Abstract
We describe a framework to model visual semantics of liver lesions in CT images in order to predict the visual semantic terms (VST) reported by radiologists in describing these lesions. Computational models of VST are learned from image data using high–order steerable Riesz wavelets and support vector machines (SVM). The organization of scales and directions that are specific to every VST are modeled as linear combinations of directional Riesz wavelets. The models obtained are steerable, which means that any orientation of the model can be synthesized from linear combinations of the basis filters. The latter property is leveraged to model VST independently from their local orientation. In a first step, these models are used to predict the presence of each semantic term that describes liver lesions. In a second step, the distances between all VST models are calculated to establish a non–hierarchical computationally–derived ontology of VST containing inter–term synonymy and complementarity. A preliminary evaluation of the proposed framework was carried out using 74 liver lesions annotated with a set of 18 VSTs from the RadLex ontology. A leave–one–patient–out cross–validation resulted in an average area under the ROC curve of 0.853 for predicting the presence of each VST when using SVMs in a feature space combining the magnitudes of the steered models with CT intensities. Likelihood maps are created for each VST, which enables high transparency of the information modeled. The computationally–derived ontology obtained from the VST models was found to be consistent with the underlying semantics of the visual terms. It was found to be complementary to the RadLex ontology, and constitutes a potential method to link the image content to visual semantics. The proposed framework is expected to foster human–computer synergies for the interpretation of radiological images while using rotation–covariant computational models of VSTs to (1) quantify their local likelihood and (2) explicitly link them with pixel–based image content in the context of a given imaging domain.
Index Terms: Computer–aided diagnosis, RadLex, visual semantic modeling, liver CT, Riesz wavelets, steerability
I. Introduction
Medical imaging aims to support decision making by providing visual information about the human body. Imaging physics has evolved to assess the visual appearance of almost every organ with both high spatial and temporal resolution and even functional information. The technologies to preprocess, transmit, store and display the images are implemented in all modern hospitals. However, clinicians rely nearly exclusively on their image perception skills for the final diagnosis [1]. The increasing variability of imaging protocols and the enormous amounts of medical image data produced per day in modern hospitals constitute a challenge for image interpretation, even for experienced radiologists [2]. As a result, errors and variations in interpretations are currently representing the weakest aspect of clinical imaging [3].
Successful interpretation of medical images relies on two distinct processes: (1) identifying important visual patterns and (2) establishing potential links among the imaging features, clinical context, and the likely diagnoses [4]. Whereas the latter requires a deep understanding and comprehensive knowledge of the radiological manifestations and clinical aspects of diseases, the former is closely related to visual perception [5]. A large–scale study on malpractice in radiology showed that the majority of errors in medical image interpretation are caused by perceptual misinterpretation [6]. Strategies for reducing perceptual errors includes the normalization of viewing conditions, sufficient training of the observers, availability of similar images and clinical data, multiple reporting, and image quantification [3]. The use of structured visual terminologies based on radiology semantics is also a promising approach to enable unequivocal definition of imaging signs [7], but is yet little used in routine practice.
Computerized assistance for image analysis and management is expected to provide solutions for the aforementioned strategies by yielding exhaustive, comprehensive and reproducible data interpretation [8]. The skills of computers and radiologists are found to be very complementary, where high–level image interpretation based on computer–generated image quantification and its clinical context remains in the hands of the human observer [9]. However, several challenges remain unsolved and require further research for a successful integration of computer–aided diagnosis (CAD) into the radiology routine [10]. A fundamental question for a seamless workflow integration of CAD is to maximize interactions between CAD outputs and human conceptual processing [9]. The latter relies both on the trust and intuition of the user of the CAD system. Trust can only be achieved when a critical performance level is achieved by the system [10]. Intuition still requires extensive research efforts to design computer–generated outputs that match human semantics in radiology [7]. The transparency of the computer algorithms should be maximized so that the users can identify errors.
A. Semantic information in radiology images
Radiologists rely on many visual terms to describe imaging signs relating to anatomy, visual features of abnormality and diagnostic interpretation [11]. The vocabulary used to communicate these visual features can vary greatly [12], which limits both clear communication between experts and the formalization of the diagnostic thought process [13]. The use of standardized terminologies of imaging signs including their relations (i.e., ontologies) has been recently recommended to unambiguously describe the content of radiology images [3]. The use of biomedical ontologies in radiology opens avenues for enabling clinicians to access, query and analyze large amounts of image data using an explicit information model of image content [14].
There are several domain–specific standardized terminologies being developed, including the breast imaging reporting and data system (BI–RADS) [15], the Fleishner society glossary of terms for thoracic imaging [16], the nomenclature of lumbar disc pathology [17], the reporting terminology for brain arteriovenous malformations [18], the computed tomography (CT) colonography reporting and data system [19], the visually accessible Rembrandt images (VASARI) for describing the magnetic resonance (MR) features of human gliomas as well as others [20]. Most of the standardized terminologies do not include relations among their terms (e.g., synonyms or hierarchical taxonomic structures), and as such, they thus do not constitute true ontologies. As a result, these terminologies cannot be used for semantic information processing leveraging inter–term similarities. Recent efforts from the Radiological Society of North America (RSNA) were undertaken to create RadLex1, an ontology unifying and regrouping several of the standardized terminologies mentioned above [21], in addition to providing terms unique to radiology that are missing from other exiting terminologies. RadLex contains more than 30,000 terms and their relations, which constitutes a very rich basis for reasoning about image features and their implications in various diseases.
B. Linking image contents to visual semantics
The importance of ontologies in radiology for knowledge representation is well established, but the importance of explicitly linking semantic terms with pixel–based image content has only recently been emphasized2 [7]. Establishing such a link constitutes a next step to computational access to exploding amounts of medical image data [22]. This also provides the opportunity to assist radiologists in the identification and localization of diagnostically meaningful visual features in images. The creation of computational models of semantic terms may also allow the establishment of distances between the terms that can be learned from data, which can add knowledge about the meaning of semantic terms within existing ontologies.
C. Related work
The bag–of–visual–words (BOVW) approach [23] aims at discovering visual terms in an unsupervised fashion to minimize the semantic gap between low–level image features alone and higher–level image understanding. The visual words (VW) are defined as the cluster centroids obtained from clustering the image instances expressed in a given low–level feature space. Its ability to enhance medical image classification and retrieval when compared to using the low–level features was demonstrated by several studies [24–28]. Attempts were made for visualizing the VWs, aiming at interpreting the visual semantics being modeled. In [24, 26], color image overlays are used to mark the local presence of VWs in image examples. In [25, 27–29], prototype image patches (those closest to the respective VWs) are displayed to visualize the information modeled. Unfortunately, VWs often do not correspond to the actual semantics in medical images, and they are therefore very difficult to interpret for radiologists.
