOXK1a-DNA complex and its implications on the diverse binding specificity of winged helix/forkhead proteins. Journal of Biological Chemistry. 2006;281(25):17400–17409. doi: 10.1074/jbc.M600478200. [DOI] [PubMed] [Google Scholar]
- 82.Kamashev D, Balandina A, Mazur AK, Arimondo PB, Rouviere-Yaniv J. HU binds and folds single-stranded DNA. Nucleic Acids Research. 2008;36(3):1026–1036. doi: 10.1093/nar/gkm667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Swinger KK, Lemberg KM, Zhang Y, Rice PA. Flexible DNA bending in HU-DNA cocrystal structures. The EMBO Journal. 2003;22(14):3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Masai H, Arai K. Initiation of lagging-strand synthesis for pBR322 plasmid DNA replication in vitro is dependent on primosomal protein i encoded by dnaT. The Journal of Biological Chemistry. 1988;263(29):15016–15023. [PubMed] [Google Scholar]
- 85.Masai H, Bond MW, Arai K. Cloning of the Escherichia coli gene for primosomal protein i: the relationship to dnaT, essential for chromosomal DNA replication. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(5):1256–1260. doi: 10.1073/pnas.83.5.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Arai K, McMacken R, Yasuda S, Kornberg A. Purification and properties of Escherichia coli protein i, a prepriming protein in ∅X174 DNA replication. The Journal of Biological Chemistry. 1981;256(10):5281–5286. [PubMed] [Google Scholar]
- 87.Masai H, Arai K. Escherichia coli dnaT gene function is required for pBR322 plasmid replication but not for R1 plasmid replication. Journal of Bacteriology. 1989;171(6):2975–2980. doi: 10.1128/jb.171.6.2975-2980.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Black SL, Dawson A, Ward FB, Allen RJ. Genes required for growth at high hydrostatic pressure in Escherichia coli K-12 identified by genome-wide screening. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073995.e73995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jang S, Sandler SJ, Harshey RM. Mu insertions are repaired by the double-strand break repair pathway of Escherichia coli . PLoS Genetics. 2012;8(4) doi: 10.1371/journal.pgen.1002642.e1002642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang YH, Lin MJ, Huang CY. DnaT is a single-stranded DNA binding protein. Genes Cells. 2013;18:1007–1019. doi: 10.1111/gtc.12095. [DOI] [PubMed] [Google Scholar]
- 91.Szymanski MR, Jezewska MJ, Bujalowski W. The Escherichia coli primosomal dnat protein exists in solution as a monomer-trimer equilibrium system. Biochemistry. 2013;52(11):1845–1857. doi: 10.1021/bi301568w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lo YH, Tsai KL, Sun YJ, Chen WT, Huang CY, Hsiao CD. The crystal structure of a replicative hexameric helicase DnaC and its complex with single-stranded DNA. Nucleic Acids Research. 2009;37(3):804–814. doi: 10.1093/nar/gkn999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annual Review of Biophysics and Biomolecular Structure. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang C, Curth U, Urbanke C, Kang CH. Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4 A resolution. Nature Structural Biology. 1997;4(2):153–157. doi: 10.1038/nsb0297-153. [DOI] [PubMed] [Google Scholar]
- 95.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 96.Chen C-C, Hwang J-K, Yang J-M. (PS)2-v2: template-based protein structure prediction server. BMC Bioinformatics. 2009;10, article 366 doi: 10.1186/1471-2105-10-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen CC, Hwang JK, Yang JM. (PS)2: protein structure prediction server. Nucleic Acids Research. 2006;34:W152–W157. doi: 10.1093/nar/gkl187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nature Reviews Molecular Cell Biology. 2008;9(5):391–401. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 99.Zhang W, Dillingham MS, Thomas CD, Allen S, Roberts CJ, Soultanas P. Directional loading and stimulation of PcrA helicase by the replication initiator protein RepD. Journal of Molecular Biology. 2007;371(2):336–348. doi: 10.1016/j.jmb.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 100.Shereda RD, Bernstein DA, Keck JL. A central role for SSB in Escherichia coli RecQ DNA helicase function. Journal of Biological Chemistry. 2007;282(26):19247–19258. doi: 10.1074/jbc.M608011200. [DOI] [PubMed] [Google Scholar]
- 101.Matson SW, Robertson AB. The UvrD helicase and its modulation by the mismatch repair protein MutL. Nucleic Acids Research. 2006;34(15):4089–4097. doi: 10.1093/nar/gkl450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]


