Summary
Zebrafish has become a widely used model for analysis of gene function. Several methods have been used to create mutations in this organism and thousands of mutant lines are available. However, all the conventional zebrafish mutations affect the gene in all cells at all time, making it difficult to determine tissue-specific functions. We have adopted a FlEx Trap approach to generate conditional mutations in zebrafish by gene trap mutagenesis. Combined with appropriate Cre or Flp lines, the insertional mutants not only allow spatial and temporal specific gene inactivation, but also permit spatial and temporal specific rescue of the disrupted gene. We provide experimental details on how to generate and use such mutations.
Keywords: zebrafish, mutagenesis, gene-trap, Cre, Flp
1. Introduction
While high-throughput methods have been developed to analyze the genome and make predictions on gene function, the gold standard for determining the function of genes has been detailed analysis of genetic mutants. In many organisms, random mutagenesis followed by functional analysis has been a preferred approach. However, these mutations often result in embryonic lethality and later functions or tissue specific functions are difficult if not impossible to assess. In contrast, engineered conditional alleles allow for stage or tissue specific functions to be investigated.
In the mouse, conditional alleles are most commonly generated by introducing two target sites of a site-specific recombinase, either Cre or Flp, in a configuration that will delete an essential portion or an entire gene (1; 2). In combination with tissue-specific expression of Cre and ligand-dependent Cre (e.g. CreER), (3; 4) gene function can be assessed in different tissues/organs and at different stages; from development to adults.
Zebrafish has proven to be a powerful vertebrate model for forward genetic analysis of vertebrate development. Several strategies have been developed to generate and identify mutations in a gene of interest after chemical mutagenesis or insertional mutagenesis (5–8). Furthermore, zinc finger nucleases (9–11) or TALE nucleases (12–15) can be used to generate targeted mutations. Although very useful in determining gene function during development, these approaches are limited since the mutation is present throughout the organism beginning at embryogenesis. Analysis of later or tissue specific processes can be complicated. Conditional mutations are more desirable because they allow inactivation of genes in somatic cells in a temporal- and/or tissue-specific manner.
1.1 Gene trap mutagenesis to generate conditional mutations
While gene targeting is not available for zebrafish because of the lack of ES cells, gene trapping using the Tol2 transposon system (16) is very efficient. Several transposon-based vectors have been developed for gene-trap mutagenesis in zebrafish (17; 18) including a “gene-breaking” transposon with removable gene-trap (19) and a “FlipTrap” transposon with invertible gene-trap (20).
Since the orientation of a gene trap determines the mutagenicity of the insertion, it is possible to make conditional alleles if the orientation of the gene trap can be stably switched (21; 22). For this utility, only intronic insertions are useful since exonic insertions are likely mutagenic regardless of the orientation of the cassette. To switch the orientation of the gene trap, a Flip and Excision (FlEx) (23; 24) approach is used that takes advantage of site-specific recombinases and strategically arranged heterodimers of heterotypic (incompatible) recombinase target sites. With a FlEx approach, incorporating targets sites for both Cre and Flp, the intervening sequence can be inverted in two successive rounds. Coupling the FlEx strategy to the efficient Tol2 transgenesis in zebrafish will allow conditional gene inactivation without the need for ES cells.
1.2 Vector design for efficient gene-trapping
A few features are desirable in an efficient conditional gene-trap vector. Importantly, the cassette needs to be highly mutagenic in one orientation and non-mutagenic in the other orientation to confer conditionality. The mutagenicity is determined by how efficiently the gene trap intercepts and prematurely terminates the transcription of the tagged genes. For this to be efficient, both a strong splice acceptor and a strong polyadenylation signal in the cassette would ensure a highly mutagenic gene trap. To achieve this, the consensus splice acceptor from zebrafish (25) was used along with tandem repeats of the well characterized bovine growth hormone gene polyadenylation and transcription termination sequence (Figure 1A) (26; 27). Incorporation of a FlEx module containing target sites for two recombinases allows two rounds of inversion, one round mediated by Flp and one round mediated by Cre (27; 28). After recombination, the remaining heterotypic recognition sites make the cassette resistant to further inversion by the same recombinase. Although not essential, fluorescent reporter genes can indicate the expression pattern of the tagged genes as well as visualization of the genotype of individual cells. However, only a fraction of insertions are in frame, and not all will lead to high enough levels for visual detection.