The link between VWs and medical VSTs is studied in [30, 31]. Liu et al. used supervised sparse auto–encoders to automatically derive several patterns (i.e., VWs) per disease. However, albeit the learned patterns were derived from the disease classes, they did not correspond to visual semantics belonging to a controlled vocabulary and therefore did not have a clear semantic interpretation [31]. In the context of endomicroscopic video retrieval, André et al. use a Fisher–based approach to learn the links between VWs learned from dense–scale–invariant feature transform (SIFT) and 8 visual semantic terms (VST) [30]. Entire videos can be summarized by star plots reflecting the presence of VSTs, but the transparency of the algorithms remains limited as the occurrence of the VSTs are not localized in the images.
Other studies have focused on the direct modeling of VSTs from application–specific semantic vocabularies [32–37]. In the context of histological image retrieval, Tang et al. built semantic label maps that localize the occurrence of VSTs from Gabor and color histogram features [33]. The authors further refined their VST maps using spatial–hidden Markov models in [38]. In [34], the link between ad–hoc low–level image features based on gray–level intensity and VSTs can be tailored for each specific user for describing high–resolution CT (HRCT) images of the lungs. Shyu et al. evaluated the discriminatory power of low–level computational features (i.e., gray–level intensity) for predicting human perceptual categories in HRCT in [39]. In [40], a comprehensive set of low–level image features (i.e., shape, size, gray–level intensity, and texture) was used to probabilistically model lung nodule image semantics. Kwitt et al. [36] defined semantic spaces by assuming that VSTs in endoscopic images are living on Riemannian manifolds in a space spanned by SIFT features. They could derive a positive–definite semantic kernel that can be used with support vector machine (SVM) classifiers. Gimenez et al. [37] used a comprehensive feature set including state–of–the–art contrast, texture, edge and shape features together with LASSO (Least Absolute Shrinkage and Selection Operator [41]) regression models to predict the presence of VST from entire ROIs.
The above–mentioned studies demonstrated the feasibility of predicting VSTs from lower–level image features. However, both transparency and performance of most systems suffer from two limitations. First, the automatic annotation of global regions of interest (ROI) can be ambiguous [32]. Local quantifications of the VSTs can increase the transparency of the system by highlighting visual features that are recognized as positive inside the ROI. Second, most of the studies do not allow for rotation–invariant detection of the VSTs by relying on low–level computational features that are analyzing images along arbitrary directions (e.g., SIFT, oriented filterbanks, gray–level co–occurrence matrices). VSTs are typically characterized by directional information (e.g., lesion boundary, nodule, vascular structure), but their local orientation may vary greatly over the ROI. Optimal modeling of VSTs requires image operators that are rotation–covariant, enabling the modeling of the local relative organization of the directions independently from the orientation of the VST [42]. The importance of the local relative orientation of directions for classifying normal liver tissue versus cancer tissue has also been highlighted by Upadhyay et al. using 3–D rigid motion invariant texture features [43].
In this work, we learn rotation–covariant computational models of RadLex VSTs from the visual appearance of liver lesions in CT. The models are built from linear combinations of N–th order steerable Riesz wavelets, which are learned using SVMs (see Section II-C). This allows local alignment of the models to maximize their response, which can be computed analytically for any order N. The scientific contribution of the VST models is twofold. First, the models can be used to predict and quantify the local likelihood of VSTs in ROIs. The latter is computed as the dot product between 12 × 12 image patch instances and one–versus–all (OVA) SVM models in a feature space spanned by the energies of the magnitudes of locally–steered VST models. Second, Euclidean distances are computed for every pair of VST models to establish a non–hierarchical computationally–derived ontology containing inter–term synonymy and complementarity. This work constitutes, to the best of our knowledge, a first attempt to establish a direct link between image contents and the visual semantics used by radiologists to interpret images.
II. Methods
A. Notations
A VST is denoted as ci, with i = 1,…, I while its likelihood of appearance in an image f is denoted ai ∈ [0, 1].
A generic d–dimensional signal f indexed by the continuous–domain space variable x = {x1, x2,…, xd} ∈ ℝd is considered. The d–dimensional Fourier transform of f is noted as:
with ω = {ω1, ω2,…, ωd} ∈ ℝd.
B. Dataset: VSTs of liver lesions in CT
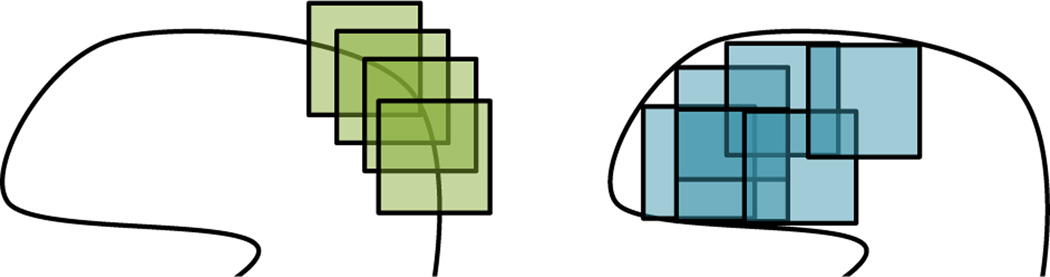
The institutional review board approved the retrospective analysis of de–identified patient images. The dataset consisted of 74 contrast–enhanced CT images of liver lesions in the portal venous phase with a slice thickness of 5mm [22, 37]. There are eight diagnoses of the lesions: metastasis (24), cyst (21), hemangioma (13), hepatocellular carcinoma (6), focal nodular hyperplasia (5), abscess (3), laceration (1), and fat deposition (1). A radiologist (C.F.B., 15 years of abdominal CT experience) used the axial slice f with the largest lesion area to circumscribe the liver lesions, producing image ROIs. Each lesion was annotated with an initial set of 72 VSTs from the RadLex ontology [22]. The presence of a term ci did not imply the absence of all others, where each lesion can contain multiple VSTs. All terms with an appearance frequency3 below 10% and above 90% were discarded, resulting in an intermediate set of 31 terms. A final set of 18 VSTs describing the margin and the internal texture of the lesions was used, which excludes terms describing the overall shape of the lesion (see Table I). Each of the 74 ROIs was divided into 12 × 12 patches4 to analyze (A) the margin of the lesion (i.e., periphery) and (B) the internal texture of the lesion. The distinction between these zones of a lesion is relevant to understanding how the algorithm performs, and also parallels how radiologists interpret lesions, taking into consideration both the boundary and internal features. The peripheral patches (A) were constrained to have their center on the ROI boundary and the internal patches (B) had to have their four corners inside the ROIs (see Fig. 1). The patches were overlapping with a minimum distance between the centers equals to one pixel. A maximum of 100 patches were randomly selected per ROI (i.e., 50 peripheral and 50 internal). Each of the 18 VST were represented by every patch extracted from the corresponding ROIs. Peripheral or internal patches were used as image instances depending on the VST’s localizations (see Table I). Peripheral patches were used for enhancing and nonenhancing to model the transition of enhancement at the boundary of the lesion in the portal venous phase.
