Abstract
A mesophilic, neutrophilic and aerobic, ammonia-oxidizing archaeon, strain EN76T, was isolated from garden soil in Vienna (Austria). Cells were irregular cocci with a diameter of 0.6–0.9 µm and possessed archaella and archaeal pili as cell appendages. Electron microscopy also indicated clearly discernible areas of high and low electron density, as well as tubule-like structures. Strain EN76T had an S-layer with p3 symmetry, so far only reported for members of the Sulfolobales. Crenarchaeol was the major core lipid. The organism gained energy by oxidizing ammonia to nitrite aerobically, thereby fixing CO2, but growth depended on the addition of small amounts of organic acids. The optimal growth temperature was 42 °C and the optimal pH was 7.5, with ammonium and pyruvate concentrations of 2.6 and 1 mM, respectively. The genome of strain EN76T had a DNA G+C content of 52.7 mol%. Phylogenetic analyses of 16S rRNA genes showed that strain EN76T is affiliated with the recently proposed phylum Thaumarchaeota, sharing 85 % 16S rRNA gene sequence identity with the closest cultivated relative ‘Candidatus Nitrosopumilus maritimus’ SCM1, a marine ammonia-oxidizing archaeon, and a maximum of 81 % 16S rRNA gene sequence identity with members of the phyla Crenarchaeota and Euryarchaeota and any of the other recently proposed phyla (e.g. ‘Korarchaeota’ and ‘Aigarchaeota’). We propose the name Nitrososphaera viennensis gen. nov., sp. nov. to accommodate strain EN76T. The type strain of Nitrososphaera viennensis is strain EN76T ( = DSM 26422T = JMC 19564T). Additionally, we propose the family Nitrososphaeraceae fam. nov., the order Nitrososphaerales ord. nov. and the class Nitrososphaeria classis nov.
Introduction
Microbes are immensely diverse and abundant, and inhabit virtually all environments on Earth. However, most of this microbial diversity remains undescribed, given that many novel organisms are fastidious and their isolation and cultivation is time-consuming or even impossible (Rappé & Giovannoni, 2003). Cultivation-independent techniques have increased our knowledge of microbial diversity and metabolism tremendously (Lane et al., 1985; Marcy et al., 2007; Rinke et al., 2013; Schmidt et al., 1991; Stein et al., 1996), and have led to the proposal of several new bacterial and archaeal phyla (e.g. Elkins et al., 2008; Gordon & Giovannoni, 1996; Huber et al., 2002; Hugenholtz et al., 1998; Nunoura et al., 2011; Rinke et al., 2013). In addition, metabolic predictions based on meta-omic studies can also support the design of media and consequently facilitate the cultivation of uncharacterized microorganisms (Tyson & Banfield, 2005). When used in combination, microbial cultivation and metagenomics represent a powerful toolbox to describe newly discovered microbial physiologies, and to simultaneously assess their ecological distribution and impact. The discovery of ammonia-oxidizing archaea (AOA) represents a successful example of such an integrated approach. These archaea, originally called ‘mesophilic Crenarchaeota’, were first identified in marine samples (DeLong, 1992; DeLong et al., 1994; Fuhrman et al., 1992) and were later detected in many more environments (Jurgens et al., 1997; Karner et al., 2001; MacGregor et al., 1997; Ochsenreiter et al., 2003; Preston et al., 1996; Schleper et al., 1997; Takai et al., 2001). The first evidence for their metabolism and potential ecological role was given by the discovery of genes encoding a putative ammonia mono-oxygenase, the key enzyme for ammonia oxidation, through metagenomics (Hallam et al., 2006b; Treusch et al., 2005; Venter et al., 2004) and through the cultivation and isolation of the first AOA, ‘Candidatus Nitrosopumilus maritimus’ SCM1 (Könneke et al., 2005). The widespread occurrence of potential archaeal ammonia oxidizers in the environment has been confirmed by a large number of qualitative and quantitative molecular surveys (Adair & Schwartz, 2008; Alves et al., 2013; de la Torre et al., 2008; Francis et al., 2005; He et al., 2007; Hershberger et al., 1996; Leininger et al., 2006; Pester et al., 2012; Reigstad et al., 2008; Shen et al., 2008; Wuchter et al., 2006; Zhang et al., 2008). Based on phylogenetic analyses of concatenated ribosomal protein sequences and full-genome comparisons with the first genome sequence of the putative AOA ‘Candidatus Cenarchaeum symbiosum’ (Hallam et al., 2006a), Brochier-Armanet et al. (2008) suggested that the AOA comprise a new archaeal phylum, the Thaumarchaeota, which was subsequently supported by Spang et al. (2010) upon the inclusion of two more genome sequences of members of the new phylum.
Due to the difficulty in cultivating and purifying members of the Thaumarchaeota in laboratory cultures, pure isolates are still rare (Könneke et al., 2005; Tourna et al., 2011) and insights into the physiology of AOA are confined to studies of two pure cultures (Könneke et al., 2005; Martens-Habbena et al., 2009; Schouten et al., 2008) or are based on enrichment cultures (de la Torre et al., 2008; Hatzenpichler et al., 2008; Jung et al., 2011; Lehtovirta-Morley et al., 2011; Santoro & Casciotti, 2011). ‘Candidatus Nitrosopumilus maritimus’ SCM1 is a representative of the marine I.1a group, which is one of the distinct lineages formed within the phylum Thaumarchaeota. With the isolation of strain EN76T, the first pure culture from soil and group I.1b was obtained, which represents the second major lineage of the Thaumarchaeota. The isolation of strain EN76T confirmed ammonia oxidation by thaumarchaeotes in soil and expanded the metabolic spectrum of AOA to include the utilization of urea and organic substrates (Tourna et al., 2011). These findings demonstrate the importance of pure cultures to the investigation of growth requirements, and will also help in future cultivation and purification experiments of other members of the Thaumarchaeota.
The open ocean, marine sediments and soil contain large numbers of microbes, with approximately 1.2×1029, 2.9×1029 and 2.6×1029 cells, respectively (Kallmeyer et al., 2012; Whitman et al., 1998). Molecular surveys based on amoA or 16S rRNA gene sequences have shown that members of the Thaumarchaeota make up a large fraction of the microbial biomass in all three habitats (Bates et al., 2011; Karner et al., 2001; Leininger et al., 2006). With up to 20 % of the picoplankton in the marine environment (Karner et al., 2001), up to 80 % of the microbiota in certain horizons of marine sediments (Durbin & Teske, 2010; Jorgensen et al., 2012) and up to 1 % of the total microbiota in soil (Bates et al., 2011; Leininger et al., 2006), the total abundance of thaumarchaeotes seems to be comparable to that of other highly abundant bacterial phyla, even to that of the ubiquitous Proteobacteria. Based on their large numbers in many environments and their ability to perform the first and rate-limiting step in nitrification, the oxidation of ammonia to nitrite, members of the Thaumarchaeota are now considered to play a major role in the global nitrogen cycle (Alves et al., 2013; Di et al., 2009; Erguder et al., 2009; Gubry-Rangin et al., 2010; Hatzenpichler, 2012; Jia & Conrad, 2009; Nicol & Schleper, 2006; Offre et al., 2009; Prosser & Nicol, 2012; Schauss et al., 2009; Schleper & Nicol, 2010; Stahl & de la Torre, 2012; Stieglmeier et al., 2014a; Wuchter et al., 2006; Xia et al., 2011; Zhalnina et al., 2012). Several factors have been suggested as drivers of environmental adaptation and niche selection of thaumarchaeotes, such as pH and ammonia concentration (reviewed by Prosser & Nicol, 2012). For example, ‘Candidatus Nitrosopumilus maritimus’ SCM1 and other marine members of the Thaumarchaeota have been shown to have a higher affinity to ammonia than cultivated ammonia-oxidizing bacteria (AOB), and all strains in pure culture or enrichment are adapted to low concentrations of ammonia (Horak et al., 2013; Martens-Habbena et al., 2009; Prosser & Nicol, 2012). However, thaumarchaeotes are widespread in diverse habitats, exposed to wide ranges of pH and ammonia concentration, and recent studies suggest that they almost certainly harbour a broader range of physiological and/or metabolic properties than is currently known (Alves et al., 2013; Durbin & Teske, 2010; Jorgensen et al., 2012; Muller et al., 2010; Mussmann et al., 2011). In order to understand their ecological impact, it is important to identify and characterize in detail the metabolic functions and physiological versatility of more representatives of these archaea.
Although AOA have been studied intensively in the last decade (reviewed by e.g. Nicol & Schleper, 2006; Stahl & de la Torre, 2012; Stieglmeier et al., 2014a), there are presently no validly published names of species, genera or higher ranks within the phylum Thaumarchaeota. Here, we extend the original characterization of strain EN76T (Tourna et al., 2011), with a special focus on its ultrastructure and growth improvements, and formally propose the species Nitrososphaera viennensis sp. nov. and propose to assign this species as the type species of the genus Nitrososphaera gen. nov. The genus Nitrososphaera is the type of the family Nitrososphaeraceae fam. nov. and the order Nitrosophaerales ord. nov., which is in turn the type of the class Nitrososphaeria classis nov.
Methods
Sample source and culture conditions.
Sample source and isolation strategy for strain EN76T were described previously by Tourna et al. (2011). In brief, strain EN76T was isolated from Viennese garden soil (48° 13′ 48.72″ N 16° 21′ 28.93″ E) by transferring 5 g soil into 50 ml freshwater medium (FWM) containing (l−1) 1 g NaCl, 0.4 g MgCl2 . 6H2O, 0.1 g CaCl2 . 2H2O, 0.2 g KH2PO4, 0.5 g KCl, 1 ml trace element mixture, 1 ml vitamin solution and 7.5 µM ferric sodium EDTA, 0.5 mM NH4Cl as energy source and 2 mM NaHCO3 as carbon source. Additionally 0.1 mM NaNO2 was supplied to the cultures. The medium was adjusted to pH 7.5 and cultures were incubated at 37 °C in the dark without shaking. Supplementation with antibiotics (carbenicillin, streptomycin) and filtration of the cultures (0.45 µm pore size) were applied to reduce bacterial and fungal contaminants. Growth was followed by measuring ammonium consumption and nitrite production photometrically. Additionally, light microscopy and quantitative PCR were used as described previously (Tourna et al., 2011).
The purified strain EN76T was routinely cultured in 20 ml FWM in sterile 30 ml polystyrene screw-capped containers (VWR; catalogue no. 216-2637) supplemented with 1 mM NH4Cl, 2 mM NaHCO3, 0.1 mM sodium pyruvate and 100 µg antibiotics ml−1 (kanamycin, streptomycin or ofloxacin in MilliQ water). The medium was buffered with HEPES/NaOH to pH 7.5. Larger volumes were cultured in glass bottles and shaken (150 r.p.m.) in darkness.
Physiological characterization and multivariate optimization of growth conditions.
To determine optimal growth parameters as well as the effect of different nitrogen and carbon substrates on the growth of EN76T, the strain was cultivated in closed batch in 20 ml FWM containing 100 µg antibiotics ml−1 as described above. Substrates were dissolved in MilliQ water and sterile filtered (0.2 µm) before usage. Table 1 gives an overview of tested substrates.
Table 1. Effect of substrates on growth of strain EN76T.
