Abstract
Objective:
The PTEN hamartoma tumor syndromes (PHTS) are a collection of disorders caused by germline mutations of the tumor suppressor gene PTEN. Eosinophilic gastrointestinal disorders (EGID) are rare diseases characterized by food-induced, eosinophil-dominant inflammation in various segments of the gastrointestinal tract. On the basis of our clinical observations of several patients with EGID-PHTS, we investigated whether there is an association between these two disorders.
Methods:
Cincinnati Children’s Hospital Medical Center (CCHMC) Informatics for Integrating Biology & the Bedside (i2b2) warehouse was queried for the years 2007-2012 using ICD 9 codes for PTEN-related diseases; the results were cross-referenced with participants enrolled in the Cincinnati Center for Eosinophilic Disorder’s EGID database to identify patients with both disorders. Similarly, the Cleveland Clinic Genomic Medicine Institute PTEN database was queried for cases between 2005 and 2012. Inclusion criteria were age ≥18 years, history of PHTS, and an esophagogastroduodenoscopy (EGD) and/or colonoscopy with at least one histologic EGID diagnosis confirmed by a CCHMC pathologist. Pearson’s Chi-square was used to determine the odds of EGID enrichment in PHTS.
Results:
Of the 1,058,260 CCHMC distinct patients identified by the i2b2 search, 53 had clinical diagnoses suggestive of PHTS. Thirteen of the 53 had PTEN mutations, with 8/13 (62%) having had an EGD and/or colonoscopy. Five of the 8 had confirmed EGID. At the Cleveland Clinic, 3/75 patients with PHTS had confirmed EGID. CCHMC i2b2 query data showed a substantial enrichment of EGID in PHTS (OR=272; CI 89-831, p<0.0001). An EGID prevalence estimate from the i2b2 query supported a marked enrichment of EGID in PHTS in the Cleveland Clinic database (p<0.0001). Among the 8 subjects with EGID and PHTS, the age at EGID and PHTS diagnosis was 7.6 ± 3.2 and 7.9 ± 5.8 years, respectively. Patients with EGID-PHTS had excess eosinophils in biopsies of the esophagus (75%), stomach (38%), and colon (13%) with a notable presence of eosinophil-rich gastrointestinal polyposis (88%).
Conclusion:
EGID is a previously unrecognized comorbid disease in pediatric patients with PHTS. These data suggest a potential role of PTEN in contributing to EGID susceptibility.
INTRODUCTION
The PTEN hamartoma tumor syndromes (PHTS) are a collection of syndromes characterized by hamartomatous lesions most commonly found in the skin and the gastrointestinal tract and by a risk of malignancy caused by the presence of germline mutations of the tumor suppressor gene PTEN(1-4). PTEN, located on 10q22-23(5), encodes for a non-redundant, plasma-membrane lipid phosphatase that negatively regulates the PI3K/AKT/mTOR signaling pathway(2,6). Cowden syndrome (CS), a prototypic PHTS, is an autosomal dominant disorder with an approximate 80% penetrance(7,8) and estimated prevalence of 1 in 200,000(9). This prevalence is likely an underestimate due to the phenotypic variability and the under-recognition of its hallmark features(1,10). Specific haplotypes and rare alleles underlying PHTS disease etiology constitute low-penetrance, modifying loci and may harbor pathogenic variants that have escaped detection by standard PTEN mutation-scanning methodologies(11).
Loss-of-function mutations in PTEN prevent the protein from regulating cell proliferation effectively, leading to uncontrolled cell division and the formation of hamartomas and cancerous tumors, especially in the breast, thyroid, endometrium, and gastrointestinal tract(3,6,12). Functional analysis of PHTS-associated PTEN mutations found in tumors and in the germline revealed distinct functional patterns ranging from partial to complete loss, which could be relevant for therapy(13).
Eosinophilic gastrointestinal disorders (EGID) are rare disorders characterized by eosinophil-dominant inflammation in various segments of the gastrointestinal tract and are almost always induced by immune sensitization to multiple foods(14). Many affected patients have an atopic disease history. EGID include eosinophilic esophagitis (EoE), eosinophilic gastritis (EG), eosinophilic gastroenteritis (EGE), eosinophilic enteritis, and eosinophilic colitis (EC). The most common type of EGID, EoE, occurs in approximately 1 in 4,000 children(15). In contrast to patients who have PTEN mutations(16), gastrointestinal tract polyps are rarely reported in patients who have EGID(17,18). It is notable that PTEN has been reported to regulate chemotactic responses of leukocytes by its effect on the Rho GTPase/Rac pathway, as this signaling is key for eosinophil chemotactic and activation responses(19).
