SUMMARY
ALIX plays a role in nucleocapsid release during viral infection, as does lysobisphosphatidic acid (LBPA). However, the mechanism remains unclear. Here we report that LBPA is recognized within an exposed site in ALIX Bro1 domain predicted by MODA, an algorithm for discovering membrane-docking areas in proteins. LBPA interactions revealed a strict requirement for a structural calcium tightly bound near the lipid interaction site. Unlike other calcium– and phospholipid-binding proteins, the all-helical triangle-shaped fold of the Bro1 domain confers selectivity for LBPA via a pair of hydrophobic residues in a flexible loop, which undergoes a conformational change upon membrane association. Both LBPA- and calcium–binding are necessary for endosome association and virus infection, as are ALIX ESCRT-binding and dimerization capacity. We conclude that LBPA recruits ALIX onto late endosomes via the calcium-bound Bro1 domain, triggering a conformational change in ALIX to mediate the delivery of viral nucleocapsids to the cytosol during infection.
INTRODUCTION
After internalization, activated signaling receptors are sorted into lumenal invaginations of the early endosome membrane, which are pinched off as free cargo-containing vesicles (Gruenberg and Stenmark, 2004). These multivesicular regions detach and become multivesicular endosomes or bodies, which are transported to and fuse with late endosomes. Eventually, intralumenal vesicles (ILVs) and their cargo are delivered to lysosomes and degraded. Viruses hijack this doorway into the cell through a poorly understood process that is mediated by ALIX, LBPA (or BMP, bis(monoacylglycero)phosphate) and the ESCRT machinery.
The sorting of activated signaling receptors into forming ILVs is achieved by ESCRT complexes, including ESCRT-I and –II, which may induce membrane invaginations and ESCRT-III, which may catalyze membrane fission (Hurley and Hanson, 2010; Raiborg and Stenmark, 2009). ESCRT-I and ESCRT-III are also involved in two processes topologically equivalent to ILV formation, cytokinesis and virus budding at the plasma membrane. As is ALIX, which interacts with the ESCRT-I subunit TSG101 (Strack et al., 2003; von Schwedler et al., 2003) and the ESCRT-III subunit CHMP4 (Fisher et al., 2007; Usami et al., 2007). Unlike ESCRTs, however, ALIX does not play a role in ubiquitin-dependent lysosome targeting of the EGF receptor (Cabezas et al., 2005; Doyotte et al., 2008; Luyet et al., 2008; Schmidt et al., 2004), but perhaps in the ubiquitin-independent, ESCRT-III-dependent sorting of the GPCR PAR1 (Dores et al., 2012).
We had found that ALIX interacts with the unconventional phospholipid LBPA (Matsuo et al., 2004), which is detected only in late, but not early, endosomes, and is abundant in ILVs (Kobayashi et al., 1999). Much like ALIX, LBPA plays no role in signaling receptor trafficking to lysosomes (Luyet et al., 2008). Instead, both ALIX and LBPA play a crucial role in the penetration of some pathogenic agents into host cells, including anthrax toxin (Abrami et al., 2004), vesicular stomatitis virus (VSV) (Le Blanc et al., 2005; Luyet et al., 2008), Lassa virus and lymphocytic choriomeningitis virus (Pasqual et al., 2011). Although the mechanism remains to be elucidated, it has been proposed that anthrax lethal factor and VSV RNA would first be released into the ILV lumen in order to reach late endosomes without encountering their degradative environment (Abrami et al., 2004; Le Blanc et al., 2005). There, ILV backfusion with the limiting membrane would ensure toxin or RNA release into the cytoplasm in a process controlled by LBPA, ALIX and ESCRTs. This LBPA- and ALIX-dependent endosomal way-out may be used by endogenous cargo in transit towards destinations other than the lysosomes (Chevallier et al., 2008; Kobayashi et al., 1999; Kobayashi et al., 1998; Le Blanc et al., 2005; Luyet et al., 2008; Pasqual et al., 2011).
Here, we elucidate the molecular mechanism by which ALIX mediates viral penetration through recognition of LBPA, a unique lipid found within late endosomes. We conclude that LBPA recruits ALIX onto late endosomes in a process controlled by calcium, which may trigger ALIX conformational changes, and that this mechanism in turn controls ALIX functions in viral infection.
RESULTS
ALIX-membrane interactions are LBPA- and calcium-dependent
Since ALIX may be involved in regulating LBPA functions (Matsuo et al., 2004), we expressed and purified to homogeneity (Fig 1A and S1A–B) ALIX’s structural domains, in order to identify the determinants of LBPA recognition. The recombinant proteins were incubated with liposomes of the same phospholipid composition as late endosomes, including 20Mol% LBPA (Kobayashi et al., 1999), and membrane binding was measured in a step sucrose gradient after liposome floatation. The N-terminal Bro1 domain, but not the middle V domain, copurified with liposomes (Fig 1B), and this interaction specifically required the presence of LBPA (Fig 1F–G). While screening for optimal binding conditions, we found that membrane interaction requires calcium — an unprecedented finding since ALIX does not contain any known calcium-binding motif. Liposome binding was enhanced with increasing calcium concentrations, with an apparent EC50 ≈ 100nM, while without calcium virtually no binding could be detected (Fig 1C–D). Calcium was specifically required for ALIX-membrane interactions, since it could not be replaced with magnesium (Fig S1C). Unless otherwise indicated, 2μM calcium was present in all subsequent experiments.