The effect of various substrates on growth of EN76T was evaluated compared with a chemolithoautotrophic culture supplemented with 1 mM ammonium and 2 mM bicarbonate (default culture). Growth is scored as: ++, positive effect; +, similar to default culture; +/−, negative effect; −, total inhibition of growth. All incubations were performed at least in duplicate. The table is an extended version of Table S1 of Tourna et al. (2011). The default ammonium concentration added to cultures was 1 mM (if not stated otherwise) and the default pyruvate concentration added to cultures was 0.5 mM (if not stated otherwise).
| Substrate | Substrate added | Growth | |
| Ammonium | Pyruvate | ||
| Carbon compounds | |||
| Bicarbonate >2 mM (up to 10 mM) | Yes | No | + |
| Organic acids (TCA cycle) | |||
| Acetate (0.05–2 mM) | Yes | No | + |
| Citrate, succinate, fumarate, malate (0.1–0.5 mM) | Yes | No | + |
| Pyruvate (0.05–10 mM) | Yes | Yes | ++ |
| Pyruvate (0.5 mM) | No | Yes | − |
| Oxaloacetate (0.5 mM) | Yes | No | ++ |
| α-Ketoglutarate (0.5 mM) | Yes | No | ++ |
| Glyoxylate (0.5 mM) | Yes | No | ++ |
| Sugars and sugar alcohols | |||
| Glucose, fructose, lactose, arabinose, sucrose, galactose, mannose (0.5–1 mM) | Yes | No | + |
| Ribose (0.5 mM) | Yes | No | − |
| Glycerol (0.1 %, v/v) | Yes | No | − |
| Complex organic compounds | |||
| Peptone, yeast extract (0.05 %, w/v) | Yes | No | − |
| Nitrogen compounds | |||
| Ammonium | |||
| 0.5–3 mM | Yes | No | + |
| 0.5–15 mM | Yes | Yes | ++ |
| Urea | |||
| 0.5–1 mM | No | No | + |
| 0.5 mM | No | Yes | ++ |
| Amino acids | |||
| l-Alanine, d-alanine, l-glutamine, l-aspartic acid (0.1 g l−1) | Yes | No | − |
| l-Glutamic acid, d-glutamic acid (0.1 g l−1) | Yes | No | + |
| Amino acid mixture (1 mM), Casamino acids (0.05 %, w/v) | Yes | No | − |
| Amines | |||
| Trimethylamine, ethanolamine (1 mM) | No | No | +/− |
| Methanolamine (1 mM) | No | No | − |
| Methylamine (0.5 mM) | Yes | No | +/− |
| Nitrate (1 mM) | No | Yes | − |
| Taurine | |||
| 0.25–0.5 mM | Yes | No | + |
| 0.25–0.5 mM | Yes | Yes | ++ |
| Nucleobases | |||
| Pyrimidine, purine (0.1–1 mM) | Yes | No | +/− |
| Cytidine (0.1–1 mM) | Yes | No | + |
In order to investigate optimal growth conditions for EN76T, a design of experiments (DoE) strategy was applied, using the factors temperature, pyruvate concentration and ammonium concentration. Based on our preliminary knowledge of the strain’s growth requirements (Tourna et al., 2011), the range for each factor (design space) was set as follows: 37–47 °C, 0.1–1.5 mM sodium pyruvate and 1–4 mM NH4Cl. As nitrite production was shown to follow biomass production (Tourna et al., 2011), it was used to calculate the growth rate (μ) and maximum growth rate (μmax), which were eventually used to develop the model (Design-Expert 8 software; Stat-Ease Inc.). Experiments were conducted in triplicate, except for the centre points of the initial two-level factorial screening design, which were set up in fivefold replicates. The two-level factorial design was applied in order to screen the design space rapidly. Due to a low model significance of data obtained from the initial two-level factorial screening design space, an augmented matrix was used in order to account for putative interactions of individual factors. Thus, the two-level factorial design space was extended by using a face-centred augmented matrix. Eventually, data points of all experiments (n = 51) were used to establish a response surface model (RSM). Data were analysed with the software Design-Expert 8. ANOVA, based on a stepwise regression elimination procedure, was used to develop the model. The desirability approach, as described elsewhere (Derringer & Suich, 1980), was used to maximize μ or μmax (variable) based on variation of quantitative factors, here c(ammonium), c(pyruvate) and temperature (within the design space). A score is given to each quantitative factor setting that can be used to maximize the variable. In this approach, desirability between 0 and 1 (corresponding to 0–100 %) can be assigned to a variable for optimization; factors identified as being outside a certain desirability function will not be considered for model generation. To verify the calculated optimal growth conditions identified by the established RSM model design space, one additional growth experiment (fivefold-replicated closed-batch cultures) was performed (Fig. 1a).
Fig. 1.
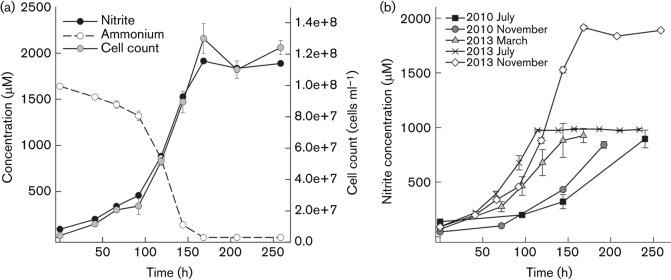
(a) Growth curve of a culture of strain EN76T grown at 42 °C with 2 mM NH4Cl and 1 mM pyruvate. Cell counts, ammonium consumption and nitrite production were used to follow growth. Data represent mean values of triplicate cultures with standard deviations plotted (sometimes smaller than symbols). (b) Acceleration of growth of strain EN76T since purification of the strain in 2010 (Tourna et al., 2011). The cultivation conditions were as follows: July 2010 and November 2010, 37 °C, 1 mM NH4Cl, 1 mM pyruvate; March 2013, 37 °C, 1 mM NH4Cl, 0.1 mM pyruvate; July 2013, 42 °C, 1 mM NH4Cl, 0.8 mM pyruvate; November 2013, 42 °C, 2 mM NH4Cl, 1 mM pyruvate. Nitrite production was used to follow growth. Data represent mean values of replicated cultures (three to five replicates) with standard deviations plotted (sometimes smaller than symbols). Data points previously published in Fig. 3(b) of Tourna et al. (2011) (i.e. July 2010) were included in the figure.
For the cultivation of strain EN76T under reduced oxygen concentrations, serum bottles were sealed with rubber stoppers. Therefore, the effect of black butyl (Glasgerätebau Ochs), grey butyl (Sigma Aldrich), blue butyl (Dunn Labortechnik), red isoprene (Sigma Aldrich), grey natural (Carl Roth) and grey–blue natural (VWR) rubber stoppers on the growth of strain EN76T was tested (Table S1, available in the online Supplementary Material).
Microscopy.
For negative staining, cells were fixed with 2.5 % glutaraldehyde in 1× PBS, transferred to carbon-coated copper grids (200 mesh) and stained with 0.5 % uranyl acetate for 2 min as described previously (Tourna et al., 2011). Images were recorded with a megaview III camera (SIS) attached to a Philips EM 208 transmission electron microscope (FEI) operated at 70 keV.
For scanning electron microscopy (SEM), poly-l-lysine-coated glass slides were added to the culture from early to late exponential phase. Attached cells were fixed for 2 h at room temperature with 2 % glutaraldehyde and 2 % formaldehyde in 0.06 × PHEM buffer (buffer based on PIPES, HEPES, EGTA and MgCl2; J. Montanaro and N. Leisch, unpublished). The cells were post-fixed with 1 % osmium tetroxide for 2 h at room temperature followed by dehydration in a graded ethanol series. After immersion in pure acetone, the slides were critical-point-dried with a CPD 300 unit (Leica). The slides were then mounted on stubs, gold-coated with an AGAR B7340 sputter-coater and imaged using an XL20 instrument (Philips) running the Microscope control program (version 7.00; FEI).
For the preparation of ultrathin sections, cells were grown in 1 l FWM supplemented with 3 mM NH4Cl and 0.15 mM sodium pyruvate until the late exponential growth phase. Cells were harvested by centrifugation and fixed as mentioned above for SEM preparation. After fixation, cells were washed twice in 100 mM PHEM buffer and covered with 1 % agar. Cells were post-fixed for 1 h in osmium tetroxide (1 %), washed three times (1×PHEM; J. Montanaro and N. Leisch, unpublished), dehydrated in a graded ethanol series and embedded in resin with acetonitrile as solvent. Polymerization was achieved by incubating the resin blocks for 1 h at 40 °C and for 48–72 h at 60 °C (Leisch et al., 2011). An Ultracut S (Leica) was used to produce ultrathin sections (70 nm), which were then transferred to copper grids (300 mesh). They were post-stained with uranyl acetate and lead citrate before visualization on a Zeiss 902 instrument equipped with an Olympus SharpEye camera at an accelerating voltage of 80 keV and a Libra120 instrument (Carl Zeiss) equipped with a slow-scan CCD camera (Tröndle) at an accelerating voltage of 120 keV. Images were post-processed with Adobe Photoshop CS5.
To determine the general morphology and properties of the potential S-layer protein of EN76T, negative staining of purified S-layer sheets as well as freeze-fracturing/freeze-etching was performed. If not otherwise mentioned, freeze-etching was carried out as described previously (Klingl et al., 2011; Rachel et al., 2010). The purification of S-layer proteins was done by breaking the cells by sonication, differential centrifugation and extraction of lipids, using a MES buffer system (Klingl, 2011; Veith et al., 2009). Subsequent negative staining of S-layer sheets for transmission electron microscopy (TEM) was performed as described previously (Rachel et al., 2010). For the investigation of freeze-etching replicas and purified S-layer proteins, a JEOL JEM 2100 TEM, equipped with a fast scan 2k×2k CCD camera F214 (TVIPS), was used at an accelerating voltage of 120 kV. Image analyses, correlation averaging and determination of S-layer symmetry and lattice constants of negatively stained S-layer proteins as well as freeze-etching replicas was performed with the animetra crystals software package (release 1.1; Animetra).
Lipid analyses.
Intact polar lipids and glycerol dibiphytanyl glycerol tetraether (GDGT) were analysed previously and described by Sinninghe Damsté et al. (2012).
DNA isolation and phylogenetic analyses.
Isolation of DNA from the enrichment culture of strain EN76T and 454 pyrosequencing on a 454/FLX-Titanium sequencer (Roche) have been described in detail previously (Tourna et al., 2011). The genome sequence was assembled and annotated on the MicroScope platform (Vallenet et al., 2006, 2009). The 16S rRNA, amoA and amoB gene sequences of EN76T have been deposited previously in GenBank under accession numbers FR773157, FR773159 and FR773160, respectively (Tourna et al., 2011). Phylogeny reconstruction of archaeal 16S rRNA genes was based on the alignment of 1202 bp gene fragments with four independent methods: clustal w (Thompson et al., 1994), muscle (Edgar, 2004), mafft (Katoh et al., 2005; Katoh & Toh, 2010), and T-Coffee (Di Tommaso et al., 2011). The final consensus multiple sequence alignment of the four methods was calculated with MergeAlign (Collingridge & Kelly, 2012). After manual curation, hypervariable positions that could not be aligned unambiguously were excluded. Maximum-likelihood phylogenetic trees and bootstrap support values were calculated with RaxML VI-HPC (Stamatakis, 2006; Stamatakis et al., 2008) based on the GTR model with invariable sites and gamma site rate variation (GTR+I+G). clustal w, muscle, mafft and RaxML analyses were performed through the CIPRES Science Gateway version 3.3 (Miller et al., 2010).