The Cincinnati Center for Eosinophilic Disorders (CCED) at the Cincinnati Children’s Hospital Medical Center (CCHMC) serves local patients and is also a referral center for affected pediatric patients who reside elsewhere in the United States and in other countries. The CCED has maintained a database approved by our Institutional Review Board for approximately a decade, and the database includes 1,673 patients who have an EGID diagnosis. In the course of our routine clinical work at the CCED, we identified several pediatric patients who had gastrointestinal polyps and excess eosinophils in the polyps and other parts of the gastrointestinal tract. Several of the patients had clinical manifestations of CS, and genetic testing confirmed PTEN abnormalities in those patients. Additionally, a recent publication from the Cleveland Clinic Genomic Medicine Institute reported that PTEN dysfunction in humans also results in autoimmunity and marked lymphoid hyperplasia, as well as severe and early onset of tonsil and thymus hypertrophy(3). These observations led us to query whether those patients with PHTS also had EGID and to explore a potential association between the two disorders. Herein, we report an enriched association of EGID in pediatric patients with PHTS.
METHODS
Study Design
This study used retrospective data from two centers, CCHMC and the Cleveland Clinic Genomic Medicine Institute. This study was approved by both Institutional Review Boards (IRB).
Participants
Participants were eligible for this study if they were aged ≥18 years at the time of EGID or PHTS diagnosis, had PHTS with a documented germline PTEN mutation, and the histologic results from an esophagogastroduodenoscopy (EGD) and/or colonoscopy met EGID diagnostic criteria. EGID diagnoses were confirmed histologically by a CCHMC pathologist (MHC) and were defined as: EoE, esophageal eosinophilia with ≥15 eosinophils (eos) per high-power field (hpf)(20,21); EG, gastric mucosal eosinophilia with ≥30 eos/hpf in at least 5 hpfs(7); ED/EJ, eosinophilic duodenitis/jejunitis with ≥26 eos/hpf(22); EI, eosinophilic ileitis with ≥28 eos/hpf; and EC, colonic mucosal eosinophilia with ≥50 eos/hpf(22). Slides obtained at the first diagnostic endoscopy performed at CCHMC were reviewed for the CCHMC subjects. If prior endoscopies had been performed elsewhere, the slides were not requested for re-review, but the pathology reports were reviewed. Slides of biopsies reported to show excess eosinophils were supplied by the Cleveland Clinic Genomic Medicine Institute and reviewed to confirm the EGID diagnosis.
CCHMC Search Strategies
Utilizing CCHMC’s Informatics for Integrating Biology & the Bedside (i2b2) research data warehouse(23), cohort identification of patients with PTEN-related diagnoses was ascertained. From November 2007 to December 2012, patients with the following PTEN-related syndromes and diseases were identified based on the following ICD9 diagnosis codes: Cowden disease (759.6DM); Cowden syndrome (759.6C); Cowden’s disease (759.6DN); Bannayan-Riley-Ruvalcaba syndrome (759.89ANQ); Hamartoma syndrome, multiple (759.6EC); Hamartoma (759.6B); Hamartomatous disease or syndrome (759.6ED); and antrum polyps (471.8). Patients identified by the i2b2 query were cross-referenced with patients who had consented to have their clinical data included in the CCED IRB-approved EGID database. All PTEN genetic test results from the CCED patients were reviewed by the Cleveland Clinic Genomic Medical Institute (CE and JN) to confirm disease-causing PTEN germline mutation. Research-based PTEN analysis was performed by the Cleveland Clinic Genomic Medicine Institute and confirmed in clinical and commercial genetic testing laboratories (CLIA or CAP certified) including the CCHMC Cytogenetics Laboratory, The Ohio State University Molecular Pathology Laboratory, GeneDx, and the Ambry Genetics Laboratory.
Cleveland Clinic Genomic Medicine Institute Search Strategies
The IRB-approved PTEN database developed by the Cleveland Clinic Genomic Medicine Institute was searched for the time interval between October 2005 and December 2012. Patients with germline, deleterious PTEN mutations and a reported EGD and/or colonoscopy and were aged ≥18 years were identified. De-identified clinical data and available gastrointestinal biopsy slides were forwarded from the Cleveland Clinic to CCHMC.
Participant Phenotypes and Gastrointestinal Biopsy Findings
Demographic and phenotypic characteristics of participants including atopy and food allergies, as well as participant clinical history and histologic information from gastrointestinal biopsies, were summarized using descriptive statistics.
EGID Enrichment Analyses in Patients with PHTS
To determine whether EGID is enriched in patients with PHTS, we used two statistical strategies. First, using CCHMC’s i2b2 query results and a 2 × 2 table, the odds (95% CI, p-value) of EGID enrichment in patients with PHTS was calculated. In this analysis, significance was determined using Pearson’s Chi Square. Second, using Cleveland Clinic data, a binomial distribution was calculated based on population prevalence estimates for EGID and EoE in order to determine the probability of EGID enrichment in PHTS. To date, the prevalence of EGID has not been reported; therefore, its estimated prevalence, 1 in 435, was calculated using the observed number of patients with EGID in the CCHMC i2b2 query (2,435 patients with EGID in 1,058,260 patient records) and the EoE prevalence of 1 in 4,000(15). The odds ratio of EGID given PHTS was not able to be determined. The Cleveland Clinic Genomic Medicine Institute PTEN database contains individuals who meet clinical or relaxed clinical criteria for CS, and we selected only those patients with pathogenic PTEN mutations. Having an EGD and/or colonoscopy was not a database eligibility requirement.