TABLE I.
VSTs from RadLex used to describe the appearance of the liver lesions in CT scans. The 18 VSTs describing the margin and the internal texture of the lesions are marked in bold.
| category | VST | frequency | patch location |
|---|---|---|---|
| lesion margin | 1) circumscribed margin | 70.3 % | peripheral |
| 2) irregular margin | 12.2 % | ||
| 3) lobulated margin | 12.2 % | ||
| 4) poorly–defined margin | 16.2 % | ||
| 5) smooth margin | 45.9 % | ||
| lesion substance | 6) internal nodules | 12.2 % | internal |
| perilesional tissue characterization | 7) normal perilesional tissue | 43.2 % | peripheral |
| lesion focality | 8) solitary lesion | 37.8 % | — |
| 9) multiple lesions 2–5 | 21.6 % | ||
| 10) multiple lesions 6–10 | 20.3 % | ||
| 11) multiple lesions > 10 | 18.9 % | ||
| lesion attenuation | 12) hypodense | 72.2 % | internal |
| 13) soft tissue density | 16.2 % | ||
| 14) water density | 14.9 % | ||
| overall lesion enhancement | 15) enhancing | 62.2 % | peripheral |
| 16) hypervascular | 14.9 % | internal | |
| 17) nonenhancing | 29.7 % | peripheral | |
| spatial pattern of enhancement | 18) heterogeneous enh. | 13.5 % | internal |
| 19) homogeneous enh. | 32.4 % | internal | |
| 20) peripheral discont. nodular enh. | 17.6 % | peripheral | |
| temporal enhancement | 21) centripetal fill–in | 17.6 % | — |
| 22) homogeneous retention | 18.9 % | ||
| 23) homogeneous fade | 21.6 % | ||
| lesion uniformity | 24) heterogeneous | 41.9 % | internal |
| 25) homogeneous | 56.8 % | ||
| overall lesion shape | 26) round | 25.7 % | — |
| 27) ovoid | 45.9 % | ||
| 28) lobular | 25.7 % | ||
| 29) irregularly shaped | 12.2 % | ||
| lesion effect on liver | 31) abuts capsule of liver | 17.6 % | — |
Fig. 1.
Location of the image patches for the characterization of the margin (left) and the internal texture (right) of the lesions.
C. Rotation–covariant VST modeling
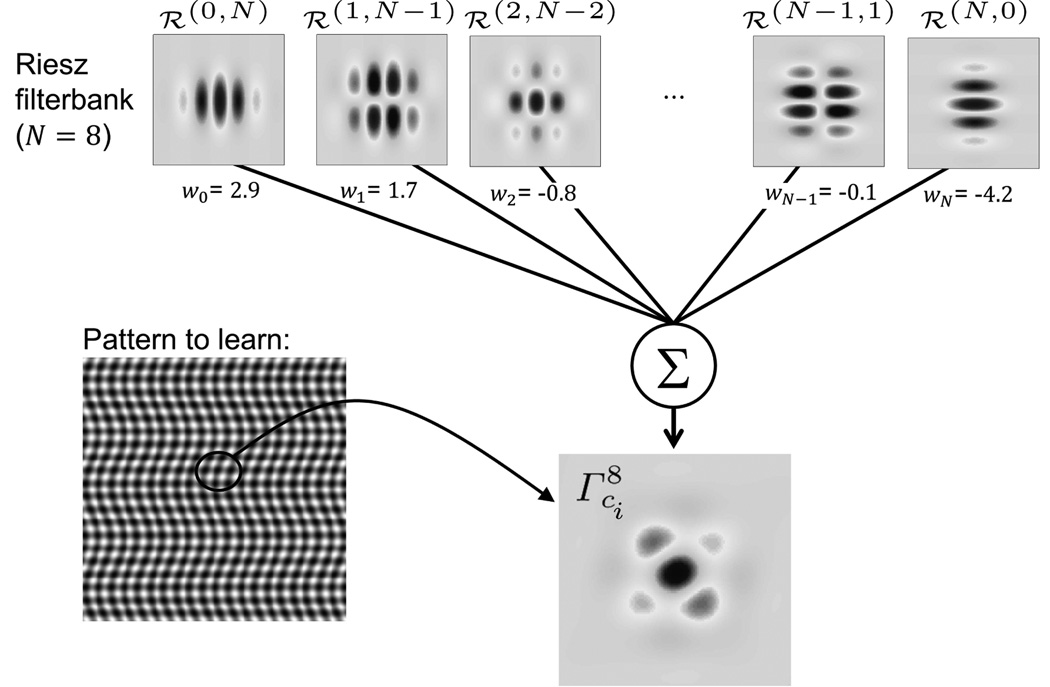
Recent work showed that the Riesz transform and its multi–scale extension constitutes a very efficient computational model of visual perception [44], since it performs multi–directional and multi–scale image analysis while fully covering the angular and spatial spectrums. This constitutes a major advantage when compared to other approaches relying on arbitrary choices of scales or directions for analysis (i.e., Gabor wavelets, gray–level co–occurrence matrices (GLCM), local binary patterns (LBP)). In a first step, we create VST models using linear combinations of N–th order steerable Riesz wavelets. Then, the VST models are steered locally to maximize their response, which can be done analytically for any order N and yields rotation–covariant results [42].
1) Steerable Riesz wavelets
Steerable multi–directional and multi–scale image analysis is obtained using the Riesz transform. Steerable Riesz wavelets are derived by coupling the Riesz transform and an isotropic multi–resolution framework5 [45]. The Nth–order Riesz transform ℛN of a 2–D function f yields N + 1 components ℛ(n,N−n), n = 0, 1,…, N that form multi–directional filterbanks. Every component ℛ(n,N−n) ∈ ℛN is defined in the Fourier domain as:
| (1) |
with ω1,2 corresponding to the frequencies along the two image axes x1,2. The multiplication with jω1,2 in the numerator corresponds to partial derivatives of f and the division by the norm of ω in the denominator ensures that only phase information (i.e., directionality) is retained. The directions of every component is defined by Nth–order partial derivatives in Eq. (1).
The Riesz filterbanks are steerable, which means that the response of every component ℛ(n,N−n) oriented with an angle θ can by synthesized from a linear combination of all components ℛ using a steering matrix Aθ as in [42]:
| (2) |
Aθ contains the respective coefficients of each component ℛ(n,N−n) to be oriented with an angle θ.