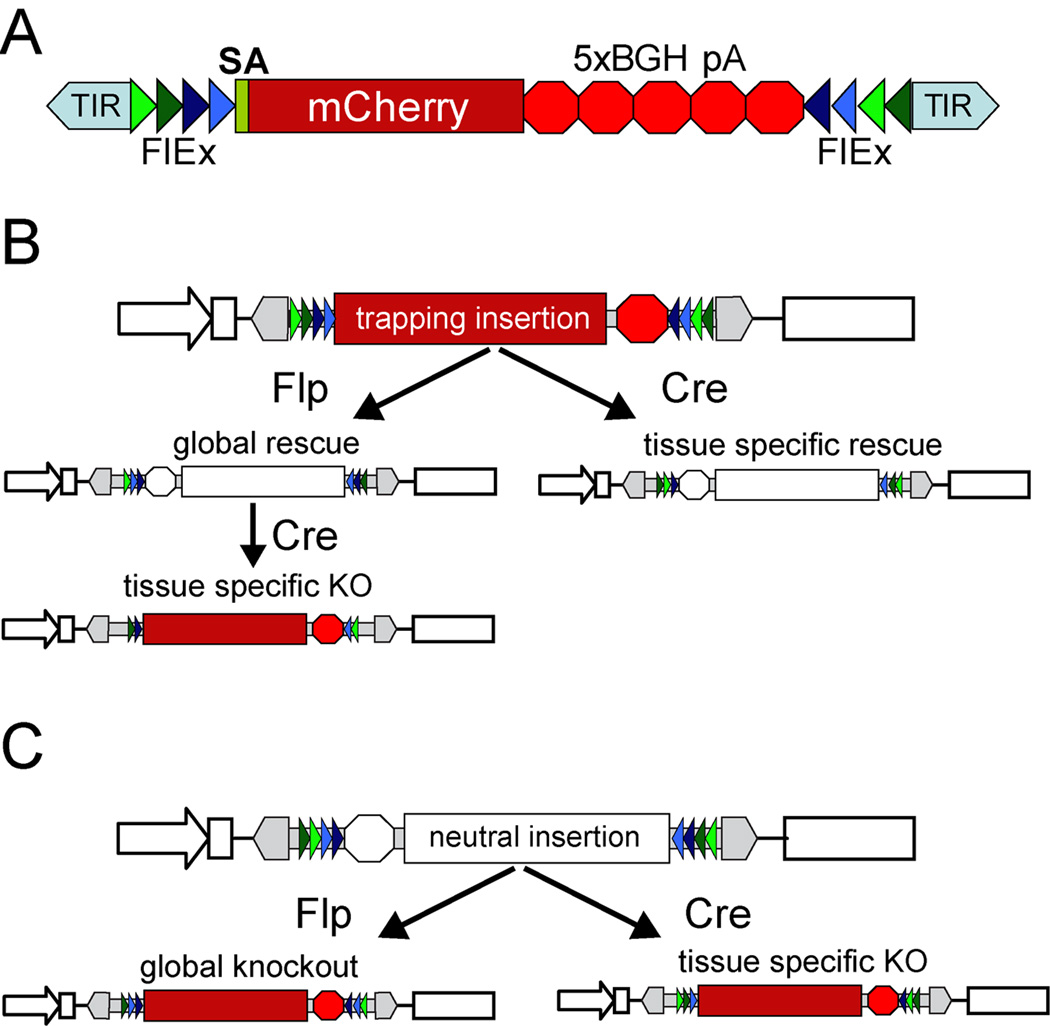
Figure 1. Schematic and utility of conditional gene-trap mutagen.
A- The gene trap mutagen contains an mCherry open reading frame (red rectangle) downstream of a strong splice acceptor (SA) and upstream of five repeats of the transcriptional stop and polyadenylation sequence derived from the BGH gene (red octagon). This mutagen is flanked by FlEx sites containing Flp recognition sites (green triangles) and Cre recognition sites (blue triangles) allowing for two rounds of stable inversion. The cassette is carried in a Tol2 transposon (grey arrow) to facilitate transgenesis. B,C – utility of the gene trap in either orientation. Examples show integration within the first intron of a gene (open arrow and rectangles). B – integration in the trapping (mutagenic) orientation. Flp or Cre mediated recombination can render the insertion neutral. Since tissue specific Cre transgenic lines are available, this allows for tissue specific rescue to be examined. After the insertion rendered neutral in the germline by Flp, Cre can induce a second round of inversion allowing for tissue specific knockout. C – integration in the neutral (non-trapping) orientation. Flp or Cre mediated recombination can render the insertion mutagenic. Since tissue specific Cre transgenic lines are available, this allows for tissue specific rescue to be examined and Flp RNA injection can be used to determine a global knockout phenotype.