Storage.
Cells of strain EN76T were harvested, suspended in 40 % (v/v) glycerol and stored at −80 °C. Growth could be restored after preservation for 12 months by carefully thawing cells on ice and removing the glycerol by centrifugation of cells prior to inoculation into fresh medium.
Results and Discussion
Metabolism
Strain EN76T is a mesophilic and neutrophilic organism, growing at 28–47 °C and pH 6–8.5 (Tourna et al., 2011). It produces energy by oxidizing ammonia aerobically to nitrite (Fig. 1a). Strain EN76T grows equally well on urea as an energy source, with production of about 2 mmol nitrite per mol urea (Tourna et al., 2011). In contrast to ‘Candidatus Nitrosopumilus maritimus’ SCM1 (group I.1a), strain EN76T and its close relative ‘Candidatus Nitrososphaera gargensis’ Ga9.2 (both associated with group I.1b) possess genes encoding urease and urea transporters (Spang et al., 2012; Stieglmeier et al., 2014a; Tourna et al., 2011; Walker et al., 2010) and, accordingly, growth on urea has also been demonstrated in enrichment cultures of ‘Candidatus Nitrososphaera gargensis’ Ga9.2 (Spang et al., 2012). Genes for urea utilization were recently also found in metagenomic analyses of marine group I thaumarchaeotes from Arctic and meso-/bathypelagic waters and marine sediments, indicating that urea is a more widespread energy (and perhaps also carbon) source for AOA (Alonso-Sáez et al., 2012; Park et al., 2012) than was previously appreciated. The NO scavenger carboxy-PTIO [2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide] (Amano & Noda, 1995) has been shown to inhibit growth and nitrite production of both strain EN76T (Shen et al., 2013) and ‘Candidatus Nitrosopumilus maritimus’ SCM1 (Yan et al., 2012), indicating an important role for NO in their energy metabolism. In addition, hydroxylamine is probably an intermediate during the oxidation of ammonia to nitrite, as shown for AOB [Arp & Stein (2003) and references therein; Hooper & Terry (1979)], and recently also shown for the marine AOA ‘Candidatus Nitrosopumilus maritimus’ SCM1 (Vajrala et al., 2013). Strain EN76T tolerated ammonium concentrations up to 15 mM and nitrite concentrations up to 10 mM, as reported previously (Tourna et al., 2011). Thus, EN76T is less sensitive to high ammonium and nitrite concentrations than, for example, its close relative ‘Candidatus Nitrososphaera gargensis’ Ga9.2 and the marine strain ‘Candidatus Nitrosopumilus maritimus’ SCM1 (Hatzenpichler et al., 2008; Könneke et al., 2005). The strain produces nitrous oxide (N2O) in amounts comparable to those of AOB under oxic conditions (4.6±0.6 amol N2O cell−1 h−1). However, in contrast to AOB, N2O production does not increase under reduced oxygen levels, and might occur via a hybrid formation mechanism (Stieglmeier et al., 2014b).
Strain EN76T is a mixotrophic organism that requires organic acids (e.g. pyruvate, oxaloacetate, α-ketoglutarate or glyoxylate) to stimulate growth (Table 1; Tourna et al., 2011). Other organic substrates, such as sugars or amines, did not have a positive effect on growth of EN76T (Table 1). Growth stimulation by organic acids has recently also been reported for the marine strain ‘Candidatus Nitrosopumilus maritimus’ SCM1 (Stahl & de la Torre, 2012; Urakawa et al., 2011). Comparative genomic analyses of strain EN76T, ‘Candidatus Nitrosopumilus maritimus’ SCM1 (Walker et al., 2010) and ‘Candidatus Nitrososphaera gargensis’ Ga9.2 (Spang et al., 2012) revealed the presence of genes encoding transporters for amino acids, sulfonates (e.g. taurine) and glycerol in all three strains (P. Offre, M. Kerou, A. Spang and C. Schleper, unpublished). In addition, EN76T encodes putative nucleobase transporters. Therefore, different amino acids, taurine, glycerol and nucleobases were tested in various concentrations as possible substrates for EN76T (Table 1). However, none of the above-mentioned compounds had a positive effect on growth under the conditions tested. Instead, glycerol, several amino acids and nucleobases inhibited growth.
Strain EN76T was initially described to grow optimally in FWM supplemented with 1 mM pyruvate and 1 mM NH4Cl at 37 °C and pH 7.5 (Tourna et al., 2011), with a doubling time of approximately 45 h (based on cell counts). However, the generation time of strain EN76T decreased progressively from 45 to 27.5 h during continuous cultivation and growth optimizations over 3 years (Fig. 1b). In order to reassess the optimal growth conditions and possible interactions between the three known factors that influence growth rate (i.e. temperature and ammonium and pyruvate concentrations) comprehensively, we used a DoE screening and optimization strategy (Box & Draper, 1987; Box & Lucas, 1959; Rittmann & Herwig, 2012). As an example for the output of those experiments, the RSM shown in Fig. S1 illustrates the effect of temperature and pyruvate concentration on the maximum specific growth rate (μmax) at a constant ammonium concentration of 2.5 mM. Based on the DoE data and the calculated model obtained after varying temperature, ammonium and pyruvate concentrations, we predicted the optimal growth conditions of EN76T (see Table S2). The highest maximum specific growth rate (μmax 0.024 h−1) should be reached at 41.83 °C, 1.05 mM pyruvate and 2.59 mM NH4Cl (with a desirability of 84 %). This corresponds to a generation time of 29.0 h. In order to verify μmax and the generation time predicted by the RSM experimentally, we grew the strain at 42 °C, 1 mM pyruvate and 2 mM NH4Cl and obtained a maximum specific growth rate (μmax) of 0.023 h−1 and a generation time of 30.1±0.6 h (based on nitrite production). Similar values for growth rate and generation time were obtained using cell counts of EN76T for the calculation (μmax 0.026 h−1; generation time 27.5±6.5 h). These experimentally determined values are close to the generation time (29.2 h) and a maximum specific growth rate (μmax 0.024 h−1) predicted by the model equation under the tested conditions (Table S2). The model equation was additionally verified by recalculating the generation time reported by Tourna et al. (2011). The generation time based on the previously used growth conditions was calculated as 42.0±2.7 h, which is close to the initial experimental determination of 45 h (Tourna et al., 2011). Enrichment cultures of the closely related strain ‘Candidatus Nitrososphaera gargensis’ Ga9.2 have been reported to grow at 46 °C with an ammonium concentration of 1 mM (Hatzenpichler et al., 2008).
Given that thaumarchaeotes have been shown to be light-sensitive (French et al., 2012; Merbt et al., 2012), strain EN76T was incubated in the dark. When cultivated in larger volumes (>100 ml), cultures were shaken at 150 r.p.m. Although the strain grows aerobically [21 % (v/v) O2 in the gas phase], it can grow at oxygen concentrations as low as 3 % (v/v) O2 in the gas phase (Stieglmeier et al., 2014b).
Tests of various rubber stoppers indicated that growth of EN76T was inhibited completely by black butyl and red isoprene rubber stoppers, although it tolerated grey and blue butyl rubber stoppers, as well as grey natural rubber stoppers (Table S1). Inhibition of the activity of methanotrophic bacteria by black butyl rubber stoppers has been reported previously (Ettwig et al., 2009).
EN76T is not affected by water-soluble antibiotics like kanamycin, streptomycin, carbenicillin, ampicillin (Tourna et al., 2011) and ofloxacin, but is inhibited by antibiotics that are soluble in ethanol or DMSO (e.g. chloramphenicol). Recently, the effects of nitrification inhibitors (e.g. nitrapyrin, allylthiourea and dicyandiamide) and the antibiotic sulfathiazole, which are commonly used in agriculture and livestock production, on the AOA strain EN76T and a strain of the ammonia-oxidizing bacterium Nitrosospira multiformis have been tested (Shen et al., 2013). Nitrapyrin had a stronger inhibitory effect on EN76T compared with the bacterium, whereas dicyandiamide, the copper chelators allylthiourea and amidinothiourea and the antibiotic sulfathiazole had a weaker inhibitory effect on EN76T (Shen et al., 2013).
Morphology
The irregular coccoid cells of strain EN76T had a diameter of 0.78±0.13 µm (n = 16) and usually occurred as single cells, although clusters comprising several cells were sometimes observed. Cells were motile and possess archaella (archaeal flagella that are homologous to type IV pili; Jarrell & Albers, 2012) with a diameter of 12.0±1.8 nm (Fig. 2g, h). Genes encoding Crenarchaeota-like type-2 flagellins and Euryarchaeota-like chemotaxis proteins (Fig. S2) were found in the genome of EN76T, supporting the conclusion that EN76T is probably motile. Crenarchaeota-like type-2 fla gene clusters have been found in ‘Candidatus Nitrososphaera gargensis’ Ga9.2 and the group I.1a-related strain ‘Candidatus Nitrosoarchaeum limnia’ SFB1, but not in the genome of ‘Candidatus Nitrosopumilus maritimus’ SCM1 (Blainey et al., 2011; Spang et al., 2012; Walker et al., 2010). In addition, pili with a diameter of 6.4±1.3 nm were observed (not shown). The diameters of both appendages are within the size ranges described for other archaea (Klingl et al., 2013).
Fig. 2.
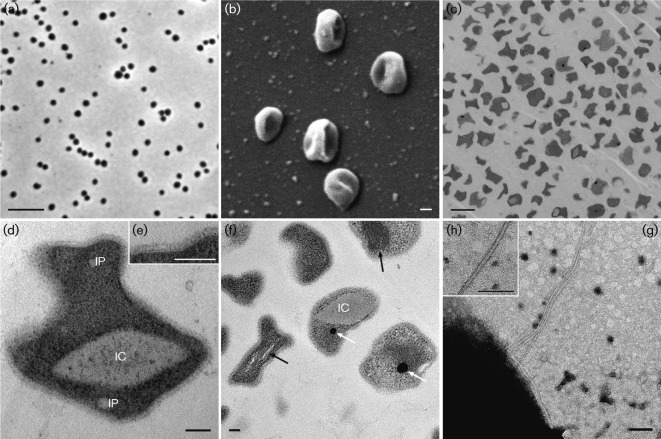
Ultrastructure of cells of strain EN76T. (a) Phase-contrast image; bar, 5 µm. (b) Scanning electron micrograph of several cells depicting the irregular coccoid shape; bar, 100 nm. (c–f) TEM images of ultrathin sections of chemically fixed cells of strain EN76T. (c) Overview displaying the irregular cell shape; bar, 1 µm. (d) Magnified cell showing intracellular features including a clearly discernible area [potential intracellular compartment (IC)] and incorporations (IP). The inset (e) illustrates the cell membrane, pseudo-periplasm and S-layer at higher magnification; bars, 100 nm. (f) Potential intracellular compartment (IC), tubule-like structures (white arrows) and electron-dense particles (black arrows) are highlighted; bar, 100 nm. (g, h) Transmission electron micrographs of a cell with an archaellum; inset (h) shows the magnified archaellum. Bars, 100 nm.