Statistical analyses were performed using PASW Statistics 18.0 software (SPSS, Inc., Chicago, IL).
RESULTS
Enrollment
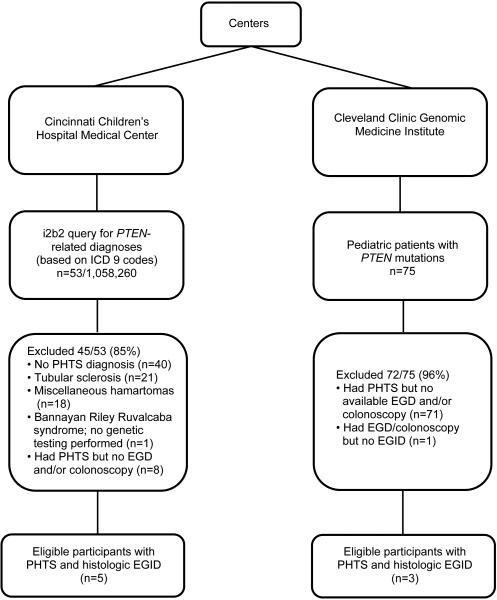
Flow diagrams showing patient selection for this study for each center are shown in Figure 1. A total of 1,058,335 patients from CCHMC (n=1,058,260) and the Cleveland Clinic Genomic Medicine Institute (n=75) were assessed for eligibility, and 8 patients met inclusion criteria (n=4 from CCHMC; n=3 from Cleveland Clinic Genomic Medicine Institute; and n=1 patient that was identified at both centers — the latter patient received medical treatment at CCHMC; however, PTEN gene test was performed at the Cleveland Clinic Genomic Medicine Institute). Inclusion criteria included the presence of PHTS, results of EGD and/or colonoscopy, age ≥18 years, and the presence of an EGID diagnosis.
Figure 1.
Flow diagram of patient selection for current study from each center
Search Results
At CCHMC, 53 of the 1,058,260 unique patient records were identified based on the PTEN-related i2b2 query. Approximately 25% (13/53) of these patients had DNA analysis to confirm PHTS (germline PTEN mutation). Of those with confirmed PHTS, 62% (8/13) had an EGD and/or colonoscopy. Of these 8 patients, 63% (5/8) had EGID. Of the 75 patients from the Cleveland Clinic who were aged ≥18 years, 5% (n=4) had an EGD and/or colonoscopy. Of these 4 patients, 75% (n=3) had a histologic EGID diagnosis. The fourth patient had an endoscopy for diarrhea; however, there was no histologic evidence of EGID on the biopsy slides that were available.
Clinical Features of EGID-PHTS Patients
Demographic and phenotypic characteristics of participants are shown in Table 1. Of the 8 enrolled patients with EGID-PHTS, 63% (n=5) were male and 88% (n=7) were white. The age at EGID and PHTS diagnosis (mean ± SD) was 7.6 ± 3.2 and 7.9 ± 5.8 years, respectively. There was no correlation between age of EGID and PHTS diagnosis (p>0.3). EGID included 75% EoE (n=6), with 2 of the 6 patients having EG in addition to EoE, 38% (n=3) EG, and 13% (n=1) EC. Atopy was identified in 63% (n=5) of patients; specifically 38% (n=3) had asthma, 25% (n=2) had allergic rhinitis, and 25% (n=2) had eczema. Food allergies were reported in 38% (n=3) patients. Two CCHMC patients had both skin prick testing (SPT) and atopy patch testing (APT) performed. Each patient responded positively to two or more foods on SPT and neither experienced positive reactions on APT.
Table 1.
Clinical features of EGID-PHTS patients
| Patient | Center* | Gender† | Ethnicity‡ | Age EGID Dx, y |
Age PHTS Dx, y |
PTEN
Germline Mutation |
EGID Phenotype | Asthma | Allergic Rhinitis |
Eczema | Food Allergies |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EoE | EG | EC | |||||||||||
| 1 | CH | M | W | 9.5 | 9.8 | c.723delT | X§ | X | X | ||||
| 2 | CH | F | W | 2.7 | 2.4 | c.278A>G | X | X | X | ||||
| 3 | CH | F | W | 6.5 | 18.7 | c.492+2delT | X§ | X | X | X | |||
| 4 | CH | M | W | 8.6 | 1.1 | 46,XY, d10 (q23.1q23.3) |
X | X | |||||
| 5 | CH/ CL |
M | W | 6.0 | 5.7 | c491delA | X | X | X | ||||
| 6 | CL | F | AA | 10.3 | 11.0 | c.202T>A | X§ | ||||||
| 7 | CL | M | W | 4.6 | 4.0 | c.388C>T | X | ||||||
| 8 | CL | M | W | 12.4 | 11.0 | c.26delT | X | X | X | ||||
Centers - CH=Cincinnati Children’s Hospital Medical Center, CL=Cleveland Clinic Genomic Medicine Institute;
Gender – M=male, F=female;
Ethnicity - W=white, AA=African American; Dx=diagnosis; EGID=Eosinophilic gastrointestinal disorders; EoE=Eosinophilic esophagitis defined as ≥15 eosinophils/high-power field; EG=Eosinophilic gastritis defined as ≥30 eosinophils/high-power field in at least 5 high-power fields; EC=Eosinophilic colitis defined as ≥50 eosinophils/high-power field;
EoE not PPI confirmed
Gastrointestinal Biopsy Findings
Table 2 outlines participant clinical history and gastrointestinal biopsy histologic findings. All patients had an EGD, and 75% (n=6) had colonoscopies. The peak esophageal eosinophil count (mean ± SD) for patients diagnosed with EoE was 53 ± 26 /hpf (range, 21-81 /hpf). Gastrointestinal polyps were detected in 88% (n=7) of patients, and 86% (n=6) had polyps in more than one gastrointestinal site (Figure 2). Polyps were esophageal in 14% (n=1); gastric in 86% (n=6); duodenal in 57% (n=4); colonic in 57% (n=4); jejunal in 29% (n=2); and rectal in 29% (n=2).