2) Steerable VST models
VST models are built using linear combinations of multi–scale Riesz components. Such models characterize the organizations of directions at various scales that are specific to each VST. At a fixed scale, a VST model is defined as:
| (3) |
where wci contains the weights of the respective Riesz components for the VST ci. An example of the construction of a model for a synthetic pattern is shown in Fig. 2. By combining Eqs. (2) and (3), the response of a model oriented by θ can still be expressed as a linear combination of the initial Riesz components as:
| (4) |
Fig. 2.
Example of the construction of a model using a linear combination of the Riesz templates ℛ(n,N−n). is visually similar to the pattern [42].
l2–norm SVMs are used to learn the optimal weights in a feature space spanned by the energies of concatenated multi–scale Riesz components to be optimally discriminant in OVA classification configurations [42, 46].
D. From VST models to a computationally–derived ontology
Once multi–scale models are learned for every VST using OVA configurations, the distance between every pair of VST can be computed as the Euclidean distance between the corresponding set of weights wci. The symmetric matrix Φ(ci, cj) can be considered as a non–hierarchical computationally–derived ontology modeling the visual inter–term synonymy and complementarity relations.
E. Rotation–covariant local quantification of VSTs
The orientation θ of each model is optimized at each position xp and for each scale sj to maximize its local magnitude as:
| (5) |
The maximum magnitude of the model steered using θdom,sj at the position xp is computed as:
| (6) |
For a given image patch, a feature vector υ can be built as the energies E of the magnitudes over the patch of every steered model as:
| (7) |
The dimensionality of υ is I × J. It worth noting that the features from Eq. (7) are not steerable anymore after using the energies of the steered model’s magnitudes. OVA SVM models ui with Gaussian kernels ϕ(υ) are used to learn the presence of every VST in the feature space spanned by the vectors υ in Eq. 7. The decision value of the SVM for the image patch υp measures the likelihood ai of a VST as:
| (8) |
where b is the bias of the SVM model. Likelihood maps are created by displaying ai values from all overlapping patches.
F. Experimental setup
The number of scales was chosen as J = ⌊log2(12)⌋ = 3 to cover the full spatial spectrum of 12 × 12 patches. The order of the Riesz transform N = 8 was used, which we found to provide an excellent trade–off between computational complexity and the degrees of freedom of the filterbanks in [42, 47]. A leave–one–patient–out (LOPO) cross–validation (CV) was used both to learn the VST models and to estimate the performance of VST detection using OVA