In order to investigate the ultrastructure of EN76T, ultrathin sections of chemically fixed and embedded cells were prepared (Fig. 2c–f). Cells often contained one or two electron-dense particles with a size of 87±15 nm (Fig. 2f). Similar electron-dense particles present in anaerobic ammonium-oxidizing (anammox) bacteria have been shown to be enriched in iron and were proposed to constitute bacterioferritins, which are known as iron storage proteins (Andrews et al., 2003). Furthermore, the genome of EN76T carries genes for proteins belonging to the ferritin/Dps domain proteins (Nvie_002890, Nvie_017250, Nvie_020180, Nvie_028750, Nvie_029390), which have also been identified in genomes of other members of the Thaumarchaeota (Spang et al., 2012). Dps proteins are known to protect cells against oxidative stress by binding iron (Haikarainen & Papageorgiou, 2010). These findings suggest that the electron-dense particles present in strain EN76T may have a role in iron storage and cellular protection. Tubule-like structures were also identified in cells of strain EN76T (Fig. 2f), and may have a cytoskeletal function, as proposed previously for the hexagonal long tubule-like structures found inside the anammoxosome of anammox bacteria (Lindsay et al., 2001; van Niftrik & Jetten, 2012). Although genomes of members of the Thaumarchaeota encode an FtsZ homologue, it is unlikely that the tubule-like structures are formed by this protein, because the marine strain ‘Candidatus Nitrosopumilus maritimus’ SCM1 was shown to recruit the Cdv mechanism primarily during cell division (Pelve et al., 2011). Small inclusions of less electron-dense material (compared with the cytoplasm) were observed in cells of strain EN76T (Fig. 2d), which might be polyphosphate or glycogen storage granules, for example (Klingl et al., 2013). Strain EN76T possesses a clearly discernible area within the cytoplasm (Fig. 2d, e). So far, we could not identify a lipid or proteinaceous layer surrounding this area, which is characteristic for intracellular microcompartments found in bacteria. Large intracellular compartments, such as the carboxysome and anammoxosome, have been described in several bacteria (Erbilgin et al., 2014; Shively et al., 1973; van Niftrik et al., 2004). However, compartmentalization has been reported in only few archaea, e.g. a two-membrane system in the crenarchaeotal genus Ignicoccus (Huber et al., 2000) and in the euryarchaeote Methanomassiliicoccus luminyensis (Dridi et al., 2012). Larger intracellular particles like polyhydroxyalkanoate granules have been found in halophilic archaea (Fernandez-Castillo et al., 1986). Raman spectroscopy analyses have indeed shown that strain EN76T also synthesizes polyhydroxyalkanoates (Spang et al., 2012). Further analyses will be necessary to show whether this region is separated by a membrane or proteinaceous layer from the cytoplasm, and to identify the function of this potential intracellular microcompartment.
S-layer proteins are one, or even the only, cell-wall component of several archaea and bacteria (Rachel et al., 1997). The S-layer usually consists of a single type of protein arranged in a regular lattice pattern, probably driven by entropic processes (Eichler, 2003; Sleytr et al., 2001, 2007). Depending on the arrangement, these pseudocrystalline areas depict highly ordered opaque p1- or p2-symmetry, square p4-symmetry or sixfold p3- or p6-symmetry. TEM analyses of both freeze-etching replicas (Fig. 3a) and negatively stained S-layer sheets (Fig. 3b) of cells of strain EN76T showed a regular pattern of a two-dimensional protein crystal with 6-fold symmetry. By further analysing the images with animetra crystals, distinction between p3- and p6-symmetry could be achieved. Correlation averaging of electron micrographs of both freeze-etched cells (Fig. 4a) and purified S-layer sheets (Fig. 4c) revealed an unexpected p3-symmetry of the S-layer protein. The relief illustration of the image in Fig. 4(a), shown in Fig. 4(b), revealed that the unit cell of the S-layer consists of a trimer of protein trimers. Additionally, the triangular shape of the pores between the protein trimers can be seen, which could function as a molecular sieve, separating the surrounding medium from the pseudo-periplasm, located between the S-layer and the cytoplasmic membrane (reviewed by Sleytr et al., 1993). Determination of the lattice constants after correlation averaging yielded slightly differing values for the two preparation methods, with 21.1 nm for freeze-etching and 20.2 nm for negative staining. This could be caused by the preparation itself, by calculation errors or by the initial choice of the reference area for correlation averaging. The range of lattice values obtained here is, nevertheless, in the same range as those for other p3-symmetry S-layers reported for members of the Sulfolobales, which are all around 21 nm (König et al., 2007; Veith et al., 2009). Up to now, p3-symmetry was thought to be unique to the order Sulfolobales, given that all investigated species from this group had this symmetry and a consistent lattice value (Baumeister & Lembcke, 1992; Deatherage et al., 1983; Grogan, 1996; Klingl et al., 2013; König et al., 2007; Lembcke et al., 1991, 1993; Prüschenk & Baumeister, 1987; Prüschenk et al., 1987; Taylor et al., 1982; Veith et al., 2009). Thus, the occurrence of p3-symmetry in the thaumarchaeote EN76T excludes this characteristic as a taxonomic marker for the order Sulfolobales (Klingl et al., 2011; König et al., 2007).
Fig. 3.
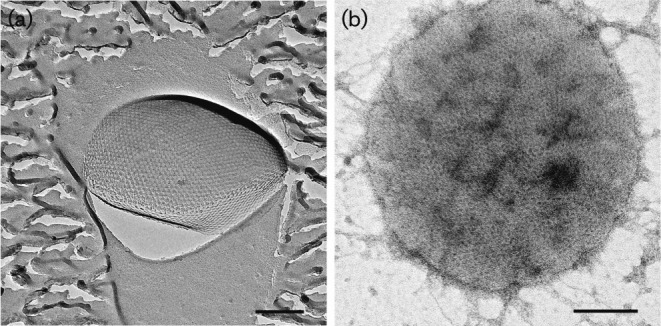
Electron micrographs of a freeze-etching replica (a) and a negatively stained purified S-layer sheet (b) of a cell of strain EN76T. Bars, 200 nm.
Fig. 4.
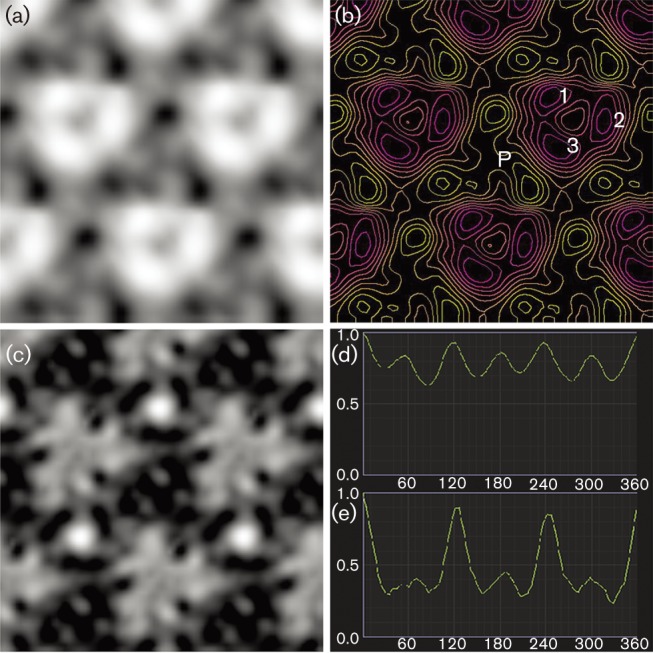
Determination of S-layer symmetry of EN76T. (a) Correlation averaging of the freeze-etched S-layer from Fig. 3(a), showing the protein subunits (white areas) and pores (grey and black areas). (b) Relief reconstruction of the averaged image from (a). The crystal unit cell probably consists of a trimer of protein-trimers, of which one is indicated (1–3). Elevated areas are labelled violet and red and depths are labelled yellow, revealing a triangular cavity or pore (P). (c) Correlation averaging of the negatively stained S-layer from Fig. 3(b). Similar to (a), the proteins are represented by white and light-grey areas and uranyl acetate-filled cavities by dark-grey and black areas. (d) and (e) show the determination of S-layer symmetry of the unit cells in (a) and (c), respectively. The images were tilted by increments of 5° and the correlation with the original, untilted image (set as 1) is plotted against the tilting angle.
As described previously (Sinninghe Damsté et al., 2012), the intact polar lipids of cells of strain EN76T consist of GDGTs bound to the polar head groups monohexose, dihexose, trihexose, phosphohexose or hexose-phosphohexose. Crenarchaeol and its regioisomer, both GDGTs with one cyclohexane and four cyclopentane rings (Sinninghe Damsté et al., 2002), were the major core lipids (80 %; Sinninghe Damsté et al., 2012). So far, crenarchaeol has been found exclusively in members of the Thaumarchaeota.
Phylogenetic analyses
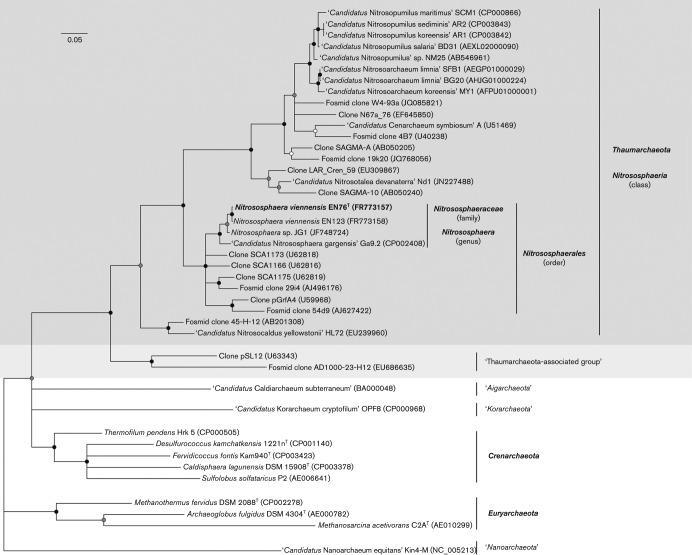
Initial phylogenetic analyses of 16S RNA gene sequences from environmental studies placed the ‘mesophilic archaea’ (i.e. Thaumarchaeota), closely related to strain EN76T, as a deep-branching group of the Crenarchaeota (DeLong, 1992; Fuhrman et al., 1992). However, later analyses based on full rRNA gene sequences, concatenated ribosomal protein sequences and full-genome sequence comparisons provided strong evidence that these mesophilic and aerobic AOA represent a distinct phylum, the Thaumarchaeota (Brochier-Armanet et al., 2008; Spang et al., 2010). Members of this phylum encode a specific set of information-processing genes that is distinct from those in the phyla Crenarchaeota and Euryarchaeota, as well as in the proposed phylum ‘Korarchaeota’ (Brochier-Armanet et al., 2008; Spang et al., 2010) and in other currently proposed phyla (Brochier-Armanet et al., 2011; Rinke et al., 2013; Spang et al., 2013). Strain EN76T is affiliated with group I.1b of the Thaumarchaeota (also known as the ‘soil group’) based on 16S rRNA gene phylogeny (Fig. 5), also showing a consistent phylogenetic clustering based on concatenated AmoAB protein sequences (Tourna et al., 2011). ‘Candidatus Nitrososphaera gargensis’ Ga9.2 shares 97 % 16S rRNA gene sequence identity with strain EN76T (Tourna et al., 2011). ‘Candidatus Nitrosopumilus maritimus’ SCM1 is currently the only described pure culture of the second major group within group I.1a of the Thaumarchaeota (Könneke et al., 2005; Walker et al., 2010), and shares 85 % 16S rRNA gene sequence identity with strain EN76T. However, the name ‘Nitrosopumilus maritimus’ has not been validly published and does not have standing in nomenclature. Based on 16S rRNA gene sequence identity, Thermofilum pendens Hrk 5 (81 % 16S rRNA gene sequence identity) and Methanothermus fervidus DSM 2088T (79 %) represent the closest related cultivated strains of species with validly published names from the phyla Crenarchaeota and Euryarchaeota, respectively (Fig. 5). Strain EN76T has a DNA base composition of 52.7 mol% G+C (Tourna et al., 2011), which is similar to that of ‘Candidatus Nitrososphaera gargensis’ Ga9.2 (48.4 mol%) and higher than that of group I.1a strains such as ‘Candidatus Nitrosopumilus maritimus’ SCM1 (34.2 mol%), ‘Candidatus Nitrosoarchaeum koreensis’ MY1 (32.7 mol%) and ‘Candidatus Nitrosoarchaeum limnia’ SFB1 (32.4 mol%) (Blainey et al., 2011; Kim et al., 2011; Spang et al., 2012; Walker et al., 2010). In conclusion, groups I.1a and I.1b differ greatly in their G+C content and form two highly supported distinct phylogenetic lineages based on both 16S rRNA and amoA gene sequences (Fig. 5).