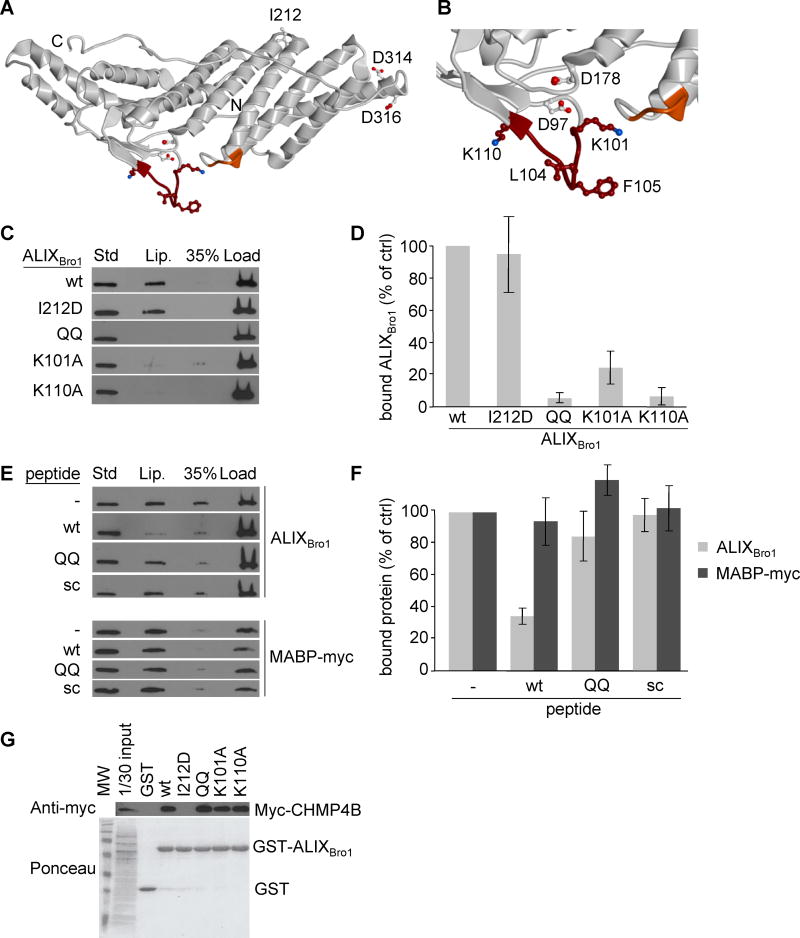
Figure 1. ALIX-membrane interactions are LBPA- and calcium-dependent.
(A) Schematic representation of ALIX protein domains. (B) Recombinant ALIXBro1 and ALIXV were incubated with liposomes (DOPC/DOPE/PI/LBPA; [5:2:1:2 mol]) for 2h at 4°C. Then, the liposome-bound protein was separated from free protein by floatation in sucrose gradients. Fractions were collected and analyzed by western blotting: (Lip) 50% of the fraction containing liposomes; (35%) 5.5% of the 35% sucrose cushion; (load) 12% of the fraction containing the load; (Std) 0.25 μg of the corresponding recombinant protein as standard. With LBPA present at the same level as in late endosomes (Kobayashi et al., 1998), 5% of total ALIXBro1 was found associated with liposomes after floatation, consistent with the characteristically weak and dynamic nature of protein-lipid interactions. (C–D) Liposome-binding of recombinant ALIXBro1 was analyzed as in (B), but in the presence of the indicated concentrations of free calcium (C), and quantified in (D). (E) A typical calcium titration curve monitored by ITC (upper panel) shows calcium-binding to ALIXBro1. The curve fit is illustrated in the lower panel, revealing that approximately 1 calcium atom was bound per ALIXBro1 molecule, and the thermodynamic values obtained from the curve fit are: n = 1.58 ± 0.06, K = 2.14*106 ± 7.33*105 M−1, ΔH = 2892 ± 143.4cal/mol, ΔS = 39.2cal/mol/deg, KD = 467 ±160nM. (F) Recombinant ALIXBro1 was incubated with liposomes containing LBPA as in (B) or not (DOPC/DOPE/PI; [7:2:1 mol]). Protein binding to liposomes was analyzed as in (B). (G) Data in (F) were quantified and are expressed as a percentage of LBPA values. (H) Binding of ALIXBro1 to liposomes containing 10mol% of the indicated phospholipids in 90mol% DOPC was analyzed as in (B). (I) Data in (H) were quantified and are expressed as in (G). All quantifications show means (±SEM) of 3 independent experiments (see also Fig S1).
Isothermal titration calorimetry (ITC) measurements showed that approximately 1 calcium atom is bound per ALIXBro1 molecule and that ALIXBro1 binds calcium with an KD of 467 ±160nM (Fig 1E) — a value in the same range as calcium-dependent ALIXBro1 binding to liposomes (Fig 1C–D). These values are in the range of cytosolic free calcium concentrations under resting conditions. Hence, calcium binding is unlikely to be controlled by calcium-dependent signaling. As calcium can bind to the Bro1 domain while free in solution (Fig 1E) and is also required for membrane interaction (Fig 1C–D), calcium may play a structural role by stabilizing the ALIX conformation competent to bind membranes.
ALIXBro1 binding to membranes required LBPA, a negatively charged lipid, and binding was reduced to background when LBPA was omitted (Fig 1F–G). However, other negatively charged phospholipids did not substitute for LBPA (Fig 1H–I). Thus, ALIX interactions with LBPA are not simply due to the presence of a negative charge on the lipid headgroup. ALIX recruitment to membranes appears to be mediated by its calcium-bound Bro1 domain specifically recognizing LBPA. As such, the ALIX Bro1 domain is fundamentally different from other membrane binding domains — none of which exhibits LBPA binding.
Identification of the membrane interaction site of ALIX
The Bro1 domain of ALIX does not share any obvious homologies to known membrane interaction sites. For de novo prediction we thus made use of a newly developed Membrane Optimal Docking Area (MODA) program. This algorithm identifies protein surface patches, which serve as privileged sites for lipid interactions, and has been trained against experimentally validated membrane binding sites (Kufareva and Abagyan, 2009) (Irina Kufareva & Ruben Abagyan, in preparation).
Using the available protein structure of ALIX, a major membrane interaction site was predicted on the convex surface of the Bro1 domain, corresponding to 101-KGSLFGGSVK-110 and 232-QYKD-235, with two hydrophobic residues L104 and F105 being prominently exposed at the extremity of the former flexible loop (Fig 2A–B). These two residues are flanked by glycine residues and by basic lysine residues at position 101 and 110 (Fig 2B) — a signature feature of some membrane insertion loops, thus further strengthening the MODA prediction. The membrane interaction site predicted in the Bro1 domain of ALIX is conserved in vertebrates, which contain LBPA, and is not found in yeast and plants (Fig S2A), in which LBPA has not been detected. Furthermore, the site is not found in functionally unrelated proteins containing a Bro1 domain (Fig S2B).
Figure 2. Identification of the membrane interaction site of ALIXBro1.
(A) Ribbon model of ALIXBro1 using the atomic coordinates derived from the X-ray diffraction analysis (PDB ID: 2R03) (Zhai et al., 2008). The N- and C-termini of the Bro1 domain are marked with N and C, respectively. The membrane interacting residues predicted by MODA are shown in brown (101-KGSLFGGSVK-110) and orange (232-QYKD-235). Side chains of residues mutagenized in this study are indicated as sticks, including i) I212 involved in CHMP4 interactions (Fisher et al., 2007; Usami et al., 2007), ii) L104, F105, K101 and K110 in membrane binding, iii) D97 and D178 in calcium coordination, and iv) D314 and D316 as controls. (B) Higher magnification view of the membrane-interacting region. (C) The liposome-binding capacity of ALIXBro1, ALIXBro1I212D, ALIXBro1QQ, ALIXBro1K101A and ALIXBro1K110A was analyzed as in Fig 1B. (D) Data in (C) were quantified and are expressed as a percentage of the wt values. (E) ALIXBro1 or the myc-tagged MABP domain of MVB12B was incubated with liposomes containing 20% LBPA or PS, respectively, in the presence of 100-fold molar excess wt, QQ or scrambled peptide. Membrane binding of ALIXBro1 was analyzed as in Fig 1B. (F) Data in (E) were quantified and are expressed as a percentage of a control without peptide. (G) Extracts prepared from cells expressing CHMP4B-myc were incubated with ALIXBro1-GST or with the same mutants as in (C-D). These were then retrieved using glutathione Sepharose beads and analyzed by western blotting using anti-myc antibody. First lane: 1/30 of the starting materials. All quantifications show means (±SEM) of at least 3 independent experiments (see also Fig S2).