1.3 Utility of conditional gene-trap insertions
With the incorporation of the two sets of heterotypic recombination sites, insertions in either orientation can be used to examine gene function either globally or in a tissue specific manner. Although both Cre and Flp catalyze efficient inversion (28) injection of Flp RNA is most useful for inverting the gene-trap in the germline because tissue-specific Cre-expressing lines are more readily available for inversion in somatic cells. Starting with an insertion in a mutagenic orientation, referred to as a trapping insertion (Figure 1B), the impact of global gene function can be determined by incrossing heterozygous carriers. To further link the trapping insertion to the phenotype, Flp RNA can be injected into these embryos to quickly determine if converting the trapping insertion to a neutral insertion alleviates the phenotype. Injection of Flp RNA is also used to convert a trapping insertion to a neutral insertion in the germline. With a neutral insertion, a transgene for tissue specific expression of Cre or tamoxifen-dependent recombinase can be used to assess spatial and temporal specific function of the tagged gene. Likewise, insertions that are in a neutral orientation initially (Figure 1C) can be used directly to determine tissue specific function or can be converted to a trapping insertion in the germline, via injection of Flp RNA, to determine the impact of global gene inactivation. With a germline trapping allele combined with an appropriate Cre transgene, the tagged gene function can be rescued in a spatial and temporal specific fashion to dissect the tissue-specific contribution to the observed phenotype(27).
2. Materials
2.1 Reagents and Solutions
Danieau Solution (50× stock): 2.9M NaCl, 20mM MgSO4, 35mM KCl, 30mM Ca(NO3)2, 250mM HEPES, pH 7.6.
Egg water: 0.3g Instant Ocean® aquarium salt in 1 liter distilled H20.
Plasmid DNA for Tol2 mediated gene trap (see Note 1), and Tol2 transposase, Flp recombinase, Cre recombinase in an expression vector.
MESAB (1.0% stock solution): 1.0g Tricaine (3-amino benzoic acid ethyl ester) in 100mL distilled H20, adjust pH to ~7.
NaOH: 50mM in sterile distilled H20
Tris-HCl: 1M, do not adjust pH which should be near 3
Reagents for PCR: 5× Taq Buffer (see Note 2), 25mM MgCl2, 10mM dNTP mix, Taq DNA polymerase (5U/µL)
Reagents for reverse transcription: 50 mM OligodT, transcription buffer, DTT (if not contained in transcription buffer), RNase inhibitor, MMLV reverse transcriptase.
Phenol Red: 0.5% in DPBS (available from Sigma-Aldrich)
Penicillin and streptomycin solution: 100× stock solution used for cell culture (10000U penicillin, 10000 µg streptomycin)
TRIzol® (Life Technologies)
2.2 Primer sequences
See Table 1
Table 1.
Primer sequences for the gene-trap that can be used for 3-primer PCR and to determine orientation of the insertion. All oligo sequences are in 5’ to 3’ orientation.
| 5’ TIR1 | CCAAAGGACCAATGAACATGTCTGAC |
| 5’ TIR2 | AACTGGGCATCAGCGCAATTCA |
| 3’ TIR1 | TCAGCCCCAAAAGAGCTAGGCTTG |
| 3’ TIR2 | GCGTGTACTGGCATTAGATTGTCTGTC |
| GT1 | CTTCCGACCGCGAAGAGTTTGTC |
| GT2 | CTTCTAACTATAACGGTCCTAAGGTAGCG |
2.3 Miscellaneous
mMESSAGE mMACHINE SP6 Kit (Ambion/Life Technologies)
NucAway™ Spin Columns (Ambion/Life Technologies)
Gel extraction columns (Qiagen)
Geneclean Spin Kit (MP Biomedicals)
Glass capillaries, 4 inch (100mm) 1/0.74 OD/ID
Flaming/Brown micropipette puller (Sutter Instruments Co.)