Fig. 5.
Maximum-likelihood 16S rRNA gene phylogeny of the Thaumarchaeota and representative strains of the Crenarchaeota, Euryarchaeota and other proposed archaeal phyla. The tree depicts Nitrososphaera viennensis EN76T (bold), the marine pure culture ‘Candidatus Nitrosopumilus maritimus’ SCM1, organisms from laboratory or natural enrichment cultures (labelled Candidatus) and a selection of environmental sequences representing major uncultured lineages. Proposed phyla and uncharacterized archaeal lineages are placed in quotes. Phylogeny reconstruction was based on 1202-bp 16S rRNA gene fragments and calculated with RaxML VI-HPC using the GTR+I+G model. Bootstrap support values (1000 replicates) are indicated by circles: filled, ≥90 %; shaded, ≥80 % but <90 %; open, ≥70 % but <80 %. Some branching points are not well supported in the displayed tree, such as the lineages of ‘Candidatus Caldiarchaeum’ and ‘Candidatus Korarchaeum’. The former was affiliated rather with Thaumarchaeota in more comprehensive phylogenetic calculations (see e.g. Eme et al., 2013).
Description of Nitrososphaera gen. nov.
Nitrososphaera (Ni.tro.so.sphae′ra. N.L. adj. nitrosus full of natron; here intended to mean nitrous; L. fem. n. sphaera a ball, sphere; N.L. fem. n. Nitrososphaera the sphere producing nitrite).
Mesophilic to moderately thermophilic, acidophilic to neutrophilic, aerobic, autotrophic or mixotrophic, ammonia-oxidizing organisms. Cells are irregular coccoid. The major lipid is crenarchaeol and its regioisomer. The type species is Nitrososphaera viennensis.
Description of Nitrososphaera viennensis sp. nov.
Nitrososphaera viennensis (vi.en.nen′sis. N.L. fem. adj. viennensis from Vienna, where the type strain was isolated and characterized).
Irregular cocci with a diameter of 0.78±0.13 µm. Occur as single cells and as clusters of several cells. Cells exhibit archaella (12.0±1.8 nm) and archaeal pili (6.4±1.3 nm) as cell appendages, and clearly discernible areas of high and low electron density and tubule-like structures in the cytoplasm. Cells possess an S-layer with p3-symmetry. Grows at pH 6–8.5, with an optimum at pH 7.5. The temperature optimum is 42 °C; grows at 28–47 °C. Energy is produced by oxidizing ammonia to nitrite with oxygen as electron acceptor. Optimal NH4Cl concentration for growth is 2.6 mM, but concentrations up to 15 mM are tolerated. Nitrite concentrations up to 10 mM are tolerated. Urea can be used as substrate. N2O is formed as a side product during ammonia oxidation. Mixotrophic growth is observed with bicarbonate and small carboxylic acids, i.e. pyruvate, α-ketoglutarate, oxaloacetate or glyoxylate, as carbon sources. The following substrates have a negative effect or inhibit growth under the conditions tested: ribose, glycerol, peptone, yeast extract, l-alanine, d-alanine, l-glutamine, l-aspartic acid, an amino acid mixture, Casamino acids, methylamine, trimethylamine, ethanolamine, methanolamine, nitrate, pyrimidine and purine.
The type strain, EN76T ( = DSM 26422T = JMC 19564T), was isolated from a garden soil in Vienna, Austria. The DNA base composition of the type strain is 52.7 mol% G+C.
Description of Nitrososphaeraceae fam. nov.
Nitrososphaeraceae (Ni.tro.so.sphae.ra′ce.ae. N.L. fem. n. Nitrososphaera type genus of the family; L. suff. -aceae ending to denote a family; N.L. fem. pl. n. Nitrososphaeraceae the family of the genus Nitrososphaera).
The description is the same as for the genus Nitrososphaera. The type genus is Nitrososphaera.
Description of Nitrososphaerales ord. nov.
Nitrososphaerales (Ni.tro.so.sphae.ra′les. N.L. fem. n. Nitrososphaera type genus of the order; N.L. suff. -ales ending to denote an order, N.L. fem. pl. n. Nitrososphaerales the order of the genus Nitrososphaera).
The name Nitrososphaerales refers to the former group I.1b (or ‘soil group’) within the phylum Thaumarchaeota. Cultivated organisms of this order have an irregular coccoid cell shape and occur predominantly in terrestrial ecosystems. By contrast, cells of all known organisms affiliated with group I.1a (order ‘Nitrosopumilales’) are rod-shaped. The order Nitrososphaerales comprises a highly supported distinct phylogenetic group based on 16S rRNA gene phylogeny (Fig. 5). The 16S rRNA genes of all members share ≥90 % sequence identity, as do all members of other phylogenetically well-defined groups, e.g. the tentative orders ‘Nitrosopumilales’ (represented by ‘Candidatus Nitrosopumilus maritimus’ SCM1; Könneke et al., 2005), ‘Nitrosotaleales’ (represented by ‘Candidatus Nitrosotalea devanaterra’ Nd1; Lehtovirta-Morley et al., 2011) and ‘Nitrosocaldales’ (represented by ‘Candidatus Nitrosocaldus yellowstonii’ HL72; de la Torre et al., 2008). The names of these orders are currently not validly published, given the lack of representative organisms in pure culture, or depositions in culture collections. The type genus is Nitrososphaera.
Description of Nitrososphaeria classis nov.
Nitrososphaeria (Ni.tro.so.sphae′ri.a. N.L. fem. n. Nitrososphaera the type genus of the type order of the class; N.L. suff. -ia ending to denote a class, N.L. neut. pl. n. Nitrososphaeria the class of the order Nitrososphaerales).
Cultivated strains within this class possess genes of both FtsZ- and Cdv-based cell division systems and have a topoisomerase IB. Similar to euryarchaeal strains, but in contrast to crenarchaeal strains, they have DNA polymerases B and D, eukaryote-like histones (H3/H4) and only one copy of the proliferating cell nuclear antigen and lack genes for RNA polymerase G (Brochier-Armanet et al., 2011; Spang et al., 2010). Crenarchaeol is the major core lipid and is not known to occur in any other bacterial or archaeal phylum (Pitcher et al., 2010; Schouten et al., 2008; Sinninghe Damsté et al., 2002, 2012). Additionally, genes encoding an ammonia monooxygenase have been found exclusively in all lineages within the class, among all archaeal taxa described, and might therefore be considered a distinctive feature. So far, all investigated genomes of members of this class contain genes encoding key enzymes of the 3-hydroxypropionate/4-hydroxybutyrate pathway, including acetyl-CoA carboxylase, 4-hydroxybutyryl-CoA dehydratase and methylmalonyl-CoA mutase, suggesting that members of the phylum Thaumarchaeota might assimilate their cellular carbon via a modified version of this pathway (Berg et al., 2007; Blainey et al., 2011; Kim et al., 2011; Mosier et al., 2012a, b; Park et al., 2012; Spang et al., 2012; Walker et al., 2010). The class comprises a highly supported monophyletic lineage in the 16S rRNA gene phylogeny of the Archaea (Fig. 5). The type order is Nitrososphaerales.
Acknowledgements
We thank Christine Moissl-Eichinger for technical assistance with electron microscopy at the University of Regensburg, Romana Bittner and Daniela Trojan for technical assistance with cultivation and Melina Kerou, Pierre Offre and Anja Spang for genome annotation. We are grateful to the Core Facility Cell Imaging and Ultrastructure Research of the University of Vienna for technical support and we thank Uwe-G. Maier for allocating the electron microscope facilities in Marburg. We thank the FWF (Austrian Science Fund) (grants P23000 and P25369) and Krajete GmbH for financial support.
Abbreviations:
- AOA
ammonia-oxidizing archaea
- AOB
ammonia-oxidizing bacteria
- DoE
design of experiment
- GDGT
glycerol dibiphytanyl glycerol tetraether
- RSM
response surface model
Footnotes
Two supplementary tables and two supplementary figures are available with the online version of this paper.