Table 2.
Clinical history and gastrointestinal biopsy histologic findings
| Patient | Age EGID Dx, y |
GI biopsy Indication |
EGID Phenotypes |
GI sites reviewed |
Esophagus | Stomach Peak Eos/hpf |
Colon Peak Eos/hpf |
GI Polyps | |
|---|---|---|---|---|---|---|---|---|---|
| Peak Distal Eos/hpf |
Peak Proximal Eos/hpf |
||||||||
| 1 | 9.5 | Constipation, Rectal bleeding |
EoE | Esophagus Stomach Duodenum Colon Rectum |
59* | 3 | NDA | NDA | Gastric Colonic |
| 2 | 2.7 | Diarrhea | EC | Esophagus Stomach Duodenum Colon Rectum** |
4 | 2 | NDA | A† 104 T/D‡ 81 |
Colonic |
| 3 | 6.5 | Rule out Crohn’s disease |
EoE, EG | Esophagus Stomach Duodenum |
53* | N/D | 108 | NDA | Gastric Duodenal |
| 4 | 8.6 | Cowden syndrome |
EoE | Esophagus Stomach Duodenum Colon Rectum |
81 | N/D | NDA | NDA | Gastric Duodenal Colonic Rectal |
| 5 | 6.0 | Cowden syndrome |
EG | Esophagus Stomach Duodenum Jejunem |
NDA | 90 | NDA | Esophageal Gastric Duodenal Jejunal |
|
| 6 | 10.3 | Rectal bleeding |
EoE | Esophagus Stomach Duodenum Colon |
26*§ | N/A | NDA | Gastric Duodenal Colonic |
|
| 7 | 4.6 | Diarrhea | EoE | Esophagus Stomach Duodenum Colon |
78§ | NDA | NDA | --- | |
| 8 | 12.4 | Abdominal pain, diarrhea |
EoE, EG | Esophagus Stomach Duodenum Jejunum Colon Rectum |
21§ | 55 | NDA | Gastric Jejunal Rectal |
|
Dx=Diagnosis; EGID=Eosinophilic gastrointestinal disorders; GI=Gastrointestinal; Eos/hpf=Eosinophils per high-power field; EoE=Eosinophilic esophagitis; EG=Eosinophilic gastritis; EC=Eosinophilic colitis;
EoE not PPI confirmed;
Rectosigmoid slide not available; NDA=no diagnostic abnormality;
A=Ascending colon;
T/D=Transverse descending colon; N/D=not determined;
Proximal esophageal eosinophil count not determined; N/A=data not available
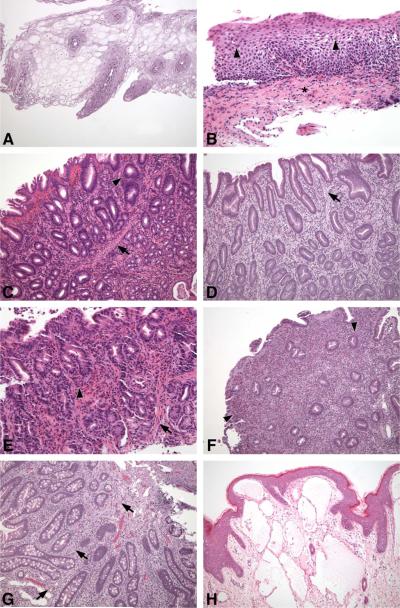
Figure 2.