Injection plate mold (TU1, Adaptive Science Tools)
Microloader tips (Eppendorf)
MPPI-3 pressure injector (ASI)
Micromanipulator, magnetic base, and steel base plate (World Precision Instruments)
3. Methods
3.1 Generating Gene-trap alleles
The details concerning construction of the Tol2 transposon mediated gene trap cassette and the elements incorporated have been presented in detail elsewhere (26; 27). The elements contained in the gene trap cassette are outlined in Figure 1A. Tol2 transgenesis is used to generate the gene-trap alleles since this is an efficient and straightforward system to use.
- Preparation of microinjection needles and plates
- Microinjection needles are prepared using glass capillaries and a Flaming/Brown micropipette puller with settings that result in desired qualities for a microinjection needle. In general, the needle should have a long taper and fine tip, but thick enough to penetrate into the embryo chorion and cells. Multiple needles can be pulled in one session and stored in a 150mm Petri dish on top of clay for stability.
- Heat to dissolve 1.5% agarose with 0.3X Danieau buffer, then pour into a 100mm Petri dish, covered with an injection plate mold that has 6 troughs with a 90° wall and a 45° slope. When solidified, remove the mold and store the plates in 4°C until use.
- RNA synthesis and purification of DNA.
- Tol2 transposase mRNA is synthesized using the mMESSAGE mMACHINE SP6 Kit. Plasmid DNA with the transposase (29) is digested by Not I, run on a 1.0% agarose gel and purified using gel extraction columns. The same method is used to synthesize Flp and Cre mRNA. The transcription reaction is assembled following the manufacturer’s instruction using the linerized plasmid as template, and incubated at 37°C for 2 hr. After synthesis, the reaction is purified by NucAway™ Spin Columns, RNA concentration is determined using a NanoDrop 1000 Spectrophotometer, diluted to 150 ng/µL and stored in 2 µL aliquots at −80°C until use.
- The plasmid containing the conditional gene-trap cassette was purified by the Geneclean Spin Kit following manufacturer protocol. The concentration of purified mRNA and DNA is measured by NanoDrop 1000 Spectrophotometer, and confirmed by running on agarose gel.
- Embryo production.
- Mating pairs of adult zebrafish fish are placed in a breeding tank in the afternoon prior to injection. Each tank has one male and one female zebrafish that are separated from each other by a divider placed in the center of the insert. Generally 6–8 pairs of fish are set up for mating, anticipating an egg yield of 300 or more.
- The morning of the injection day, the insert is transferred to a new tank with fresh water and the divider removed allowing the fish to mate. The bottom of the tank is checked for embryos after about 20 minutes. If there are no embryos, the divider for another pair of fish will be removed. All tanks should be checked for embryos in regular intervals. Embryos are collected with a strainer and transferred to a 100mm Petri dish with egg water.
- Microinjection procedure
- An injection solution of Tol2 transposase mRNA, plasmid DNA and phenol red are mixed in sterile water so the final concentration of the mRNA and plasmid are 25 ng/µL and 0.05% for phenol red which is used as a tracer to ensure accurate injection.
- The microinjection needle is prepared by using a razor blade to cut a sharp angle in the end of the needle, taking care not to leave a ragged edge. 1–2µL of the injection solution is loaded into the microinjection needle using microloader tips, taking care to load solution to the end of the glass needle and to avoid air bubbles. Turn on the air supply and a MPPI-3 pressure injector. The pressure injector is connected to a foot switch. Adjust the pressure to 40 PSI and select pulse release. Carefully place the needle in the pipette holder with the end passing the O-ring. Make sure the O-ring is tightened and the outer ring is secure so the needle does not wobble. Attach the pipette holder to a micromanipulator that is set at the desired working angle. The micromanipulator has a magnetic base and is mounted on a steel base plate (Figure 2A).
- To determine the injection volume, a drop of mineral oil is placed on a micrometer. Carefully lower the needle with the micromanipulator until the tip is submerged in the mineral oil. Using the foot switch to trigger an air pulse, dispense a drop of working solution into the mineral oil. Adjust the pulse duration so the diameter of the liquid drop is between 0.1mm–0.15mm, which results in a volume of about 1–2nL (see Note 3).