References
- Adair K. L., Schwartz E. (2008). Evidence that ammonia-oxidizing archaea are more abundant than ammonia-oxidizing bacteria in semiarid soils of northern Arizona, USA. Microb Ecol 56, 420–426 10.1007/s00248-007-9360-9 [DOI] [PubMed] [Google Scholar]
- Alonso-Sáez L., Waller A. S., Mende D. R., Bakker K., Farnelid H., Yager P. L., Lovejoy C., Tremblay J. E., Potvin M. & other authors (2012). Role for urea in nitrification by polar marine archaea. Proc Natl Acad Sci U S A 109, 17989–17994 10.1073/pnas.1201914109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves R. J. E., Wanek W., Zappe A., Richter A., Svenning M. M., Schleper C., Urich T. (2013). Nitrification rates in Arctic soils are associated with functionally distinct populations of ammonia-oxidizing archaea. ISME J 7, 1620–1631 10.1038/ismej.2013.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano F., Noda T. (1995). Improved detection of nitric oxide radical (NO·) production in an activated macrophage culture with a radical scavenger, carboxy PTIO and Griess reagent. FEBS Lett 368, 425–428 10.1016/0014-5793(95)00700-J [DOI] [PubMed] [Google Scholar]
- Andrews S. C., Robinson A. K., Rodríguez-Quiñones F. (2003). Bacterial iron homeostasis. FEMS Microbiol Rev 27, 215–237 10.1016/S0168-6445(03)00055-X [DOI] [PubMed] [Google Scholar]
- Arp D. J., Stein L. Y. (2003). Metabolism of inorganic N compounds by ammonia-oxidizing bacteria. Crit Rev Biochem Mol Biol 38, 471–495 10.1080/10409230390267446 [DOI] [PubMed] [Google Scholar]
- Bates S. T., Berg-Lyons D., Caporaso J. G., Walters W. A., Knight R., Fierer N. (2011). Examining the global distribution of dominant archaeal populations in soil. ISME J 5, 908–917 10.1038/ismej.2010.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W., Lembcke G. (1992). Structural features of archaebacterial cell envelopes. J Bioenerg Biomembr 24, 567–575 10.1007/BF00762349 [DOI] [PubMed] [Google Scholar]
- Berg I. A., Kockelkorn D., Buckel W., Fuchs G. (2007). A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in archaea. Science 318, 1782–1786 10.1126/science.1149976 [DOI] [PubMed] [Google Scholar]
- Blainey P. C., Mosier A. C., Potanina A., Francis C. A., Quake S. R. (2011). Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS ONE 6, e16626 10.1371/journal.pone.0016626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box G. E. P., Draper N. R. (1987). Empirical Model-Building and Response Surfaces. Oxford: Wiley [Google Scholar]
- Box G. E. P., Lucas H. L. (1959). Design of experiments in non-linear situations. Biometrika 46, 77–90 10.1093/biomet/46.1-2.77 [DOI] [Google Scholar]
- Brochier-Armanet C., Boussau B., Gribaldo S., Forterre P. (2008). Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol 6, 245–252 10.1038/nrmicro1852 [DOI] [PubMed] [Google Scholar]
- Brochier-Armanet C., Forterre P., Gribaldo S. (2011). Phylogeny and evolution of the Archaea: one hundred genomes later. Curr Opin Microbiol 14, 274–281 10.1016/j.mib.2011.04.015 [DOI] [PubMed] [Google Scholar]
- Collingridge P. W., Kelly S. (2012). MergeAlign: improving multiple sequence alignment performance by dynamic reconstruction of consensus multiple sequence alignments. BMC Bioinformatics 13, 117 10.1186/1471-2105-13-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre J. R., Walker C. B., Ingalls A. E., Könneke M., Stahl D. A. (2008). Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol 10, 810–818 10.1111/j.1462-2920.2007.01506.x [DOI] [PubMed] [Google Scholar]
- Deatherage J. F., Taylor K. A., Amos L. A. (1983). Three-dimensional arrangement of the cell wall protein of Sulfolobus acidocaldarius. J Mol Biol 167, 823–848 10.1016/S0022-2836(83)80113-2 [DOI] [PubMed] [Google Scholar]
- DeLong E. F. (1992). Archaea in coastal marine environments. Proc Natl Acad Sci U S A 89, 5685–5689 10.1073/pnas.89.12.5685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong E. F., Wu K. Y., Prézelin B. B., Jovine R. V. M. (1994). High abundance of archaea in Antarctic marine picoplankton. Nature 371, 695–697 10.1038/371695a0 [DOI] [PubMed] [Google Scholar]
- Derringer G., Suich R. (1980). Simultaneous optimization of several response variables. J Qual Technol 12, 214–219 [Google Scholar]
- Di H. J., Cameron K. C., Shen J. P., Winefield C. S., O’Callaghan M., Bowatte S., He J. Z. (2009). Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci 2, 621–624 10.1038/ngeo613 [DOI] [Google Scholar]
- Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J. M., Taly J. F., Notredame C. (2011). T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res 39 (Web Server issue), W13–W17 10.1093/nar/gkr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi B., Fardeau M.-L., Ollivier B., Raoult D., Drancourt M. (2012). Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol 62, 1902–1907 10.1099/ijs.0.033712-0 [DOI] [PubMed] [Google Scholar]
- Durbin A. M., Teske A. (2010). Sediment-associated microdiversity within the marine group I Crenarchaeota. Environ Microbiol Rep 2, 693–703 10.1111/j.1758-2229.2010.00163.x [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler J. (2003). Facing extremes: archaeal surface-layer (glyco)proteins. Microbiology 149, 3347–3351 10.1099/mic.0.26591-0 [DOI] [PubMed] [Google Scholar]
- Elkins J. G., Podar M., Graham D. E., Makarova K. S., Wolf Y., Randau L., Hedlund B. P., Brochier-Armanet C., Kunin V. & other authors (2008). A korarchaeal genome reveals insights into the evolution of the Archaea. Proc Natl Acad Sci U S A 105, 8102–8107 10.1073/pnas.0801980105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eme L., Reigstad L. J., Spang A., Lanzén A., Weinmaier T., Rattei T., Schleper C., Brochier-Armanet C. (2013). Metagenomics of Kamchatkan hot spring filaments reveal two new major (hyper)thermophilic lineages related to Thaumarchaeota. Res Microbiol 164, 425–438 10.1016/j.resmic.2013.02.006 [DOI] [PubMed] [Google Scholar]
- Erbilgin O., McDonald K. L., Kerfeld C. A. (2014). Characterization of a planctomycetal organelle: a novel bacterial microcompartment for the aerobic degradation of plant saccharides. Appl Environ Microbiol 80, 2193–2205 10.1128/AEM.03887-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erguder T. H., Boon N., Wittebolle L., Marzorati M., Verstraete W. (2009). Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev 33, 855–869 10.1111/j.1574-6976.2009.00179.x [DOI] [PubMed] [Google Scholar]
- Ettwig K. F., van Alen T., van de Pas-Schoonen K. T., Jetten M. S. M., Strous M. (2009). Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl Environ Microbiol 75, 3656–3662 10.1128/AEM.00067-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Castillo R., Rodriguez-Valera F., Gonzalez-Ramos J., Ruiz-Berraquero F. (1986). Accumulation of poly(β-hydroxybutyrate) by halobacteria. Appl Environ Microbiol 51, 214–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis C. A., Roberts K. J., Beman J. M., Santoro A. E., Oakley B. B. (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102, 14683–14688 10.1073/pnas.0506625102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French E., Kozlowski J. A., Mukherjee M., Bullerjahn G., Bollmann A. (2012). Ecophysiological characterization of ammonia-oxidizing archaea and bacteria from freshwater. Appl Environ Microbiol 78, 5773–5780 10.1128/AEM.00432-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman J. A., McCallum K., Davis A. A. (1992). Novel major archaebacterial group from marine plankton. Nature 356, 148–149 10.1038/356148a0 [DOI] [PubMed] [Google Scholar]
- Gordon D. A., Giovannoni S. J. (1996). Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific oceans. Appl Environ Microbiol 62, 1171–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan D. W. (1996). Organization and interactions of cell envelope proteins of the extreme thermoacidophile Sulfolobus acidocaldarius. Can J Microbiol 42, 1163–1171 10.1139/m96-148 [DOI] [Google Scholar]
- Gubry-Rangin C., Nicol G. W., Prosser J. I. (2010). Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol 74, 566–574 10.1111/j.1574-6941.2010.00971.x [DOI] [PubMed] [Google Scholar]
- Haikarainen T., Papageorgiou A. C. (2010). Dps-like proteins: structural and functional insights into a versatile protein family. Cell Mol Life Sci 67, 341–351 10.1007/s00018-009-0168-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam S. J., Konstantinidis K. T., Putnam N., Schleper C., Watanabe Y., Sugahara J., Preston C., de la Torre J., Richardson P. M., DeLong E. F. (2006a). Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci U S A 103, 18296–18301 10.1073/pnas.0608549103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam S. J., Mincer T. J., Schleper C., Preston C. M., Roberts K., Richardson P. M., DeLong E. F. (2006b). Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol 4, e95 10.1371/journal.pbio.0040095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzenpichler R. (2012). Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea. Appl Environ Microbiol 78, 7501–7510 10.1128/AEM.01960-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzenpichler R., Lebedeva E. V., Spieck E., Stoecker K., Richter A., Daims H., Wagner M. (2008). A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci U S A 105, 2134–2139 10.1073/pnas.0708857105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J. Z., Shen J. P., Zhang L. M., Zhu Y. G., Zheng Y. M., Xu M. G., Di H. (2007). Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol 9, 2364–2374 10.1111/j.1462-2920.2007.01358.x [DOI] [PubMed] [Google Scholar]
- Hershberger K. L., Barns S. M., Reysenbach A. L., Dawson S. C., Pace N. R. (1996). Wide diversity of Crenarchaeota. Nature 384, 420 10.1038/384420a0 [DOI] [PubMed] [Google Scholar]
- Hooper A. B., Terry K. R. (1979). Hydroxylamine oxidoreductase of Nitrosomonas. Production of nitric oxide from hydroxylamine. Biochim Biophys Acta 571, 12–20 10.1016/0005-2744(79)90220-1 [DOI] [PubMed] [Google Scholar]
- Horak R. E. A., Qin W., Schauer A. J., Armbrust E. V., Ingalls A. E., Moffett J. W., Stahl D. A., Devol A. H. (2013). Ammonia oxidation kinetics and temperature sensitivity of a natural marine community dominated by Archaea. ISME J 7, 2023–2033 10.1038/ismej.2013.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H., Burggraf S., Mayer T., Wyschkony I., Rachel R., Stetter K. O. (2000). Ignicoccus gen. nov., a novel genus of hyperthermophilic, chemolithoautotrophic Archaea, represented by two new species, Ignicoccus islandicus sp. nov. and Ignicoccus pacificus sp. nov. Int J Syst Evol Microbiol 50, 2093–2100 10.1099/00207713-50-6-2093 [DOI] [PubMed] [Google Scholar]
- Huber H., Hohn M. J., Rachel R., Fuchs T., Wimmer V. C., Stetter K. O. (2002). A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417, 63–67 10.1038/417063a [DOI] [PubMed] [Google Scholar]
- Hugenholtz P., Pitulle C., Hershberger K. L., Pace N. R. (1998). Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol 180, 366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell K. F., Albers S.-V. (2012). The archaellum: an old motility structure with a new name. Trends Microbiol 20, 307–312 10.1016/j.tim.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Jia Z., Conrad R. (2009). Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol 11, 1658–1671 10.1111/j.1462-2920.2009.01891.x [DOI] [PubMed] [Google Scholar]