A, Endoscopy of subject 5 at age 5 years identified esophageal polyps that consisted of esophageal squamous epithelial cells containing large amounts of glycogen imparting a clear appearance to the cytoplasm. This condition is known as glycogen acanthosis and is a typical esophageal lesion of patients who have PTEN mutations; Hematoxylin and eosin (H&E) stain, 100x magnification. B, Initial endoscopy of subject 2 at age 5 years included esophageal biopsies that demonstrated numerous intraepithelial eosinophils (arrowheads), basal layer expansion, dilated intercellular spaces, and lamina propria fibrosis (asterisk), all features typically found in EoE; H&E stain, 200x magnification. C, At the same endoscopy of subject 2, gastric polyps showed marked eosinophilic inflammation including intraepithelial eosinophils in gland epithelium (arrowhead) and smooth muscle bundles in the lamina propria (arrow), features found in inflammatory and hamartomatous polyps, respectively; H&E stain, 100x magnification. D, Biopsy of gastric antral polyps from subject 5 showed expansion of the lamina propria, mostly by smooth muscle (arrow), and also numerous eosinophils; H&E stain, 100x magnification. E, Biopsy of non-polypoid gastric mucosa of subject 2 also showed marked eosinophilic inflammation (arrowhead) and smooth muscle hyperplasia in the lamina propria (arrow); H&E stain, 200x magnification. F, A duodenal polyp from subject 5 shows sheets of eosinophils in the lamina propria and reduced numbers of crypts; H&E stain, 100x magnification. G, A jejunal polyp from subject 5 includes numerous ganglion cells in the lamina propria (arrows), which is typical of gastrointestinal polyps in individuals with CS; H&E stain, 100x magnification. H, At age 2.5 years, a soft tissue lymphangioma was removed from subject 2. Soft tissue tumors are not uncommon in patients with PHTS and often have myriad histological findings. The photomicrographs are representative of the biopsies from the CCHMC patients for whom complete slide sets and medical records were available.
Esophageal polyps consisted of esophageal squamous epithelial cells containing large amounts of glycogen imparting a clear appearance to the cytoplasm (Figure 2A), representing glycogenic acanthosis, which is typically seen in the esophageal lesions of patients who have PTEN mutations(24). Esophageal biopsies contained other features that are characteristic of EoE, including numerous intraepithelial eosinophils, basal layer expansion, dilated intercellular spaces, and lamina propria fibrosis (Figure 2B). When found, gastric polyps showed marked eosinophilic inflammation including intraepithelial eosinophils in gland epithelium and smooth muscle bundles in the lamina propria, features typically found in inflammatory and hamartomatous polyps, respectively(16) (Figure 2C). Gastric antral polyps showed that the lamina propria was expanded mostly by smooth muscle (Figure 2D). Non-polypoid gastric mucosa showed marked eosinophilic inflammation and smooth muscle hyperplasia in the lamina propria (Figure 2E). Duodenal polyps showed sheets of eosinophils in the lamina propria and reduced numbers of crypts (Figure 2F).
EGID Enrichment in Patients with PHTS
Using CCHMC-based i2b2 data, we identified 13 patients with PTEN germline mutations (Table 2). Of those, 38% (n=5) also had confirmed EGID, 38% (n=5) had uncertain EGID status due to lack of endoscopy, and 23% (n=3) did not have EGID. The finding of 38% EGID-PHTS of patients with PTEN mutations is in contrast to the only 0.23% EGID of patients without PTEN mutations. Thus, there was a significant enrichment of EGID in patients with PHTS (OR = 272; CI 89-831, p< 0.0001). Using the Cleveland Clinic Genomic Medicine Institute data, 3 of 75 patients with PHTS had an EGID. The probability of 3/75 patients with PHTS having EGID was 0.0005 based on a prevalence estimate that used data from a CCHMC i2b2 query that identified 2,435 in 1,058,260 patients with an EGID, or 1 in 435 individuals. Further, assuming that EGID occur at a rate of 1 in 4,000, as with EoE(22), the probability of 3 individuals of 75 being affected is less than 0.0001.
DISCUSSION
Herein, we demonstrate a highly significant enrichment of EGID in pediatric patients with PHTS using two independent cohorts. This unique clinical observation was based on CCHMC’s i2b2 data warehouse repository, which is comprised of greater than one million unique patient records, and the PTEN database at the Cleveland Clinic Genomic Medicine Institute. Our study is limited by its retrospective nature, resulting in potential ascertainment and reporting biases.
PTEN is the principal and non-redundant negative regulator of the PI3K signaling pathway, and nearly all organisms are highly sensitive to PTEN dosage(25), which may be mirrored in the human syndrome(26). There are multiple mechanisms for the regulation of PTEN, including transcription, mRNA stability, miRNA targeting, translation, and protein stability(2). One possible robust mechanism that precisely coordinates PTEN expression with the transcriptional programs that promote lineage specification and differentiation in the immune system is regulation by microRNAs (miRNAs), which have the capacity to regulate the PI3K-PTEN axis(27-29). Specifically, miR-181 was identified as an essential regulator of PI3K signaling strength through PTEN modulation(27). In glioma, lung, and prostate cancers that are prevalent in PHTS, PTEN expression is decreased by overexpression of miR-21, miR-25a, miR-22, or the miR106b-25 cluster(29-31). Interestingly, comprehensive analysis of global miRNA expression in the esophageal tissues of patients with EoE has identified 21 upregulated and 11 downregulated miRNAs in patients with active EoE(32). miR-21 is one of the most upregulated miRNAs in EoE(32) and notably causes reduced PTEN expression, perhaps explaining the occurrence of epithelial hyperplasia, a hallmark histologic characteristic seen in gastrointestinal biopsies of patients with EGID with and without PHTS. Of relevance, miR-21 is overexpressed in individuals with CS with germline PTEN mutations compared to those without germline PTEN mutations(33). Our results highlight the potentially important role of PTEN in the pathogenesis of EoE and related EGID and raise the possibility that targeting PTEN activity or its downstream mediators, such as AKT and/or mammalian target of rapamycin (mTOR) with drugs such as rapamycin and its derivatives(34,35), may have efficacy in EGID.