Mutation of the exposed hydrophobic residues L104 and F105 (Fig 2B) to glutamines (QQ mutant) abolished membrane interactions in our liposome-binding assay (Fig 2C–D). As did alanine mutations of the two lysine residues 101 and 110 (Fig 2C–D), which flank L104 and F105 on either side of the loop and therefore seem well positioned to interact with the negatively charged head groups of membrane phospholipids (Fig 2B). The observed effects of the mutations were not due to improper folding, since the QQ, K101A and K110A mutants still interacted with CHMP4 (Fig 2G). Conversely, the I212D mutation in the CHMP4 binding site abrogated ALIX-CHMP4 interaction (Fisher et al., 2007; Usami et al., 2007) (Fig 2G), but not liposome binding, indicating that the functional effect of the QQ, K101A and K110A mutations were specific (Fig 2C–D).
Moreover, a peptide of ALIX’s wildtype membrane-interacting loop (93-FTWKDAFDKGSLFGGSVK-110) competed with ALIXBro1 for membrane interaction, whereas the corresponding QQ peptide (93-FTWKDAFDKGSQQGGSVK-110) or a scrambled version of the wildtype peptide (GDKVKSTKWFSAFGDFGL) had no effect (Fig 2E–F). By contrast, the peptide of ALIX’s wildtype membrane-interacting loop had no effect on the membrane association of the MABP-myc (MVB12-associated beta-prism) domain of MVB12B (Fig 2E–F), which is found on the same membranes as ALIX in vivo and which binds negatively charged phospholipids (Boura and Hurley, 2012), but shows no preference for LBPA (Fig S1D). These data indicate that the loop identified by the MODA software in ALIXBro1 is the dominant contributor to direct binding of LBPA-containing membranes.
Identification of the calcium interaction site
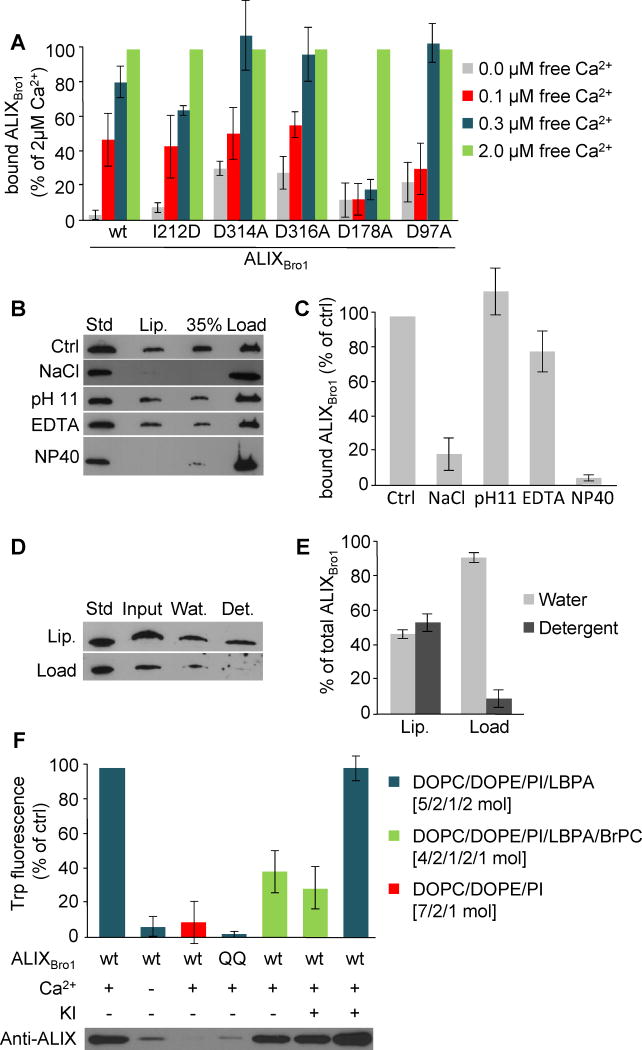
Comparison with the structures of 563 calcium-binding proteins in the PDB yielded a possible calcium coordination site involving D97 and D178, which are located close to the LBPA interaction site in ALIXBro1 (Fig 2A–B). Mutation of residue D178 to alanine significantly raised the calcium concentration required for membrane binding (Fig 3A), since membrane interactions were reduced to background even at 0.3μM free calcium — a concentration at which wildtype ALIXBro1 showed almost complete saturation (Fig 1D and 3A). A similar although less pronounced effect was observed with ALIXBro1 carrying the D97A mutation (Fig 3A). The role of D97 and D178 seems specific, since membrane binding was not affected by mutation of D314 and D316 (Fig 3A), which are also exposed on the protein surface (Fig 2A). Furthermore, the I212D mutant in the CHMP4 binding site (Fig 2G) retains calcium-dependent membrane binding capacity (Fig 3A).
Figure 3. Characterization of ALIX-membrane interactions.
(A) Liposome binding was analyzed as in Fig 1B for ALIXBro1, ALIXBro1I212D, ALIXBro1D97A, ALIXBro1D178A, ALIXBro1D314A and ALIXBro1D316A in the presence of 0., 0.1, 0.3 and 2μM free calcium. Binding is expressed as a percentage of the value obtained with 2μM free calcium. (B) ALIXBro1 was first pre-incubated with liposomes for 2h as in Fig 1B. The reaction mixture was then adjusted to 2M NaCl, 0.1M carbonate pH 11, 10mM EDTA or 2% NP40. ALIXBro1 remaining on the membrane was analyzed after floatation as in Fig 1B. (C) Quantification of membrane-bound ALIXBro1 in (B). Binding is expressed as a percentage of the control (Ctrl). (D) After binding to liposomes, membrane-bound and free ALIXBro1 were separated as in Fig 1B, and each was extracted with TX-114. Lanes 1–4: 0.25μg ALIXBro1 as standard; complete mixture before phase separation (input); water (Wat) and detergent (Det) phases after extraction. (E) Quantification of TX-114 extraction in (D), expressed as a percentage of the total protein in each condition. (F) Recombinant ALIXBro1 and ALIXBro1QQ were incubated in the presence or absence of 2.5μM calcium and liposomes lacking LBPA or containing 20mol% LBPA with or without 10% BrPC. Liposomes with membrane-bound protein were recovered by floatation and Trp fluorescence was measured. If indicated, 150 mM KI was added prior to the measurements. All quantifications show means (±SEM) of 3 independent experiments (see also Fig S3).