configurations. For each fold, the training set was used both for learning the models and to train the SVMs in the feature space spanned by the vectors υ in Eq. (7). No multi–class classification is performed since the VSTs are not considered as mutually exclusive [37]. The random selection of the patches was repeated five times to assess the robustness of the approach. The decision values ai (see Eq. (8)) of the test patches were averaged over each ROI and used to build receiver operating characteristic (ROC) curves for each VST. The values ai of the test patches were also used to locally quantify the presence of a VST and were color–coded to create VST–wise likelihood maps. CT intensities were used as additional features to model gray–level distributions in the patches, which we found to be complementary to Riesz models in [46]. The distribution of CT intensities in [−60, 220] Hounsfield Units (HU) were divided into 20 histogram bins that were directly concatenated with the features obtained from Eq. (7).
III. Results
A. VST models and computationally–derived ontology
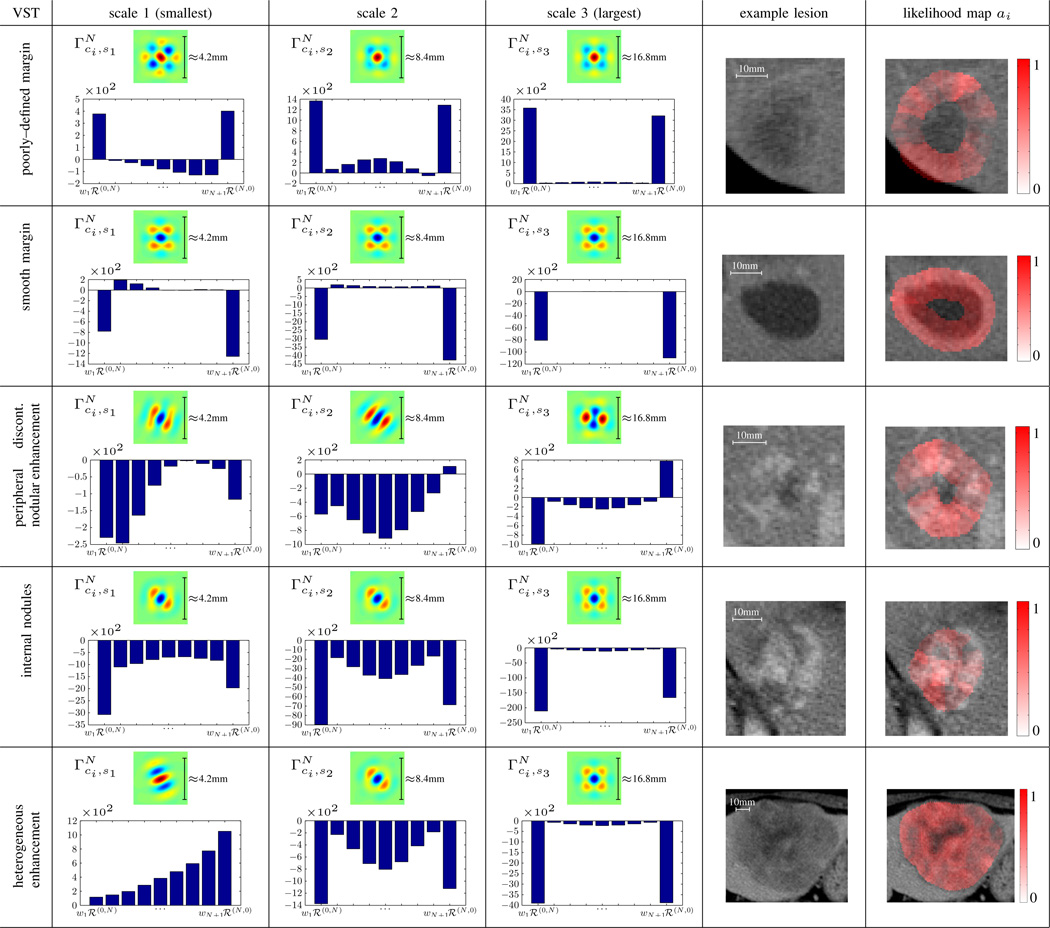
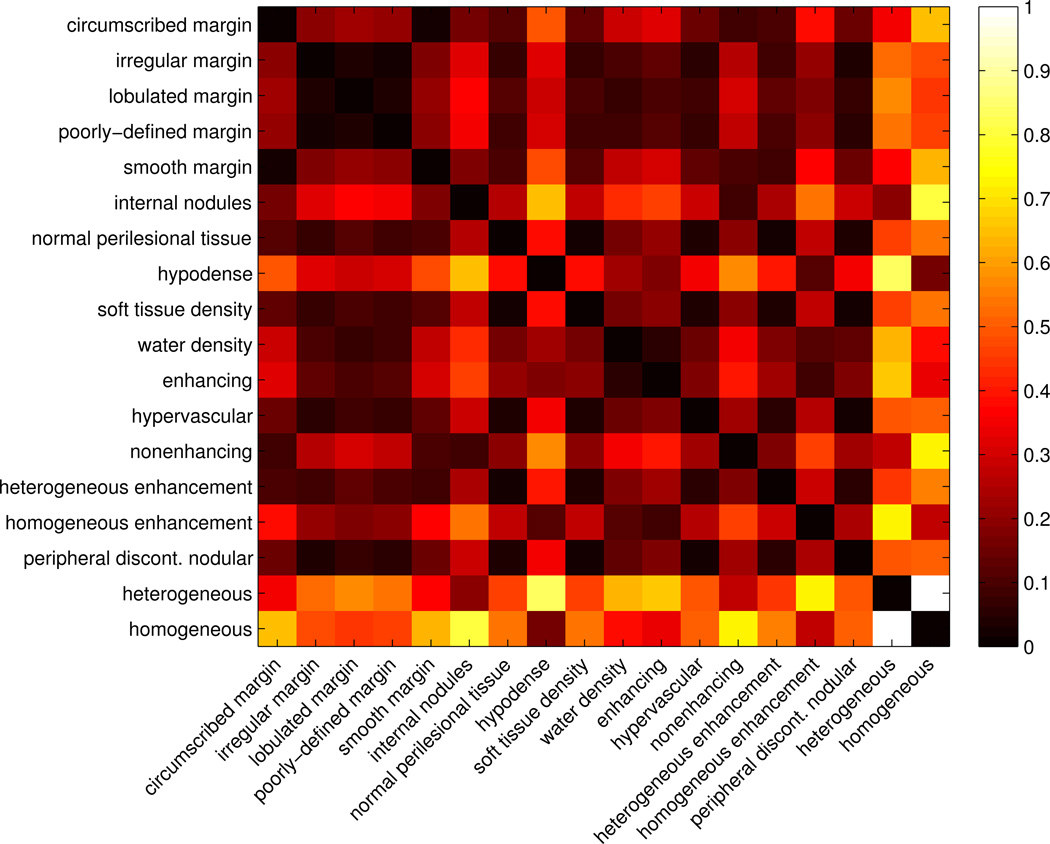
Examples of scale–wise models are shown in Fig. 5 for six VSTs. The distributions of the weights wci,sj for every N + 1 Riesz component are shown with bar plots. The normalized computationally–derived ontology matrix Φ(ci, cj) containing the Euclidean distances between every pair of VST models is represented as a heatmap in Fig. 3.
Fig. 5.
Scale–wise models for 5 VSTs and N = 8. The distributions of the weights wci,j learned by OVA SVMs are represented for each scale with bar plots. The red regions in the last column of the table are showing examples of likelihood maps ai computed from Eq. (8) for every VSTs.
Fig. 3.
Computationally–derived ontology matrix Φ(ci, cj) containing the normalized Euclidean distances for every pair of VST models. Values closer to zero (black) indicate the shortest distances, or most similar terms. Clear relationships between groups of VSTs are revealed both for terms describing lesion margin and internal texture.
B. Automated VST detection and quantification
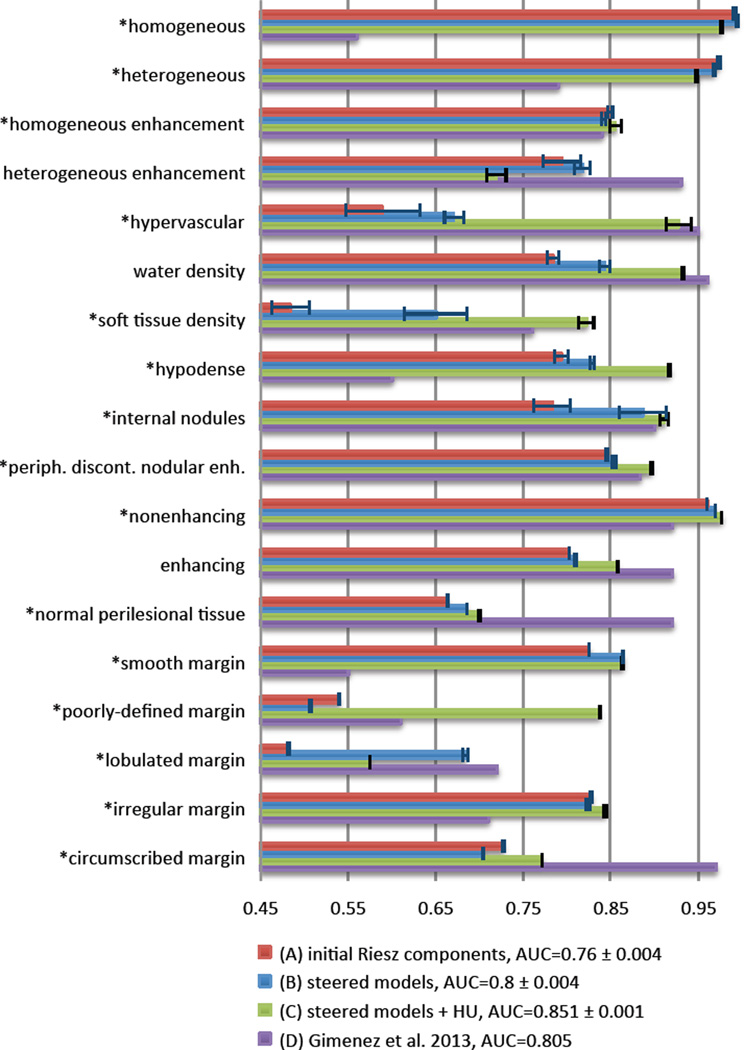
ROC curves were built by varying thresholds on the decision values ai. Fig. 4 compares the detection performance using the energies of the magnitudes of steered models (i.e., Eq. (7)) versus the energies of the initial Riesz components. A paired T–test on ai values is used to compare the two approaches. We also compared the performance of steered models combined with HU histogram bins with the best results obtained by Gimenez et al. on the same dataset as used here [37]. Examples of local quantification of the local presence ai of VSTs (i.e., likelihood maps) are shown in the last column of Fig. 5.