- Jorgensen S. L., Hannisdal B., Lanzén A., Baumberger T., Flesland K., Fonseca R., Ovreås L., Steen I. H., Thorseth I. H. & other authors (2012). Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc Natl Acad Sci U S A 109, E2846–E2855 10.1073/pnas.1207574109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M. Y., Park S. J., Min D., Kim J. S., Rijpstra W. I. C., Sinninghe Damsté J. S., Kim G. J., Madsen E. L., Rhee S. K. (2011). Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic crenarchaeal group I.1a from an agricultural soil. Appl Environ Microbiol 77, 8635–8647 10.1128/AEM.05787-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens G., Lindström K., Saano A. (1997). Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl Environ Microbiol 63, 803–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallmeyer J., Pockalny R., Adhikari R. R., Smith D. C., D’Hondt S. (2012). Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci U S A 109, 16213–16216 10.1073/pnas.1203849109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karner M. B., DeLong E. F., Karl D. M. (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510 10.1038/35054051 [DOI] [PubMed] [Google Scholar]
- Katoh K., Toh H. (2010). Parallelization of the mafft multiple sequence alignment program. Bioinformatics 26, 1899–1900 10.1093/bioinformatics/btq224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T. (2005). mafft version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33, 511–518 10.1093/nar/gki198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. K., Jung M. Y., Yu D. S., Park S. J., Oh T. K., Rhee S. K., Kim J. F. (2011). Genome sequence of an ammonia-oxidizing soil archaeon, “Candidatus Nitrosoarchaeum koreensis” MY1. J Bacteriol 193, 5539–5540 10.1128/JB.05717-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingl A. (2011). Struktur und Funktion von S-Layern acidophiler Bakterien und Archaeen, ihre Rolle bei der Pyrit-Oxidation sowie die Adhäsion an Oberflächen. Dissertation, University of Regensburg, Regensburg, Germany [Google Scholar]
- Klingl A., Moissl-Eichinger C., Wanner G., Zweck J., Huber H., Thomm M., Rachel R. (2011). Analysis of the surface proteins of Acidithiobacillus ferrooxidans strain SP5/1 and the new, pyrite-oxidizing Acidithiobacillus isolate HV2/2, and their possible involvement in pyrite oxidation. Arch Microbiol 193, 867–882 10.1007/s00203-011-0720-y [DOI] [PubMed] [Google Scholar]
- Klingl A., Flechsler J., Heimerl T., Rachel R. (2013). Archaeal cells. In eLS. Chichester: Wiley; 10.1002/9780470015902.a0000383.pub2 [DOI] [Google Scholar]
- König H., Rachel R., Claus H. (2007). Proteinaceous surface layers of archaea: ultrastructure and biochemistry. In Archaea: Molecular and Cellular Biology, pp. 315–340 Edited by Cavicchioli R. Washington, DC: American Society for Microbiology; 10.1128/9781555815516.ch14 [DOI] [Google Scholar]
- Könneke M., Bernhard A. E., de la Torre J. R., Walker C. B., Waterbury J. B., Stahl D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546 10.1038/nature03911 [DOI] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. (1985). Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A 82, 6955–6959 10.1073/pnas.82.20.6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtovirta-Morley L. E., Stoecker K., Vilcinskas A., Prosser J. I., Nicol G. W. (2011). Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci U S A 108, 15892–15897 10.1073/pnas.1107196108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leininger S., Urich T., Schloter M., Schwark L., Qi J., Nicol G. W., Prosser J. I., Schuster S. C., Schleper C. (2006). Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442, 806–809 10.1038/nature04983 [DOI] [PubMed] [Google Scholar]
- Leisch N., Dirks U., Gruber-Vodicka H. R., Schmid M., Sterrer W., Ott J. A. (2011). Microanatomy of the trophosome region of Paracatenula cf. polyhymnia (Catenulida, Platyhelminthes) and its intracellular symbionts. Zoomorphology 130, 261–271 10.1007/s00435-011-0135-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembcke G., Dürr R., Hegerl R., Baumeister W. (1991). Image analysis and processing of an imperfect two-dimensional crystal: the surface layer of the archaebacterium Sulfolobus acidocaldarius re-investigated. J Microsc 161, 263–278 10.1111/j.1365-2818.1991.tb03089.x [DOI] [Google Scholar]
- Lembcke G., Baumeister W., Beckmann E., Zemlin F. (1993). Cryo-electron microscopy of the surface protein of Sulfolobus shibatae. Ultramicroscopy 49, 397–406 10.1016/0304-3991(93)90245-S [DOI] [Google Scholar]
- Lindsay M. R., Webb R. I., Strous M., Jetten M. S., Butler M. K., Forde R. J., Fuerst J. A. (2001). Cell compartmentalisation in planctomycetes: novel types of structural organisation for the bacterial cell. Arch Microbiol 175, 413–429 10.1007/s002030100280 [DOI] [PubMed] [Google Scholar]
- MacGregor B. J., Moser D. P., Alm E. W., Nealson K. H., Stahl D. A. (1997). Crenarchaeota in Lake Michigan sediment. Appl Environ Microbiol 63, 1178–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy Y., Ouverney C., Bik E. M., Lösekann T., Ivanova N., Martin H. G., Szeto E., Platt D., Hugenholtz P. & other authors (2007). Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci U S A 104, 11889–11894 10.1073/pnas.0704662104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens-Habbena W., Berube P. M., Urakawa H., de la Torre J. R., Stahl D. A. (2009). Ammonia oxidation kinetics determine niche separation of nitrifying archaea and bacteria. Nature 461, 976–979 10.1038/nature08465 [DOI] [PubMed] [Google Scholar]
- Merbt S. N., Stahl D. A., Casamayor E. O., Martí E., Nicol G. W., Prosser J. I. (2012). Differential photoinhibition of bacterial and archaeal ammonia oxidation. FEMS Microbiol Lett 327, 41–46 10.1111/j.1574-6968.2011.02457.x [DOI] [PubMed] [Google Scholar]
- Miller M. A., Pfeiffer W., Schwartz T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Gateway Computing Environments Workshop (GCE) 2010, 14 November 2010, pp. 1–8 Chicago: IEEE [Google Scholar]
- Mosier A. C., Allen E. E., Kim M., Ferriera S., Francis C. A. (2012a). Genome sequence of “Candidatus Nitrosoarchaeum limnia” BG20, a low-salinity ammonia-oxidizing archaeon from the San Francisco Bay estuary. J Bacteriol 194, 2119–2120 10.1128/JB.00007-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosier A. C., Allen E. E., Kim M., Ferriera S., Francis C. A. (2012b). Genome sequence of “Candidatus Nitrosopumilus salaria” BD31, an ammonia-oxidizing archaeon from the San Francisco Bay estuary. J Bacteriol 194, 2121–2122 10.1128/JB.00013-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F., Brissac T., Le Bris N., Felbeck H., Gros O. (2010). First description of giant archaea (Thaumarchaeota) associated with putative bacterial ectosymbionts in a sulfidic marine habitat. Environ Microbiol 12, 2371–2383 10.1111/j.1462-2920.2010.02309.x [DOI] [PubMed] [Google Scholar]
- Mussmann M., Brito I., Pitcher A., Sinninghe Damsté J. S., Hatzenpichler R., Richter A., Nielsen J. L., Nielsen P. H., Müller A. & other authors (2011). Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc Natl Acad Sci U S A 108, 16771–16776 10.1073/pnas.1106427108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol G. W., Schleper C. (2006). Ammonia-oxidising Crenarchaeota: important players in the nitrogen cycle? Trends Microbiol 14, 207–212 10.1016/j.tim.2006.03.004 [DOI] [PubMed] [Google Scholar]
- Nunoura T., Takaki Y., Kakuta J., Nishi S., Sugahara J., Kazama H., Chee G. J., Hattori M., Kanai A. & other authors (2011). Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res 39, 3204–3223 10.1093/nar/gkq1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenreiter T., Selezi D., Quaiser A., Bonch-Osmolovskaya L., Schleper C. (2003). Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol 5, 787–797 10.1046/j.1462-2920.2003.00476.x [DOI] [PubMed] [Google Scholar]
- Offre P., Prosser J. I., Nicol G. W. (2009). Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol Ecol 70, 99–108 10.1111/j.1574-6941.2009.00725.x [DOI] [PubMed] [Google Scholar]
- Park S. J., Kim J. G., Jung M. Y., Kim S. J., Cha I. T., Ghai R., Martín-Cuadrado A. B., Rodríguez-Valera F., Rhee S. K. (2012). Draft genome sequence of an ammonia-oxidizing archaeon, “Candidatus Nitrosopumilus sediminis” AR2, from Svalbard in the Arctic Circle. J Bacteriol 194, 6948–6949 10.1128/JB.01869-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelve E. A., Lindås A. C., Martens-Habbena W., de la Torre J. R., Stahl D. A., Bernander R. (2011). Cdv-based cell division and cell cycle organization in the thaumarchaeon Nitrosopumilus maritimus. Mol Microbiol 82, 555–566 10.1111/j.1365-2958.2011.07834.x [DOI] [PubMed] [Google Scholar]
- Pester M., Rattei T., Flechl S., Gröngröft A., Richter A., Overmann J., Reinhold-Hurek B., Loy A., Wagner M. (2012). amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol 14, 525–539 10.1111/j.1462-2920.2011.02666.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher A., Rychlik N., Hopmans E. C., Spieck E., Rijpstra W. I. C., Ossebaar J., Schouten S., Wagner M., Sinninghe Damsté J. S. (2010). Crenarchaeol dominates the membrane lipids of Candidatus Nitrososphaera gargensis, a thermophilic group I.1b archaeon. ISME J 4, 542–552 10.1038/ismej.2009.138 [DOI] [PubMed] [Google Scholar]
- Preston C. M., Wu K. Y., Molinski T. F., DeLong E. F. (1996). A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc Natl Acad Sci U S A 93, 6241–6246 10.1073/pnas.93.13.6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser J. I., Nicol G. W. (2012). Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol 20, 523–531 10.1016/j.tim.2012.08.001 [DOI] [PubMed] [Google Scholar]
- Prüschenk R., Baumeister W. (1987). Three-dimensional structure of the surface protein of Sulfolobus solfataricus. Eur J Cell Biol 45, 185–191 [Google Scholar]
- Prüschenk R., Baumeister W., Zillig W. (1987). Surface structure variants in different species of Sulfolobus. FEMS Microbiol Lett 43, 327–330 10.1016/0378-1097(87)90421-6 [DOI] [Google Scholar]
- Rachel R., Pum D., Šmarda J., Šmajs D., Komrska J., Krzyzánek V., Rieger G., Stetter K. O. (1997). II. Fine structure of S-layers 1. FEMS Microbiol Rev 20, 13–23 10.1016/S0168-6445(97)00040-5 [DOI] [Google Scholar]
- Rachel R., Meyer C., Klingl A., Gürster S., Heimerl T., Wasserburger N., Burghardt T., Küper U., Bellack A. & other authors (2010). Analysis of the ultrastructure of archaea by electron microscopy. Methods Cell Biol 96, 47–69 10.1016/S0091-679X(10)96003-2 [DOI] [PubMed] [Google Scholar]
- Rappé M. S., Giovannoni S. J. (2003). The uncultured microbial majority. Annu Rev Microbiol 57, 369–394 10.1146/annurev.micro.57.030502.090759 [DOI] [PubMed] [Google Scholar]
- Reigstad L. J., Richter A., Daims H., Urich T., Schwark L., Schleper C. (2008). Nitrification in terrestrial hot springs of Iceland and Kamchatka. FEMS Microbiol Ecol 64, 167–174 10.1111/j.1574-6941.2008.00466.x [DOI] [PubMed] [Google Scholar]