Hence, calcium is likely to be coordinated by residues D97 and D178, at a site positioned to form a structured linker or hinge for the membrane interacting loop where it extends from the single β sheet. The structure of this site, together with the observations that ALIX binds calcium with a relatively high affinity in solution (Fig 1E) and while on membranes (Fig 1C–D), suggests that calcium induces a conformation in the β1–β2 loop which is favorably predisposed for membrane interactions.
ALIX interacts tightly with membranes
A treatment with 2M NaCl released bound ALIXBro1 from membranes (Fig 3B–C), indicating that electrostatic interactions are involved in membrane association. This is consistent with the role of the positively charged lysine residues 101 and 110 flanking the LBPA interaction site (Fig 2C–D) and the negative charge of LBPA. However, as other negatively charged lipids do not substitute for LBPA (Fig 1H–I), electrostatic interactions may enhance but do not determine binding. ALIXBro1 was not released by a carbonate wash at pH 11 (Fig 3B–C), a classic test that favours hydrophobic interactions (Fujiki et al., 1982), suggesting that the protein is associated to membranes via a combination of electrostatic and hydrophobic interactions. While calcium chelation prevented ALIXBro1 membrane recruitment (Fig 1C–D), it dislodged little membrane-bound ALIXBro1 (Fig 3B–C), presumably because of the stability of the membrane complex. Moreover, when liposomes with bound ALIXBro1 were extracted with Triton X-114, 50% of membrane-bound ALIXBro1 fractionated like a membrane protein into the detergent phase (Bordier, 1981), whereas 90% of the free protein was found in the water phase (Fig 3D–E). In contrast, ALIXBro1 was readily released after membrane solubilization with the detergent NP40 (Fig 3B–C). Our results show that ALIXBro1 exhibits two distinct states, i.e. free in solution or membrane-bound (Fig 3), and that membrane association involves both electrostatic and hydrophobic interactions — consistent with the critical role of exposed hydrophobic residues and their flanking basic residues (Fig 2B). We conclude that a conformational change may allow ALIXBro1 to insert into the bilayer during LBPA engagement.
Membrane-bound ALIX penetrates the lipid bilayer
We used Trp fluorescence to measure membrane association, since Trp 95 is near the interaction site. The signal was abolished without calcium, as it was with calcium but with liposomes lacking LBPA (Fig 3F). In addition, no signal was detected with the QQ mutant, consistent with its inability to bind LBPA-containing liposomes (Fig 3F; see also Fig 2C–D). The fluorescence intensity correlated well with ALIXBro1 membrane-association (Fig 3F, western blot), indicating that the signals reflect the calcium-dependent association of ALIXBro1 with liposomes containing LBPA. Strikingly, ALIXBro1 fluorescence was significantly quenched when LBPA-containing liposomes were spiked with brominated lipids, without affecting ALIXBro1 membrane-association (Fig 3F). The remaining signal was presumably emitted by a second Trp residue at position 6 of the mouse recombinant ALIXBro1 protein. Quenching likely occurred from within the bilayer, since no quenching was observed with a buffer containing KI (Fig 3F). This lack of effect could not be due to the Trp residue being hidden from the solution on the side of the protein bound to the bilayer, since KI had no effect on Trp fluorescence in solution (Fig S3A).
Altogether, our data indicated that calcium induced a conformation predisposed for membrane binding and that the conformational change was complete upon interactions with LBPA. Calcium binding, however, did not alter the overall structure of ALIXBro1, since it had little, if any, effect on the protein shape analyzed by SAXS (Fig S3E), Trp fluorescence (Fig S3B) or circular dichroism (Fig S3C–D). We conclude that binding to LBPA-containing membranes causes partial bilayer penetration of the mobile hydrophobic loop, resulting in tight membrane association of ALIXBro1.
Endosome association is impaired by mutations in the LBPA- and calcium-interaction sites
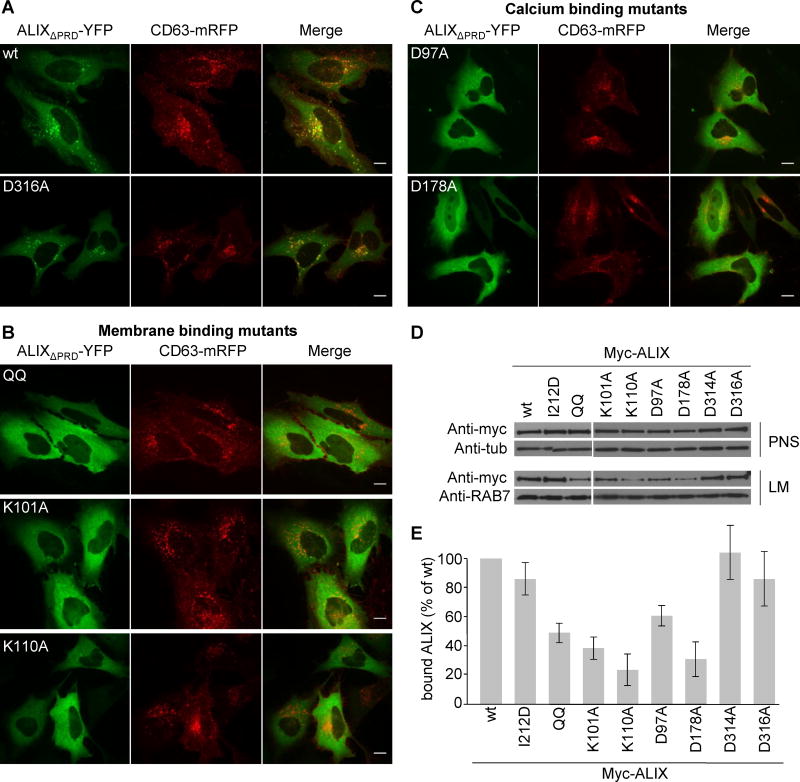
Conventional immunofluorescence is of limited use to visualize ALIX on endosomes since the protein is predominantly cytosolic (Vito et al., 1999) (Fig S3F). ALIX, however, was shown to undergo dimerization (Pires et al., 2009). We thus combined live-cell microscopy with split YFP complementation (Remy and Michnick, 2007), where N- and C-terminal halves of YFP are fused to ALIX lacking the proline-rich domain (Pires et al., 2009). After transfection of the split ALIXΔPRD-YFP constructs, the staining was cytosolic (Fig 4A) as expected (Fig S3F). But at low expression levels, ALIXΔPRD-YFP also co-localized with the late endosomal marker CD63 (Fig 4A), in agreement with the distribution of its partner phospholipid LBPA (Kobayashi et al., 2002). Strikingly, the ALIXΔPRDQQ-YFP mutant showed a diffuse, typically cytosolic staining pattern, as did the constructs carrying the K101A or K110A mutation as well as the D97A or D178A mutation – but not the control D316A mutation (Fig 4A–C).