Fig. 4.
Comparison of the automated detection performance between (A) initial Riesz components ℛN, (B) steered models , (C) steered models combined with HU histogram bins, and (D) the best results obtained by Gimenez et al. [37] on the same dataset in terms of AUCs. (*) denotes p–values below 0.05 for the comparison between (A) and the (B). (B) are always higher or close to the best performance of the (A), which highlights the importance of rotation–covariance. The difference between the global AUCs of (A) and (B) (i.e., 0.76 versus 0.8) is associated with a p–value of 7.5446e-152. (C) and (D) were shown to be complementary.
IV. Discussions and Conclusions
We built computational models of human perceptual semantics in radiology. The framework identifies relevant organizations of image scales and directions related to VSTs in a rotation–covariant fashion using steerable Riesz wavelets and SVMs. The models were used both to (1) detect and quantify the local presence of VST and (2) to create a computationally–derived ontology from actual image content. In these early applications, our techniques for detection and quantification of VST enable automatic prediction of relevant semantic terms for radiologist consideration. This is an important step towards improving the accuracy of lesion description and diagnosis, with the aim of reducing overall interpretation errors. Our approach generates VST likelihood maps (see Fig. 5), which provide insights on the information modeled by the system, making it possible for the user to evaluate the amount of trust that can be put in its outputs and maximizes the transparency of the methods. Avoiding a “black box” type approach, this feature of our system is likely to be more intuitive to the ultimate end users. The automated detection and quantification of visual semantics grants access to large amounts of similar images by enabling interoperability with semantic indexing [22, 48]. The creation of a computationally–derived ontology from VST models can be used to refine and complement existing ontologies, where relations between terms are solely encoded by their hierarchical linguistic organization. The proposed computationally–derived ontology allows measuring inter–term synonymy and complementarity in the context of a given medical application adding additional useful knowledge to existing ontologies [49].
The visualization of the computationally–derived ontology matrix Φ(ci, cj) in Fig. 3 reveals clear relationships between groups of VSTs. Two homogeneous groups are found to model antonymous terms concerning the lesion margin: well– versus poorly–defined margin. Irregular, lobulated and poorly–defined margin are very close, and they are all distant from circumscribed and smooth margin. The lack of distinction between the shape of the lesion and the type of lesion margin shows the inability of the proposed models to characterize overall lesion shape. For instance, lobulated margin and poorly–defined margin are not expected to be close, as it is possible to have lesions with margins that are both lobulated and circumscribed. Shared synonymy and antonymy is also observed in VSTs describing the internal texture of the lesions. Heterogeneous and internal nodules are found to share synonymy, and they are both opposed to homogeneous, homogeneous enhancement and hypodense. It can also be observed that hypervascular is close to heterogeneous enhancement and peripheral discontinuous nodular enhancement, which all relate to the pattern of lesion enhancement. A few erroneous associations are observed (e.g., soft tissue density and hypervascular, or water density and enhancing). Overall, the computationally–derived ontology is found to be complementary to the RadLex ontology, because it allows connecting semantic concepts with their actual appearance in CT images. For instance, heterogeneous and homogeneous are very close to each other in RadLex because they both describe the uniformity of lesion enhancement, but they are opposed to each other in the computationally–derived ontology since they are visually antonymous in terms of texture characterization. The combination of the RadLex ontology and the computationally–derived ontology was shown to significantly improve image retrieval performance in [49]. Likewise, we would expect our computationally–derived ontology to be useful in combination with existing ontologies like RadLex in image retrieval and other applications.
Figure 4 details the VST detection performance. It shows that although the steered models are not improving the results for all VSTs, they are always higher or close to the best performance of the initial Riesz components (i.e., global AUC equals to 0.8 versus 0.76, p = 7:5446e-152). An overall complementarity of the features based on steerable VST models and HU intensities is observed (e.g., water density, soft tissue density, hypervascular, hypodense). However, the detection performance is little improved or even harmed for texture–related terms when compared to using only steerable VST models (e.g., heterogeneous, heterogeneous enhancement, homogeneous). The proposed approach appears to be complementary to Gimenez et al. [37], where the errors are occurring for different VSTs. This suggests that the inclusion of other feature types can improve our approach.
Fig. 5 displays the relevance of the information modeled by a subset of VST models. The visualization of models reveals dominant scale–wise VSTs. The largest scale model of peripheral discontinuous nodular enhancement implements a detector of nodules surrounding the lesion boundary. The two smallest scale models of internal nodules are implementing circular dot detectors. These cases illustrate that the models of VSTs correspond to image features that actually describe the intended semantics. In general, the scale–wise distributions of the weights reveal the importance of Riesz components ℛ(0;N) and ℛ(N;0), which can be explained by their ability to model strongly directional structures (see Fig. 2). The stability of the models over the folds of the LOPO–CV is measured by the trace of the covariance matrices of VST–wise sets of weights wci over the CV folds. The values of VST–wise traces are all below 0.33% of the trace of the global covariance matrix of all models from all folds. This demonstrates the stability, and hence the generalization ability, of the learned models.
We recognize several limitations of the current work, including a narrow imaging domain, a relatively small number of cases, and the use of somewhat thick, 5mm CT sections. In future work, we plan to include additional cases to limit the risk of finding erroneous associations between terms caused by fortuitous co–occurrences of them. We plan to include additional image features that model the lesion shape. This will allow us to create separate computationally–derived ontologies based on the type of information modeled (i.e., intensity, texture, margin and shape).
Acknowledgments
This work was supported by the Swiss National Science Foundation (PBGEP2_142283), and the National Cancer Institute, National Institutes of Health (U01–CA–142555 and R01–CA–160251).
Footnotes
https://www.rsna.org/RadLex.aspx, as of March 2014.
Liver annotation Task at ImageCLEF, http://www.imageclef.org/2014/liver/, as of March 2014.
The appearance frequency of a VST is defined as the percentage of lesions in the database in which the term was present.
Patches larger than 12 × 12 did not fit in the smallest lesion.
Simoncelli’s multi–resolution framework is used with a dyadic scale progression. The scaling function is not used.
Contributor Information
Adrien Depeursinge, Email: adepeurs@stanford.edu, Department of Radiology of the School of Medicine, Stanford University, CA, USA.
Camille Kurtz, Department of Radiology of the School of Medicine, Stanford University, CA, USA; C. Kurtz is also with the LIPADE (EA2517), University Paris Descartes, France.
Christopher F. Beaulieu, Department of Radiology of the School of Medicine, Stanford University, CA, USA
Sandy Napel, Department of Radiology of the School of Medicine, Stanford University, CA, USA.