- Rinke C., Schwientek P., Sczyrba A., Ivanova N. N., Anderson I. J., Cheng J. F., Darling A., Malfatti S., Swan B. K. & other authors (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 10.1038/nature12352 [DOI] [PubMed] [Google Scholar]
- Rittmann S., Herwig C. (2012). A comprehensive and quantitative review of dark fermentative biohydrogen production. Microb Cell Fact 11, 115 10.1186/1475-2859-11-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro A. E., Casciotti K. L. (2011). Enrichment and characterization of ammonia-oxidizing archaea from the open ocean: phylogeny, physiology and stable isotope fractionation. ISME J 5, 1796–1808 10.1038/ismej.2011.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauss K., Focks A., Leininger S., Kotzerke A., Heuer H., Thiele-Bruhn S., Sharma S., Wilke B. M., Matthies M. & other authors (2009). Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ Microbiol 11, 446–456 10.1111/j.1462-2920.2008.01783.x [DOI] [PubMed] [Google Scholar]
- Schleper C., Nicol G. W. (2010). Ammonia-oxidising archaea – physiology, ecology and evolution. Adv Microb Physiol 57, 1–41 10.1016/B978-0-12-381045-8.00001-1 [DOI] [PubMed] [Google Scholar]
- Schleper C., Holben W., Klenk H. P. (1997). Recovery of crenarchaeotal ribosomal DNA sequences from freshwater-lake sediments. Appl Environ Microbiol 63, 321–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T. M., DeLong E. F., Pace N. R. (1991). Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol 173, 4371–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten S., Hopmans E. C., Baas M., Boumann H., Standfest S., Könneke M., Stahl D. A., Sinninghe Damsté J. S. (2008). Intact membrane lipids of “Candidatus Nitrosopumilus maritimus,” a cultivated representative of the cosmopolitan mesophilic group I Crenarchaeota. Appl Environ Microbiol 74, 2433–2440 10.1128/AEM.01709-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J. P., Zhang L. M., Zhu Y. G., Zhang J. B., He J. Z. (2008). Abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea communities of an alkaline sandy loam. Environ Microbiol 10, 1601–1611 10.1111/j.1462-2920.2008.01578.x [DOI] [PubMed] [Google Scholar]
- Shen T. L., Stieglmeier M., Dai J. L., Urich T., Schleper C. (2013). Responses of the terrestrial ammonia-oxidizing archaeon Ca. Nitrososphaera viennensis and the ammonia-oxidizing bacterium Nitrosospira multiformis to nitrification inhibitors. FEMS Microbiol Lett 344, 121–129 10.1111/1574-6968.12164 [DOI] [PubMed] [Google Scholar]
- Shively J. M., Ball F., Brown D. H., Saunders R. E. (1973). Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182, 584–586 10.1126/science.182.4112.584 [DOI] [PubMed] [Google Scholar]
- Sinninghe Damsté J. S., Schouten S., Hopmans E. C., van Duin A. C. T., Geenevasen J. A. J. (2002). Crenarchaeol: the characteristic core glycerol dibiphytanyl glycerol tetraether membrane lipid of cosmopolitan pelagic crenarchaeota. J Lipid Res 43, 1641–1651 10.1194/jlr.M200148-JLR200 [DOI] [PubMed] [Google Scholar]
- Sinninghe Damsté J. S., Rijpstra W. I., Hopmans E. C., Jung M. Y., Kim J. G., Rhee S. K., Stieglmeier M., Schleper C. (2012). Intact polar and core glycerol dibiphytanyl glycerol tetraether lipids of group I.1a and I.1b thaumarchaeota in soil. Appl Environ Microbiol 78, 6866–6874 10.1128/AEM.01681-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleytr U. B., Messner P., Pum D., Sára M. (1993). Crystalline bacterial cell surface layers. Mol Microbiol 10, 911–916 10.1111/j.1365-2958.1993.tb00962.x [DOI] [PubMed] [Google Scholar]
- Sleytr U. B., Sara M., Pum D., Schuster B. (2001). Characterization and use of crystalline bacterial cell surface layers. Prog Surf Sci 68, 231–278 10.1016/S0079-6816(01)00008-9 [DOI] [Google Scholar]
- Sleytr U. B., Huber C., Ilk N., Pum D., Schuster B., Egelseer E. M. (2007). S-layers as a tool kit for nanobiotechnological applications. FEMS Microbiol Lett 267, 131–144 10.1111/j.1574-6968.2006.00573.x [DOI] [PubMed] [Google Scholar]
- Spang A., Hatzenpichler R., Brochier-Armanet C., Rattei T., Tischler P., Spieck E., Streit W., Stahl D. A., Wagner M., Schleper C. (2010). Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol 18, 331–340 10.1016/j.tim.2010.06.003 [DOI] [PubMed] [Google Scholar]
- Spang A., Poehlein A., Offre P., Zumbrägel S., Haider S., Rychlik N., Nowka B., Schmeisser C., Lebedeva E. V. & other authors (2012). The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol 14, 3122–3145 10.1111/j.1462-2920.2012.02893.x [DOI] [PubMed] [Google Scholar]
- Spang A., Martijn J., Saw J. H., Lind A. E., Guy L., Ettema T. J. (2013). Close encounters of the third domain: the emerging genomic view of archaeal diversity and evolution. Archaea 2013, 202358 10.1155/2013/202358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., de la Torre J. R. (2012). Physiology and diversity of ammonia-oxidizing archaea. Annu Rev Microbiol 66, 83–101 10.1146/annurev-micro-092611-150128 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. (2008). A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol 57, 758–771 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Stein J. L., Marsh T. L., Wu K. Y., Shizuya H., DeLong E. F. (1996). Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J Bacteriol 178, 591–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglmeier M., Alves R. J. E., Schleper C. (2014a). Thaumarchaeota. In The Prokaryotes, 4th edn Edited by Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F. Berlin & Heidelberg: Springer; (in press). [Google Scholar]
- Stieglmeier M., Mooshammer M., Kitzler B., Wanek W., Zechmeister-Boltenstern S., Richter A., Schleper C. (2014b). Aerobic nitrous oxide production through N-nitrosating hybrid formation in ammonia-oxidizing archaea. ISME J 8, 1135–1146 10.1038/ismej.2013.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K., Moser D. P., DeFlaun M., Onstott T. C., Fredrickson J. K. (2001). Archaeal diversity in waters from deep South African gold mines. Appl Environ Microbiol 67, 5750–5760 10.1128/AEM.67.21.5750-5760.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K. A., Deatherage J. F., Amos L. A. (1982). Structure of the S-layer of Sulfolobus acidocaldarius. Nature 299, 840–842 10.1038/299840a0 [DOI] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. (1994). clustal w: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22, 4673–4680 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourna M., Stieglmeier M., Spang A., Könneke M., Schintlmeister A., Urich T., Engel M., Schloter M., Wagner M. & other authors (2011). Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A 108, 8420–8425 10.1073/pnas.1013488108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch A. H., Leininger S., Kletzin A., Schuster S. C., Klenk H. P., Schleper C. (2005). Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol 7, 1985–1995 10.1111/j.1462-2920.2005.00906.x [DOI] [PubMed] [Google Scholar]
- Tyson G. W., Banfield J. F. (2005). Cultivating the uncultivated: a community genomics perspective. Trends Microbiol 13, 411–415 10.1016/j.tim.2005.07.003 [DOI] [PubMed] [Google Scholar]
- Urakawa H., Martens-Habbena W., Stahl D. A. (2011). Physiology and genomics of ammonia-oxidizing archaea. In Nitrification, pp. 117–155 Edited by Ward B. B., Arp D. J., Klotz M. G. Washington, DC: American Society for Microbiology [Google Scholar]
- Vajrala N., Martens-Habbena W., Sayavedra-Soto L. A., Schauer A., Bottomley P. J., Stahl D. A., Arp D. J. (2013). Hydroxylamine as an intermediate in ammonia oxidation by globally abundant marine archaea. Proc Natl Acad Sci U S A 110, 1006–1011 10.1073/pnas.1214272110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D., Labarre L., Rouy Z., Barbe V., Bocs S., Cruveiller S., Lajus A., Pascal G., Scarpelli C., Médigue C. (2006). MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res 34, 53–65 10.1093/nar/gkj406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D., Engelen S., Mornico D., Cruveiller S., Fleury L., Lajus A., Rouy Z., Roche D., Salvignol G. & other authors (2009). MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford) 2009, bap021 10.1093/database/bap021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niftrik L., Jetten M. S. M. (2012). Anaerobic ammonium-oxidizing bacteria: unique microorganisms with exceptional properties. Microbiol Mol Biol Rev 76, 585–596 10.1128/MMBR.05025-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niftrik L. A., Fuerst J. A., Sinninghe Damsté J. S., Kuenen J. G., Jetten M. S. M., Strous M. (2004). The anammoxosome: an intracytoplasmic compartment in anammox bacteria. FEMS Microbiol Lett 233, 7–13 10.1016/j.femsle.2004.01.044 [DOI] [PubMed] [Google Scholar]
- Veith A., Klingl A., Zolghadr B., Lauber K., Mentele R., Lottspeich F., Rachel R., Albers S. V., Kletzin A. (2009). Acidianus, Sulfolobus and Metallosphaera surface layers: structure, composition and gene expression. Mol Microbiol 73, 58–72 10.1111/j.1365-2958.2009.06746.x [DOI] [PubMed] [Google Scholar]
- Venter J. C., Remington K., Heidelberg J. F., Halpern A. L., Rusch D., Eisen J. A., Wu D., Paulsen I., Nelson K. E. & other authors (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 10.1126/science.1093857 [DOI] [PubMed] [Google Scholar]
- Walker C. B., de la Torre J. R., Klotz M. G., Urakawa H., Pinel N., Arp D. J., Brochier-Armanet C., Chain P. S. G., Chan P. P. & other authors (2010). Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci U S A 107, 8818–8823 10.1073/pnas.0913533107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman W. B., Coleman D. C., Wiebe W. J. (1998). Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A 95, 6578–6583 10.1073/pnas.95.12.6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchter C., Abbas B., Coolen M. J. L., Herfort L., van Bleijswijk J., Timmers P., Strous M., Teira E., Herndl G. J. & other authors (2006). Archaeal nitrification in the ocean. Proc Natl Acad Sci U S A 103, 12317–12322 10.1073/pnas.0600756103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W. W., Zhang C. X., Zeng X. W., Feng Y. Z., Weng J. H., Lin X. G., Zhu J. G., Xiong Z. Q., Xu J. & other authors (2011). Autotrophic growth of nitrifying community in an agricultural soil. ISME J 5, 1226–1236 10.1038/ismej.2011.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Haaijer S. C., Op den Camp H. J., van Niftrik L., Stahl D. A., Könneke M., Rush D., Sinninghe Damsté J. S., Hu Y. Y., Jetten M. S. (2012). Mimicking the oxygen minimum zones: stimulating interaction of aerobic archaeal and anaerobic bacterial ammonia oxidizers in a laboratory-scale model system. Environ Microbiol 14, 3146–3158 10.1111/j.1462-2920.2012.02894.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhalnina K., de Quadros P. D., Camargo F. A., Triplett E. W. (2012). Drivers of archaeal ammonia-oxidizing communities in soil. Front Microbiol 3, 210 10.3389/fmicb.2012.00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. L., Ye Q., Huang Z. Y., Li W. J., Chen J. Q., Song Z. Q., Zhao W. D., Bagwell C., Inskeep W. P. & other authors (2008). Global occurrence of archaeal amoA genes in terrestrial hot springs. Appl Environ Microbiol 74, 6417–6426 10.1128/AEM.00843-08 [DOI] [PMC free article] [PubMed] [Google Scholar]