Figure 4. Endosome association is impaired in membrane- and calcium-binding mutants.
(A–C) HeLa cells were cotransfected with CD63-mRFP (red) and with split ALIXΔPRD-YFP containing the indicated mutations (green) and visualized by live-cell microscopy 16h after transfection; (A) wildtype (wt) and control D316A mutant; (B) membrane binding mutants, QQ, K101A and K110A; (C) calcium binding mutants D97A and D178A. Bars: 10 μm. (D) Myc-tagged ALIX full-length mutants were transiently overexpressed in HeLa cells and post nuclear supernatant (PNS) and light membrane fractions (LM) containing endosomes were prepared and analyzed by western blotting using the indicated antibodies. Antibodies to tubulin (Tub) and RAB7 were used as equal loading marker. (E) Quantification of myc-ALIX mutants in LM (D) expressed as a percentage of wt protein. Means (±SEM) of at least 3 independent experiments are shown (see also Fig S5).
Like the ESCRT-I and ESCRT-III complexes, ALIX is not only present on endosomes but is also found at the midbody during cytokinesis, where it is recruited by CEP55 (Carlton et al., 2008; Morita et al., 2007). As expected, ALIX labelled the midbody of mock-treated but not CEP55-depleted cells (Fig S4G). The QQ mutant was also found at the midbody (Fig S4G), as was the I212D mutant in the ESCRT-III binding site (Carlton et al., 2008). Hence, mutation of the hydrophobic L104 and F105 residues, which are involved in association with LBPA-containing membranes in vitro and late endosomes in vivo, does not affect the LBPA-independent association of ALIX to the midbody.
After subcellular fractionation by floatation in step sucrose gradients (Huber et al., 2000), we found that membrane association of the full-length protein with a defective LBPA-interaction site (ALIXQQ, ALIXK101A and ALIXK110A) was severely impaired, when compared to wildtype (Fig 4D–E). Similarly, membrane association of full-length proteins harboring the D97A or D178A mutation, but not the control mutations D314A and D316A, was also strongly impaired compared to wildtype (Fig 4D–E). These differences in membrane association did not reflect different expression levels (Fig 4D–E). The effects were specific, since mutation of the CHMP4-binding site (ALIXI212D) only marginally, if at all, affected membrane association (Fig 4D–E). Altogether, these observations indicate that the flexible loop identified in ALIX with the MODA software is necessary for binding to LBPA-containing bilayers in vitro and to late endosomes in vivo, as are the flanking basic residues K101 and K110.
Moreover, our data with the D97A and D178A mutants (Fig 4D–E) show that the putative calcium-coordinating residues next to the LBPA site play a consistent role in calcium-dependent liposome binding in vitro and endosome localization in vivo. This also defines the Bro1 domain’s calcium-dependent LBPA recognition function as being responsible for the selective recruitment to the late endosome.
ALIX functions in viral infection require intact LBPA-, calcium- and ESCRT-interaction sites
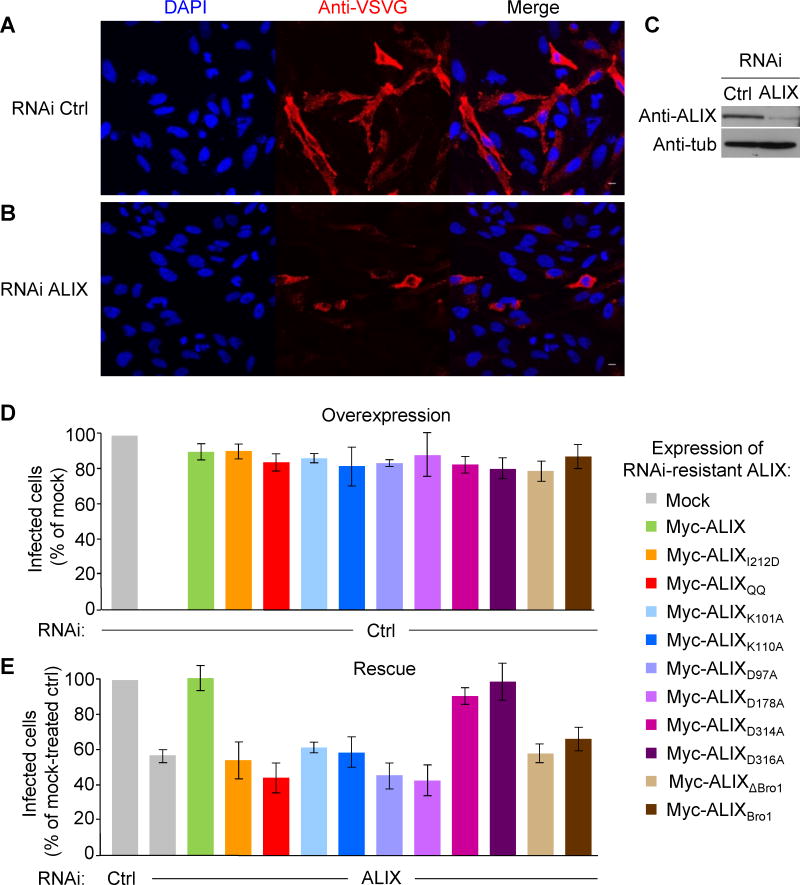
Given the involvement of ALIX and LBPA in the delivery of viral nucleocapsids to the cytoplasm during viral infection (Le Blanc et al., 2005; Luyet et al., 2008; Pasqual et al., 2011), we investigated whether ALIX binding to LBPA is necessary for VSV infection. After virus binding to the cell surface on ice at low multiplicity of infection (1.0 MOI), followed by a 3h incubation at 37°C, ≈50% of the cells were infected, as quantified by measuring viral G-glycoprotein expression (Fig 5A). A partial knockdown of ALIX (≈80%) with siRNAs (Fig 5C) reduced infection to ≈60% of the control (Fig 5B quantification in E), in agreement with previous observations (Le Blanc et al., 2005; Luyet et al., 2008). This inhibition was specific, since infection could be restored by overexpression of RNAi-resistant wildtype ALIX (Fig 5E) — an effect not due to some gain-of-function phenotype, since overexpression alone did not affect infection (Fig 5D).