Daniel L. Rubin, Department of Radiology of the School of Medicine, Stanford University, CA, USA
References
- 1.Krupinski EA. The role of perception in imaging: Past and future. Seminars in Nuclear Medicine. 2011;41(6):392–400. doi: 10.1053/j.semnuclmed.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Andriole K, Wolfe J, Khorasani R. Optimizing analysis, visualization and navigation of large image data sets: One 5000–section CT scan can ruin your whole day. Radiology. 2011;259:346–362. doi: 10.1148/radiol.11091276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson P. Radiology’s Achilles’ heel: error and variation in the interpretation of the Rontgen image. British Journal of Radiology. 1997;70(839):1085–1098. doi: 10.1259/bjr.70.839.9536897. [DOI] [PubMed] [Google Scholar]
- 4.Tourassi G, Voisin S, Paquit V, Krupinski E. Investigating the link between radiologists’ gaze, diagnostic decision, and image content. Journal of the American Medical Informatics Association. 2013 doi: 10.1136/amiajnl-2012-001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood B. Visual expertise. Radiology. 1999;211(1):1–3. doi: 10.1148/radiology.211.1.r99ap431. [DOI] [PubMed] [Google Scholar]
- 6.Berlin L. Malpractice issues in radiology. Perceptual errors. American Journal of Roentgenology. 1996;167(3):587–590. doi: 10.2214/ajr.167.3.8751657. [DOI] [PubMed] [Google Scholar]
- 7.Rubin DL. Finding the meaning in images: Annotation and image markup. Philosophy, Psychiatry, and Psychology. 2012;18(4):311–318. [Google Scholar]
- 8.Wagner RF, Insana MF, Brown DG, Garra BS, Jennings RJ. Texture discrimination: Radiologist, machine and man. In: Blakemore C, Adler K, Pointon M, editors. Vision. Cambridge University Press; 1991. pp. 310–318. [Google Scholar]
- 9.Krupinski EA. The future of image perception in radiology: Synergy between humans and computers. Academic radiology. 2003;10(1):1–3. doi: 10.1016/s1076-6332(03)80781-x. [DOI] [PubMed] [Google Scholar]
- 10.van Ginneken B, Schaefer-Prokop CM, Prokop M. Computer–aided diagnosis: How to move from the laboratory to the clinic. Radiology. 2011;261(3):719–732. doi: 10.1148/radiol.11091710. [DOI] [PubMed] [Google Scholar]
- 11.Rubin DL, Napel S. Imaging informatics: toward capturing and processing semantic information in radiology images. Yearbook of medical informatics. 2010:34–42. [PubMed] [Google Scholar]
- 12.Sobel JL, Pearson ML, Gross K, Desmond KA, Harrison ER, Rubenstein LV, Rogers WH, Kahn KL. Information content and clarity of radiologists’ reports for chest radiography. Academic Radiology. 1996;3(9):709–717. doi: 10.1016/s1076-6332(96)80407-7. [DOI] [PubMed] [Google Scholar]
- 13.Burnside ES, Davis J, Chhatwal J, Alagoz O, Lindstrom MJ, Geller BM, Littenberg B, Shaffer KA, Kahn CE, Page CD. Probabilistic computer model developed from clinical data in national mammography database format to classify mammographic findings. Radiology. 2009;251(3):663–672. doi: 10.1148/radiol.2513081346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubin DL, Mongkolwat P, Kleper V, Supekar K, Channin DS. Annotation and image markup: Accessing and interoperating with the semantic content in medical imaging. IEEE Intelligent Systems. 2009;24(1):57–65. [Google Scholar]
- 15.D’Orsi CJ, Newell MS. BI–RADS decoded: Detailed guidance on potentially confusing issues. Radiologic Clinics of North America. 2007;45(5):751–763. doi: 10.1016/j.rcl.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Hansell DM, Bankier AA, MacMahon H, McLoud TC, Müller NL, Remy J. Fleischner society: Glossary of terms for thoracic imaging. Radiology. 2008;246(3):697–722. doi: 10.1148/radiol.2462070712. [DOI] [PubMed] [Google Scholar]
- 17.Fardon DF. Nomenclature and classification of lumbar disc pathology. Spine. 2001;26(5) doi: 10.1097/00007632-200103010-00007. [DOI] [PubMed] [Google Scholar]
- 18.Atkinson RP, Awad IA, Batjer HH, Dowd CF, Furlan A, Giannotta SL, Gomez CR, Gress D, Hademenos G, Halbach V, et al. Reporting terminology for brain arteriovenous malformation clinical and radiographic features for use in clinical trials. Stroke. 2001;32(6):1430–1442. doi: 10.1161/01.str.32.6.1430. [DOI] [PubMed] [Google Scholar]
- 19.Zalis M, Barish M, Choi J, Dachman A, Fenlon H, Ferrucci J, Glick S, Laghi A, Macari M, McFarland E, Morrin M, Pickhardt P, Soto J, Yee J. CT colonography reporting and data system: A consensus proposal. Radiology. 2005;236(1):3–9. doi: 10.1148/radiol.2361041926. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg S, Grassi C, Cardella J, Charboneau J, Dupuy GD, III, Gervais D, Gillams A, Kane R, Livraghi FT, Jr, McGahan J, Phillips D, Rhim H, Silverman S, Solbiati L, Vogl T, Wood B, Vedantham S, Sacks D. Image–guided tumor ablation: Standardization of terminology and reporting criteria. Journal of Vascular and Interventional Radiology. 2009;20(7) Supplement:377–390. doi: 10.1097/01.RVI.0000170858.46668.65. [DOI] [PubMed] [Google Scholar]
- 21.Rubin DL. Creating and curating a terminology for radiology: Ontology modeling and analysis. Journal of Digital Imaging. 2008;21(4):355–362. doi: 10.1007/s10278-007-9073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Napel S, Beaulieu CF, Rodriguez C, Cui J, Xu J, Gupta A, Korenblum D, Greenspan H, Ma Y, Rubin DL. Automated retrieval of CT images of liver lesions on the basis of image similarity: Method and preliminary results. Radiology. 2010;256(1):243–252. doi: 10.1148/radiol.10091694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sivic J, Zisserman A. Proceedings of the Ninth IEEE International Conference on Computer Vision - Volume 2. Washington, DC, USA: IEEE Computer Society; 2003. Video google: A text retrieval approach to object matching in videos; pp. 1470–1477. [Google Scholar]
- 24.André B, Vercauteren T, Buchner AM, Wallace MB, Ayache N. Endomicroscopic video retrieval using mosaicing and visual words. IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 2010:1419–1422. ser. ISBI 2010. [Google Scholar]