There exist a number of intrinsic risk variants in proinflammatory and epithelial cell genes associated with the most commonly occurring EGID, EoE(36). Microarray analysis of patients’ esophageal biopsy specimens has defined an EoE transcriptome with the identification of CCL26 (eotaxin-3) as the most highly induced gene in the transcriptome(36). The EoE risk variants, CCL26, TGFB1, TSLP, and FLG, provide insight into upstream mechanisms that regulate the expression of genes that direct multiple aspects of EoE pathogenesis(36). Notably, transduction of PTEN into eosinophils attenuates cellular survival and chemotaxis, demonstrating the potential direct effects of PTEN on eosinophils(37). Furthermore, PTEN and TGF-β have already been shown to cooperate together(38,39), adding further plausibility for a unified mechanism, especially since TGF-β has been shown to be involved in EoE pathogenesis(40,41).
A clinical feature of both CS and EGID is the occurrence of gastrointestinal polyps. Gastrointestinal polyps in patients who have PTEN mutations are heterogeneous and include inflammatory polyps(16). However, inflammatory polyps are among the least common types of gastrointestinal polyps in patients with CS and occur with polyps of other types(16). In contrast, gastrointestinal polyps in EGID are eosinophil rich and may be classified as pseudopolyps; they lack hamartomatous features, such as mucosal ganglion cells, that are characteristic of some of the polyps in CS. For patients with each disorder, a potentially helpful clinical sign is that the pseudopolyps in EGID may diminish or even regress following appropriate EGID therapy(18), whereas non-inflammatory polyps in CS should not change with EGID therapy.
Herein, we report a novel association between germline PTEN mutations that cause PHTS and susceptibility to EGID. Whereas PHTS is autosomal dominant, EGID is a complex trait with 4-9% prevalence in PTEN mutation–positive pediatric patients with PHTS. This observed enrichment of EGID in PHTS compared to in the general population deserves further prospective data collection. Pathologists and clinicians should be aware of the possible presence of EGID in patients with PHTS, and conversely PHTS in patients with EGID, particularly when gastrointestinal polyps are identified. We observed the frequent occurrence of gastrointestinal polyps in patients who have either disorder and determined that polyp pathology may lead to clinical investigations that identify a comorbid disease affecting therapy and surveillance.
These results highlight the potentially important role of PTEN in the pathogenesis of EoE and related EGID and raise the possibility that targeting PTEN activity and downstream mediators (e.g. with rapamycin) may be efficacious in EGID. Further research may elucidate pathways common to both disorders and may foster the development of further treatment modalities.
Acknowledgements
This work is funded in part by PHS grants DK078392, AI083450, AI045898, DK076893, and CA124570 (to M.E.R. and C.E.), the Buckeye Foundation, the Food Allergy Research & Education Foundation, the Campaign Urging Research for Eosinophilic Disease (http://www.curedfoundation.org) Foundation, the National Institutes of Health UL1 TR000077 (to K.M.), and the Breast Cancer Research Foundation (to C.E.). J.N. is an Ambrose Monell Foundation Cancer Genomic Medicine Clinical Fellow and was partially funded by SingHealth and NMRC (Singapore) Fellowships. C.E. is the Sondra J. and Stephen R. Hardis Endowed Chair of Cancer Genomic Medicine at the Cleveland Clinic Genomic Medicine Institute and is an American Cancer Society Clinical Research Professor. We thank Dr. John J. Bissler, MD, Clark D. West Chair of Nephrology, CCHMC for his insights and manuscript review and Shawna Hottinger for editorial assistance. We also thank members and patients of the CCED (www.cchmc.org/cced) for their participation.