Figure 5. ALIX functions in VSV infection require active LBPA, calcium and ESCRT interaction sites.
(A–B) HeLa cells that had been pre-treated with control siRNAs (A) or anti-ALIX siRNAs (B) were incubated for 1h on ice with VSV at low MOI (1.0) and then for 3h at 37°C to allow infection to proceed. After fixation and permeabilization, infected cells were visualized using anti-VSV-G antibody (red) and nuclei using DAPI (blue). Data are quantified in (D). (C) Knockdown efficiency of RNAi treatment was analyzed by western blotting. Antibodies to tubulin (Tub) were used as equal loading marker. (D–E) HeLa cells were first transfected with control (D) or anti-ALIX siRNAs (E), as in A and B respectively, and 56h later with DNA coding for the indicated RNAi-resistant versions of myc-tagged ALIX mutants or wildtype (mock: empty vector). Then, 16h later the cells were infected with VSV as in (A–B), and processed for immunofluorescence using anti-VSV-G antibody and anti-myc antibody to visualize infected and ALIX-myc expressing cells, respectively. Virus infection was scored in cells expressing wildtype ALIX or each mutant, and the data are expressed as a percentage of the value obtained with control (mock transfected) cells. Means (±SEM) of more than 3 independent experiments are shown. Samples from the same experiments were analyzed by SDS gel electrophoresis followed by western blotting with anti-myc antibodies to determine the levels of expression of each protein (see also Fig S5).
In the ALIX knockdown background, infection could not be restored by re-expression of the ALIXI212D mutant (Fig 5E), which binds LBPA-containing membranes (Fig 2 and 4), but not ESCRT-III (Fig 2G). Neither was infection efficiently restored by ALIX Bro1 domain, which binds bilayers containing LBPA (Fig 1), but cannot dimerize or interact with the ESCRT-I subunit TSG101 or other partners. Similarly re-expression of ALIXΔBro1, which lacks both membrane interaction and CHMP4-binding sites did not rescue infection (Fig 5E). These observations cannot be accounted for by differences in the expression levels (Fig S4C–D). Similarly, none of the mutants affected VSV infection when overexpressed much like the wildtype protein (Fig 5D). Remarkably, we find that a monomeric mutant ALIXmono (Pires et al., 2009) failed to restore infection after ALIX knockdown (Fig S4B), while, as a control, another mutant in the same region that can still form dimers (ALIXdimer (Pires et al., 2009)) partially restored viral infection (Fig S4B). Again, VSV infection was not affected after overexpression of any of the mutant (Fig S4A) and effects are not due to differences in expression levels (Fig S4C–D). Membrane association of ALIXdimer, like ALIXmono, was essentially not impaired (Fig S4E–F).
In marked contrast to the wildtype protein, mutants defective in LBPA recognition (ALIXQQ) failed to restore viral infection after ALIX knockdown (Fig 5E). As did mutants of the lysine residues that flank the LBPA recognition site (ALIXK101A and ALIXK110A) or of the calcium binding site (ALIXD97A and ALIXD178A). Overexpression of these mutants had no effect (Fig 5D) and expression levels were comparable (Fig S4C–D). Altogether these data demonstrate that ALIX calcium-dependent LBPA recognition is necessary for the function of the protein in viral infection. As are ALIX interactions with ESCRT subunits, in good agreement with findings from us (Luyet et al., 2008) and others (Pasqual et al., 2011) that viral infection depends on both ALIX and ESCRTs. Our observations also suggest that ALIX dimerization plays an important role not only in HIV budding (Pires et al., 2009), but also in VSV infection. Finally, our data argue that ALIX binding to LBPA-containing membranes is required upstream from ALIX dimerization and engagement of the ESCRT machinery.
DISCUSSION
Mechanism of ALIX calcium-dependent lipid-binding
We show that ALIX interacts with LBPA-containing membranes via a lipid-binding motif within a flexible loop on the convex side of the protein flanked by two lysine residues, and that these interactions require calcium-binding, presumably to an unconventional site in the vicinity of the lipid-binding site.
This calcium-dependence is unexpected, as the Bro1 domain does not contain any known calcium-binding motif. Our data suggest that calcium is coordinated by D97 and D178, two residues in the vicinity of the LBPA-interaction loop with orientations consistent with those found in calcium-binding sites. ALIXBro1 interactions with membranes appear to be reminiscent of annexins and C2 domain-containing proteins (Gerke et al., 2005; Lemmon, 2008), which bind negatively charged membranes with a calcium EC50 value in the low micromolar range (Evans and Nelsestuen, 1994; Guerrero-Valero et al., 2007). Annexins (Glenney, 1986) and some C2 domains (Radhakrishnan et al., 2009), however, exhibit a reduced capacity to bind calcium in the absence of membranes, because calcium ions serve as a bridge between protein and membrane. In contrast, ALIXBro1 appears to bind calcium with a similar high affinity (EC50 < 1 μM), whether free in solution (Fig 1E) or membrane-bound (Fig 1D), arguing against the notion that calcium ions bound to ALIXBro1 link protein and membrane. The two residues D97 and D178 are well positioned to coordinate calcium at a site close to, but not in immediate contact with the lipid headgroups — and calcium chelation does not release membrane-bound ALIXBro1. Interactions with the lipid headgroups likely involve the basic residues on either side of the lipid-binding motif.
We conclude that calcium by binding close to the LBPA-interaction loop may stabilize or induce a change in the conformation of the flexible loop favorable for membrane interaction (Fig S5). Docking of LBPA headgroup using the flexible docking algorithm within the ICM molecular modeling package (Abagyan and Totrov, 1994; Totrov and Abagyan, 1997, 2008) reveals that it can be lounged in a groove immediately above the LBPA-interaction loop (Fig S5B–C). Upon membrane interaction, ALIX would then undergo a local conformational change leading to partial insertion of the hydrophobic loop into the cytoplasmic leaflet of the bilayer.