- 25.Avni U, Greenspan H, Konen E, Sharon M, Goldberger J. X–ray categorization and retrieval on the organ and pathology level, using patch–based visual words. IEEE Transactions on Medical Imaging. 2011;30(3):733–746. doi: 10.1109/TMI.2010.2095026. [DOI] [PubMed] [Google Scholar]
- 26.Foncubierta-Rodríguez A, Depeursinge A, Müller H. Using multiscale visual words for lung texture classification and retrieval. Medical Content–based Retrieval for Clinical Decision Support. 2012;7075:69–79. Lecture Notes in Computer Sciences (LNCS) [Google Scholar]
- 27.Burner A, Donner R, Mayerhoefer M, Holzer M, Kainberger F, Langs G. Texture bags: Anomaly retrieval in medical images based on local 3D–texture similarity. Medical Content-based Retrieval for Clinical Decision Support. 2012;7075:116–127. Lecture Notes in Computer Sciences (LNCS) [Google Scholar]
- 28.Yang W, Lu Z, Yu M, Huang M, Feng Q, Chen W. Content–based retrieval of focal liver lesions using bag–of–visual–words representations of single– and multiphase contrast-enhanced CT images. Journal of Digital Imaging. 2012;25(6):708–719. doi: 10.1007/s10278-012-9495-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Mei T, Gong S, Hua X-S. Combining global, regional and contextual features for automatic image annotation. Pattern Recognition. 2009;42(2):259–266. [Google Scholar]
- 30.André B, Vercauteren T, Buchner AM, Wallace MB, Ayache N. Learning semantic and visual similarity for endomicroscopy video retrieval. IEEE Transactions on Medical Imaging. 2012;31(6):1276–1288. doi: 10.1109/TMI.2012.2188301. [DOI] [PubMed] [Google Scholar]
- 31.Sidong L, Weidong C, Yang S, Sonia P, Ron K, Dagan F. MICCAI workshop MCBR–CDS 2013. 2013. A bag of semantic words model for medical content–based retrieval. ser. Springer LNCS. [Google Scholar]
- 32.Liu Y, Dellaert F, Rothfus WE. Classification driven semantic based medical image indexing and retrieval. Carnegie Mellon University, The Robotics Institute; 1998. [Google Scholar]
- 33.Tang LH, Hanka R, Ip HHS, Lam R. Extraction of semantic features of histological images for content–based retrieval of images. Medical Imaging. 1999;3662:360–368. [Google Scholar]
- 34.Barb AS, Shyu C-R, Sethi YP. Knowledge representation and sharing using visual semantic modeling for diagnostic medical image databases. IEEE Transactions on Information Technology in Biomedicine. 2005;9(4):538–553. doi: 10.1109/titb.2005.855563. [DOI] [PubMed] [Google Scholar]
- 35.Mueen A, Zainuddin R, Baba MS. Automatic multilevel medical image annotation and retrieval. Journal of Digital Imaging. 2008;21(3):290–295. doi: 10.1007/s10278-007-9070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwitt R, Vasconcelos N, Rasiwasia N, Uhl A, Davis B, Häfner M, Wrba F. Endoscopic image analysis in semantic space. Medical Image Analysis. 2012;16(7):1415–1422. doi: 10.1016/j.media.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gimenez F, Xu J, Liu Y, Liu T, Beaulieu C, Rubin DL, Napel S. AMIA Annual Symposium Proceedings. Vol. 2012. American Medical Informatics Association; 2012. Automatic annotation of radiological observations in liver CT images; pp. 257–263. [PMC free article] [PubMed] [Google Scholar]
- 38.Yu F, Ip HHS. Semantic content analysis and annotation of histological images. Computers in Biology and Medicine. 2008;38(6):635–649. doi: 10.1016/j.compbiomed.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Shyu C-R, Pavlopoulou C, Kak AC, Brodley CE, Broderick LS. Using human perceptual categories for content–based retrieval from a medical image database. Computer Vision and Image Understanding. 2002;88(3):119–151. [Google Scholar]
- 40.Raicu DS, Varutbangkul E, Furst JD, Armato SG., III Modelling semantics from image data: opportunities from LIDC. International Journal of Biomedical Engineering and Technology. 2010;3(1):83–113. [Google Scholar]
- 41.Tibshirani R. Regression shrinkage and selection via the Lasso. Journal of the Royal Statistical Society. Series B (Methodological) 1996;58(1):267–288. [Google Scholar]
- 42.Depeursinge A, Foncubierta A, Van De Ville D, Müller H. Rotation–covariant texture learning using steerable Riesz wavelets. IEEE Transactions on Image Processing. 2014;23:898–908. doi: 10.1109/TIP.2013.2295755. [DOI] [PubMed] [Google Scholar]
- 43.Upadhyay S, Papadakis M, Jain S, Gladish G, Kakadiaris IA, Azencott R. Abdominal Imaging. Computational and Clinical Applications. Vol. 7601. Berlin Heidelberg: Springer; 2012. Semi–automatic discrimination of normal tissue and liver cancer lesions in contrast enhanced X–ray CT–scans; pp. 158–167. [Google Scholar]
- 44.Langley K, Anderson SJ. The Riesz transform and simultaneous representations of phase, energy and orientation in spatial vision. Vision Research. 2010;50(17):1748–1765. doi: 10.1016/j.visres.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 45.Unser M, Van De Ville D. Wavelet steerability and the higher–order Riesz transform. IEEE Transactions on Image Processing. 2010;19(3):636–652. doi: 10.1109/TIP.2009.2038832. [DOI] [PubMed] [Google Scholar]
- 46.Depeursinge A, Foncubierta A, Van De Ville D, Müller H. Medical Image Computing and Computer–Assisted Intervention MICCAI 2012. Vol. 7512. Berlin / Heidelberg: Springer; 2012. Multiscale lung texture signature learning using the Riesz transform; pp. 517–524. [DOI] [PubMed] [Google Scholar]
- 47.Depeursinge A, Foncubierta A, Müller H, Van De Ville D. Rotation–covariant visual concept detection using steerable riesz wavelets and bags of visual words. SPIE Wavelets and Sparsity XV. 2013;8858:885, 816–11. [Google Scholar]
- 48.Gerstmair A, Daumke P, Simon K, Langer M, Kotter E. Intelligent image retrieval based on radiology reports. European Radiology. 2012;22(12):2750–2758. doi: 10.1007/s00330-012-2608-x. [DOI] [PubMed] [Google Scholar]
- 49.Kurtz C, Depeursinge A, Napel S, Beaulieu CF, Rubin DL. On combining image–based and ontological dissimilarities for medical image retrieval applications. Medical Image Analysis. doi: 10.1016/j.media.2014.06.009. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]