Abbreviations used:
- APT
Atopy patch testing
- CCED
Cincinnati Center for Eosinophilic Disorders
- CCHMC
Cincinnati Children’s Hospital Medical Center
- CS
Cowden syndrome
- EC
Eosinophilic colitis
- ED
Eosinophilic duodenitis
- EGD
Esophagogastroduodenoscopy
- EG
Eosinophilic gastritis
- EGE
Eosinophilic gastroenteritis
- EGID
Eosinophilic gastrointestinal disorder
- EI
Eosinophilic ileitis
- EJ
Eosinophilic jejunitis
- EoE
Eosinophilic esophagitis
- H&E
Hematoxylin & eosin
- hpf
High-power field
- i2b2
Informatics for Integrating Biology & the Bedside
- miRNA
MicroRNA
- PHTS
PTEN hamartoma tumor syndromes
- PTEN
Phosphatase and tensin homolog
- SPT
Skin prick testing
BIBLIOGRAPHY
- 1.Pilarski R, Eng C. Will the real Cowden syndrome please stand up (again)? Expanding mutational and clinical spectra of the PTEN hamartoma tumour syndrome. J Med Genet. 2004;41:323–6. doi: 10.1136/jmg.2004.018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heindl M, Handel N, Ngeow J, et al. Autoimmunity, intestinal lymphoid hyperplasia, and defects in mucosal B-cell homeostasis in patients with PTEN hamartoma tumor syndrome. Gastroenterology. 2012;142:1093–1096. doi: 10.1053/j.gastro.2012.01.011. e6. [DOI] [PubMed] [Google Scholar]
- 4.Zbuk KM, Eng C. Cancer phenomics: RET and PTEN as illustrative models. Nat Rev Cancer. 2007;7:35–45. doi: 10.1038/nrc2037. [DOI] [PubMed] [Google Scholar]
- 5.Nelen MR, Padberg GW, Peeters EA, et al. Localization of the gene for Cowden disease to chromosome 10q22-23. Nat Genet. 1996;13:114–6. doi: 10.1038/ng0596-114. [DOI] [PubMed] [Google Scholar]
- 6.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–14. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 7.Lwin T, Melton SD, Genta RM. Eosinophilic gastritis: histopathological characterization and quantification of the normal gastric eosinophil content. Mod Pathol. 2011;24:556–63. doi: 10.1038/modpathol.2010.221. [DOI] [PubMed] [Google Scholar]
- 8.Marsh DJ, Coulon V, Lunetta KL, et al. Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum Mol Genet. 1998;7:507–15. doi: 10.1093/hmg/7.3.507. [DOI] [PubMed] [Google Scholar]
- 9.Nelen MR, Kremer H, Konings IB, et al. Novel PTEN mutations in patients with Cowden disease: absence of clear genotype-phenotype correlations. Eur J Hum Genet. 1999;7:267–73. doi: 10.1038/sj.ejhg.5200289. [DOI] [PubMed] [Google Scholar]
- 10.Eng C. Will the real Cowden syndrome please stand up: revised diagnostic criteria. J Med Genet. 2000;37:828–30. doi: 10.1136/jmg.37.11.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pezzolesi MG, Li Y, Zhou XP, et al. Mutation-positive and mutation-negative patients with Cowden and Bannayan-Riley-Ruvalcaba syndromes associated with distinct 10q haplotypes. Am J Hum Genet. 2006;79:923–34. doi: 10.1086/508943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan MH, Mester JL, Ngeow J, et al. Lifetime cancer risks in individuals with germline PTEN mutations. Clin Cancer Res. 2012;18:400–7. doi: 10.1158/1078-0432.CCR-11-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez-Escudero I, Oliver MD, Andres-Pons A, et al. A comprehensive functional analysis of PTEN mutations: implications in tumor- and autism-related syndromes. Hum Mol Genet. 2011;20:4132–42. doi: 10.1093/hmg/ddr337. [DOI] [PubMed] [Google Scholar]
- 14.Rothenberg ME. Eosinophilic gastrointestinal disorders (EGID) J Allergy Clin Immunol. 2004;113:11–28. doi: 10.1016/j.jaci.2003.10.047. quiz 29. [DOI] [PubMed] [Google Scholar]
- 15.Noel RJ, Putnam PE, Rothenberg ME. Eosinophilic esophagitis. N Engl J Med. 2004;351:940–1. doi: 10.1056/NEJM200408263510924. [DOI] [PubMed] [Google Scholar]
- 16.Heald B, Mester J, Rybicki L, et al. Frequent gastrointestinal polyps and colorectal adenocarcinomas in a prospective series of PTEN mutation carriers. Gastroenterology. 2010;139:1927–33. doi: 10.1053/j.gastro.2010.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jimenez-Rivera C, Ngan B, Jackson R, et al. Gastric pseudopolyps in eosinophilic gastroenteritis. J Pediatr Gastroenterol Nutr. 2005;40:83–6. doi: 10.1097/00005176-200501000-00015. [DOI] [PubMed] [Google Scholar]
- 18.Chehade M, Sicherer SH, Magid MS, et al. Multiple exudative ulcers and pseudopolyps in allergic eosinophilic gastroenteritis that responded to dietary therapy. J Pediatr Gastroenterol Nutr. 2007;45:354–7. doi: 10.1097/MPG.0b013e31803219d5. [DOI] [PubMed] [Google Scholar]
- 19.Li Z, Dong X, Wang Z, et al. Regulation of PTEN by Rho small GTPases. Nat Cell Biol. 2005;7:399–404. doi: 10.1038/ncb1236. [DOI] [PubMed] [Google Scholar]
- 20.Liacouras CA, Furuta GT, Hirano I, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20. doi: 10.1016/j.jaci.2011.02.040. e6; quiz 21-2. [DOI] [PubMed] [Google Scholar]