ALIX in viral infection and backfusion
Our data indicate that ALIX is recruited onto endosomes via LBPA, where it can engage ESCRT-I and -III, and thereby regulates viral nucleocapsid release. We also have evidence that ALIX may dimerize upon membrane interaction, and that this process is crucial for the role of ALIX in virus infection, as it is for HIV budding (Pires et al., 2009). Interestingly, F105 in the LBPA-interacting loop is also necessary for HIV release (Sette et al., 2011; Zhai et al., 2011). It is attractive to speculate that the trigger role of LBPA in causing ALIX conformational change and dimerization is played by another component, presumably a viral protein, during HIV budding. Importantly, ALIX-CHMP4 interactions are dispensable for membrane targeting, but are necessary for virus infection (this study), abscission during cytokinesis (Carlton et al., 2008; Morita et al., 2007) and virus budding (Usami et al., 2007). Thus one may hypothesize that ALIX coordinates the assembly of CHMP4 filaments, which in turn contribute to remodel the membrane.
This raises some key-issues related to endosome topology, in particular how ALIX, a cytosolic protein that is targeted to the limiting membrane of late endosomes, controls the release of viral RNA by regulating ILV backfusion. Although future work will be necessary to determine precisely how this process is regulated, our study already provides insights into the mechanism at the molecular level.
ALIX and LBPA may regulate backfusion indirectly, by controlling ILV formation (Falguieres et al., 2008; Matsuo et al., 2004), as both processes have to be tightly coupled in order to maintain endosomal membrane homeostasis. Alternatively, it is also possible that ALIX controls the backfusion process more directly. We find that upon membrane binding ALIX undergoes a conformation change that leads to the partial insertion of its LBPA-interacting loop into the membrane. Insertion may affect membrane symmetry, providing a mechanistic explanation for ALIX effects on ILV formation in liposomes (Matsuo et al., 2004) and endosomes (Falguieres et al., 2008) in vitro. More important, by affecting membrane symmetry, the partial insertion of the hydrophobic loop into the cytoplasmic leaflet may cause local perturbations of the bilayer organization, in concert with ALIX dimerization, CHMP4-binding and assembly. The local perturbation may in turn render the membrane more prone to undergo fusion with ILVs on the lumenal side. This mechanism would explain how ALIX, when binding to membranes, can control ILV backfusion and virus infection. These two models are not mutually exclusive. In fact, one may speculate that ALIX and LBPA form micro-domains or hot spots on the endosomal limiting membrane, where ILVs undergo kiss-and-run fusion and fission cycles.
EXPERIMENTAL PROCEDURES
The protocols used for cell culture and transfection, as well as the list of reagents, cDNA constructs, siRNAs, peptides and recombinant proteins are indicated in the supplementary experimental procedures available online, as are the detailed protocols we used in this study.
Production and purification of recombinant proteins
Recombinant GST-ALIXBro1 and GST-MABP-myc were produced and purified as described in the supplementary experimental procedures.
Liposome-binding assays
Liposomes were prepared after lipid hydration and sonication at 0.3–3μM free calcium concentrations using calcium buffers. Liposomes containing LBPA (DOPC/DOPE/PI/LBPA; [5:2:1:2 mol]) were incubated with ALIXBro1 in 25mM Hepes-NaOH pH 7.4 containing 2μM free calcium for 2h at 4°C. In some experiments, the mixture was supplemented with 100μM peptide, or with 50mM or 100mM KCl. When indicated, liposomes without LBPA (DOPC/DOPE/PI at [7:2:1 mol]) or liposomes containing 90mol% PC and 10mol% of the designated lipid were used. The liposome-bound protein was separated from free protein by floatation in sucrose gradients. The liposome-protein mixture was adjusted to 40.6 % sucrose, overlaid with 35% and 8.5% sucrose and centrifuged for 1h at 55000rpm. Fractions of the 8.5% interphase, containing the liposomes, the 35% sucrose cushion and the load were collected and analyzed by western blotting.
Biophysical methods
The protocols used to measure isothermal titration calorimetry, Trp fluorescence, circular dichroism, and SAXS are available online as supplementary experimental procedures.
Immunofluorescence analysis and vesicular stomatitis virus infection
To study viral infection, cells were pre-incubated with VSV (1 MOI) for 1h at 4°C, and then for 3h at 37°C. After fixation and permeabiliz ation, cells were analyzed by immunofluorescence using anti-VSV antibody, anti-myc antibody (9E10) and DAPI for nuclei staining. In rescue experiments, siRNAs were transfected 72h before infection and DNA was transfected 16h before infection.
Supplementary Material
A lipid-binding mechanism is found in the protein ALIX
This mechanism requires a module specific for the late endosome lipid LBPA
Membrane binding involves a structural calcium close to the lipid binding site
Viral infection needs intact LBPA- and calcium-binding capacity
Bissig et al. show that the late-endosomal phospholipid LBPA recruits the ESCRT component ALIX onto endosomes via an exposed flexible loop near a tightly bound calcium ion. Both LBPA and calcium binding are necessary for ALIX/ESCRT-dependent virus infection, arguing that this mechanism for membrane manipulation has been exploited by pathogens.
Acknowledgments
We are grateful to members of our laboratory for fruitful discussion. We also thank Marie-Hélène Beuchat and Brigitte Bernadets for their technical help and the Bio-molecular Analysis Platform for help with ITC. We also wish to thank Paul Nielsen (Echelon Biosciences, Salt Lake City, USA) for his help and expertise with LBPA synthesis, and the protein analysis facility of Zurich University for amino acid analysis. Support to MO and JG was from the PRISM EU Sixth Framework Program, to MO from BBSRC, and to JG from the Swiss National Science Foundation, the Swiss Sinergia program, the Polish-Swiss Research Programme (PSPB-094/2010), the NCCR in Chemical Biology and LipidX from the Swiss SystemsX.ch initiative, evaluated by the Swiss National Science Foundation. IK and RA were supported by the NIH grants U01 GM094612, U54 GM094618, and R01 GM071872.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. Journal of molecular biology. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- Abrami L, Lindsay M, Parton RG, Leppla SH, van der Goot FG. Membrane insertion of anthrax protective antigen and cytoplasmic delivery of lethal factor occur at different stages of the endocytic pathway. J Cell Biol. 2004;166:645–651. doi: 10.1083/jcb.200312072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- Boura E, Hurley JH. Structural basis for membrane targeting by the MVB12-associated beta-prism domain of the human ESCRT-I MVB12 subunit. Proc Natl Acad Sci U S A. 2012;109:1901–1906. doi: 10.1073/pnas.1117597109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas A, Bache KG, Brech A, Stenmark H. Alix regulates cortical actin and the spatial distribution of endosomes. J Cell Sci. 2005;118:2625–2635. doi: 10.1242/jcs.02382. [DOI] [PubMed] [Google Scholar]