- 21.Dellon ES, Gonsalves N, Hirano I, et al. ACG Clinical Guideline: Evidenced Based Approach to the Diagnosis and Management of Esophageal Eosinophilia and Eosinophilic Esophagitis (EoE) Am J Gastroenterol. 2013;108:679–92. doi: 10.1038/ajg.2013.71. [DOI] [PubMed] [Google Scholar]
- 22.DeBrosse CW, Case JW, Putnam PE, et al. Quantity and distribution of eosinophils in the gastrointestinal tract of children. Pediatr Dev Pathol. 2006;9:210–8. doi: 10.2350/11-05-0130.1. [DOI] [PubMed] [Google Scholar]
- 23.Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010;17:124–30. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGarrity TJ, Wagner Baker MJ, Ruggiero FM, et al. GI polyposis and glycogenic acanthosis of the esophagus associated with PTEN mutation positive Cowden syndrome in the absence of cutaneous manifestations. Am J Gastroenterol. 2003;98:1429–34. doi: 10.1111/j.1572-0241.2003.07496.x. [DOI] [PubMed] [Google Scholar]
- 25.Alimonti A, Carracedo A, Clohessy JG, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42:454–8. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan MH, Mester J, Peterson C, et al. A clinical scoring system for selection of patients for PTEN mutation testing is proposed on the basis of a prospective study of 3042 probands. Am J Hum Genet. 2011;88:42–56. doi: 10.1016/j.ajhg.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henao-Mejia J, Williams A, Goff LA, et al. The MicroRNA miR-181 Is a Critical Cellular Metabolic Rheostat Essential for NKT Cell Ontogenesis and Lymphocyte Development and Homeostasis. Immunity. 2013;38:984–97. doi: 10.1016/j.immuni.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–24. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huse JT, Brennan C, Hambardzumyan D, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–37. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang JG, Wang JJ, Zhao F, et al. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clin Chim Acta. 2010;411:846–52. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 31.Poliseno L, Salmena L, Riccardi L, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3 doi: 10.1126/scisignal.2000594. ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu TX, Sherrill JD, Wen T, et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J Allergy Clin Immunol. 2012;129:1064–75. doi: 10.1016/j.jaci.2012.01.060. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pezzolesi MG, Platzer P, Waite KA, et al. Differential expression of PTEN-targeting microRNAs miR-19a and miR-21 in Cowden syndrome. Am J Hum Genet. 2008;82:1141–9. doi: 10.1016/j.ajhg.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das F, Ghosh-Choudhury N, Dey N, et al. Unrestrained mammalian target of rapamycin complexes 1 and 2 increase expression of phosphatase and tensin homolog deleted on chromosome 10 to regulate phosphorylation of Akt kinase. J Biol Chem. 2012;287:3808–22. doi: 10.1074/jbc.M111.246397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galoian K, Temple HT, Galoyan A. mTORC1 inhibition and ECM-cell adhesion-independent drug resistance via PI3K-AKT and PI3K-RAS-MAPK feedback loops. Tumour Biol. 2012;33:885–90. doi: 10.1007/s13277-011-0315-x. [DOI] [PubMed] [Google Scholar]
- 36.Sherrill JD, Rothenberg ME. Genetic dissection of eosinophilic esophagitis provides insight into disease pathogenesis and treatment strategies. J Allergy Clin Immunol. 2011;128:23–32. doi: 10.1016/j.jaci.2011.03.046. quiz 33-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adachi T, Hanaka S, Masuda T, et al. Transduction of phosphatase and tensin homolog deleted on chromosome 10 into eosinophils attenuates survival, chemotaxis, and airway inflammation. J Immunol. 2007;179:8105–11. doi: 10.4049/jimmunol.179.12.8105. [DOI] [PubMed] [Google Scholar]
- 38.Yu M, Trobridge P, Wang Y, et al. Inactivation of TGF-beta signaling and loss of PTEN cooperate to induce colon cancer in vivo. Oncogene. 2013 Apr;:22. doi: 10.1038/onc.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li DM, Sun H. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res. 1997;57:2124–9. [PubMed] [Google Scholar]
- 40.Abonia JP, Franciosi JP, Rothenberg ME. TGF-beta1: Mediator of a feedback loop in eosinophilic esophagitis--or should we really say mastocytic esophagitis? J Allergy Clin Immunol. 2010;126:1205–7. doi: 10.1016/j.jaci.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aceves SS, Chen D, Newbury RO, et al. Mast cells infiltrate the esophageal smooth muscle in patients with eosinophilic esophagitis, express TGF-beta1, and increase esophageal smooth muscle contraction. J Allergy Clin Immunol. 2010;126:1198–204. doi: 10.1016/j.jaci.2010.08.050. e4. [DOI] [PubMed] [Google Scholar]