- Carlton JG, Agromayor M, Martin-Serrano J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc Natl Acad Sci U S A. 2008;105:10541–10546. doi: 10.1073/pnas.0802008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallier J, Chamoun Z, Jiang G, Prestwich G, Sakai N, Matile S, Parton RG, Gruenberg J. Lysobisphosphatidic acid controls endosomal cholesterol levels. J Biol Chem. 2008;283:27871–27880. doi: 10.1074/jbc.M801463200. [DOI] [PubMed] [Google Scholar]
- Dores MR, Chen B, Lin H, Soh UJ, Paing MM, Montagne WA, Meerloo T, Trejo J. ALIX binds a YPX3L motif of the GPCR PAR1 and mediates ubiquitin-independent ESCRT-III/MVB sorting. The Journal of cell biology. 2012;197:407–419. doi: 10.1083/jcb.201110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyotte A, Mironov A, McKenzie E, Woodman P. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc Natl Acad Sci U S A. 2008;105:6308–6313. doi: 10.1073/pnas.0707601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans TC, Jr, Nelsestuen GL. Calcium and membrane-binding properties of monomeric and multimeric annexin II. Biochemistry. 1994;33:13231–13238. doi: 10.1021/bi00249a009. [DOI] [PubMed] [Google Scholar]
- Falguieres T, Luyet PP, Bissig C, Scott CC, Velluz MC, Gruenberg J. In vitro budding of intralumenal vesicles into late endosomes is regulated by Alix and Tsg101. Mol Biol Cell. 2008;19:4942–4955. doi: 10.1091/mbc.E08-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. The Journal of cell biology. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- Glenney J. Phospholipid-dependent Ca2+ binding by the 36-kDa tyrosine kinase substrate (calpactin) and its 33-kDa core. J Biol Chem. 1986;261:7247–7252. [PubMed] [Google Scholar]
- Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- Guerrero-Valero M, Marin-Vicente C, Gomez-Fernandez JC, Corbalan-Garcia S. The C2 domains of classical PKCs are specific PtdIns(4,5)P2-sensing domains with different affinities for membrane binding. J Mol Biol. 2007;371:608–621. doi: 10.1016/j.jmb.2007.05.086. [DOI] [PubMed] [Google Scholar]
- Huber LA, Fialka I, Paiha K, Hunziker W, Sacks DB, Bahler M, Way M, Gagescu R, Gruenberg J. Both calmodulin and the unconventional myosin Myr4 regulate membrane trafficking along the recycling pathway of MDCK cells. Traffic. 2000;1:494–503. doi: 10.1034/j.1600-0854.2000.010607.x. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Beuchat MH, Chevallier J, Makino A, Mayran N, Escola JM, Lebrand C, Cosson P, Kobayashi T, Gruenberg J. Separation and characterization of late endosomal membrane domains. J Biol Chem. 2002;277:32157–32164. doi: 10.1074/jbc.M202838200. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Beuchat MH, Lindsay M, Frias S, Palmiter RD, Sakuraba H, Parton RG, Gruenberg J. Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat Cell Biol. 1999;1:113–118. doi: 10.1038/10084. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Stang E, Fang KS, de Moerloose P, Parton RG, Gruenberg J. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 1998;392:193–197. doi: 10.1038/32440. [DOI] [PubMed] [Google Scholar]
- Kufareva I, Abagyan R. Predicted Molecular Interactions in Structural Proteomics. In: Nussinov R, Schreiber G, editors. Computational Protein-Protein Interactions. CRC Press; 2009. pp. 185–209. [Google Scholar]
- Le Blanc I, Luyet PP, Pons V, Ferguson C, Emans N, Petiot A, Mayran N, Demaurex N, Faure J, Sadoul R, et al. Endosome-to-cytosol transport of viral nucleocapsids. Nat Cell Biol. 2005;7:653–664. doi: 10.1038/ncb1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Luyet PP, Falguieres T, Pons V, Pattnaik AK, Gruenberg J. The ESCRT-I subunit TSG101 controls endosome-to-cytosol release of viral RNA. Traffic. 2008;9:2279–2290. doi: 10.1111/j.1600-0854.2008.00820.x. [DOI] [PubMed] [Google Scholar]
- Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, Faure J, Blanc NS, Matile S, Dubochet J, Sadoul R, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303:531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung HY, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. Embo J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqual G, Rojek JM, Masin M, Chatton JY, Kunz S. Old world arenaviruses enter the host cell via the multivesicular body and depend on the endosomal sorting complex required for transport. PLoS pathogens. 2011;7:e1002232. doi: 10.1371/journal.ppat.1002232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires R, Hartlieb B, Signor L, Schoehn G, Lata S, Roessle M, Moriscot C, Popov S, Hinz A, Jamin M, et al. A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure. 2009;17:843–856. doi: 10.1016/j.str.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Stein A, Jahn R, Fasshauer D. The Ca2+ affinity of synaptotagmin 1 is markedly increased by a specific interaction of its C2B domain with phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2009;284:25749–25760. doi: 10.1074/jbc.M109.042499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Remy I, Michnick SW. Application of protein-fragment complementation assays in cell biology. Biotechniques. 2007;42:137, 139, 141. doi: 10.2144/000112396. passim. [DOI] [PubMed] [Google Scholar]
- Schmidt MH, Hoeller D, Yu J, Furnari FB, Cavenee WK, Dikic I, Bogler O. Alix/AIP1 antagonizes epidermal growth factor receptor downregulation by the Cbl-SETA/CIN85 complex. Mol Cell Biol. 2004;24:8981–8993. doi: 10.1128/MCB.24.20.8981-8993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette P, Mu R, Dussupt V, Jiang J, Snyder G, Smith P, Xiao TS, Bouamr F. The Phe105 loop of Alix Bro1 domain plays a key role in HIV-1 release. Structure. 2011;19:1485–1495. doi: 10.1016/j.str.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Totrov M, Abagyan R. Flexible protein-ligand docking by global energy optimization in internal coordinates. Proteins. 1997;(Suppl 1):215–220. doi: 10.1002/(sici)1097-0134(1997)1+<215::aid-prot29>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- Totrov M, Abagyan R. Flexible ligand docking to multiple receptor conformations: a practical alternative. Current opinion in structural biology. 2008;18:178–184. doi: 10.1016/j.sbi.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami Y, Popov S, Gottlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol. 2007;81:6614–6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vito P, Pellegrini L, Guiet C, D’Adamio L. Cloning of AIP1, a novel protein that associates with the apoptosis-linked gene ALG-2 in a Ca2+-dependent reaction. J Biol Chem. 1999;274:1533–1540. doi: 10.1074/jbc.274.3.1533. [DOI] [PubMed] [Google Scholar]
- von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat Struct Mol Biol. 2008;15:43–49. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. Structure of the Bro1 domain protein BROX and functional analyses of the ALIX Bro1 domain in HIV-1 budding. PLoS One. 2011;6:e27466. doi: 10.1371/journal.pone.0027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.