- Using a transfer pipet, the embryos are loaded into the slots of the microinjection plates, using care not to crush the embryos. Excess egg water is removed so the embryos are just covered with water. Carefully turn the embryos using a micropipette so the cytoplasm of the embryos can be easily observed. Using the micromanipulator, gently advance the needle to penetrate the chorion and continue until the tip of the needle is in the center of the cytoplasm. Use the foot switch to dispense the injection solution. This should be easily visible due to the phenol red (Figure 2B). Care should be taken to avoid injection into the large yolk sac. Once the solution has been successfully injected, retract the needle with the micromanipulator and proceed to the next embryo (see Note 4). Transfer the injected embryos to a 100mm Petri dish with 0.3× Danieau solution and incubate at 28.5°C. Keep no more than 70 embryos per dish.
- Four to six hours after injection remove unfertilized and dead embryos and add 0.3× Danieau solution with 1% penicillin and streptomycin solution and continue to incubate at 28.5°C. Twenty-four hours after injection remove any dead embryos. At 3 days after injection the embryos should hatch from the chorions. At this point change the water to fresh 0.3× Danieau solution without antibiotics, removing the chorions and any abnormal embryos. Begin feeding the larvae at 5 days after injection and raise to maturity.
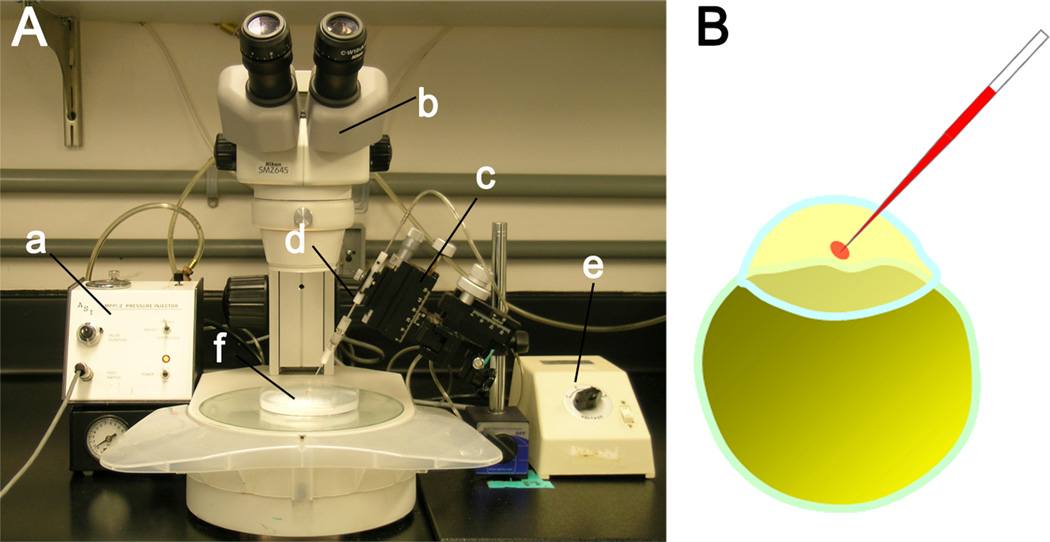
Figure 2. injection setup.
A- example of micro-injection equipment including pressure injector (a), microscope (b), micromanipulator (c), needle holder (d), microscope power supply (e) and microinjection plate (f). B – the solution should be injected into the cytoplasm of the embryo at the 1-cell stage and is visualized by phenol red added to the injection solution.
3.2 Identification of gene trap events
For this vector, mCherry is used as a marker for the tagged alleles (see Note 5). However, it is important to consider that expression will only result if the insertion is in a gene in the mutagenic, or trapping, orientation and only if the fusion is in frame with the gene transcript. While some insertions may not be identified with this approach, insertions with mCherry expression are advantageous as they reveal the expression pattern of the tagged genes. Figure 3 outlines the workflow for screening for these events as well as examples of mCherry expression that have been identified using this approach.
Cross individual gene-trap injected fish to a wild-type fish. Collect eggs from successful matings and hold the injected fish in a separate small tank while the clutch is screened for fluorescence.
Check for mCherry expression at 24hpf, 48hpf and 5 dpf. Keep embryos with expression and raise them to maturity. The fraction of mCherry-expressing progeny with a specific pattern varies from less than 1% to 50%. At least 10 embryos with identical expression pattern should be raised, making it more likely to have both sexes of adult heterozygous carriers for phenotype analysis in F2. Obtain additional progeny from the injected parent until enough mCherry-expressing embryos are identified. At this point, the injected parent may be euthanized. If more than 10 embryos with the identical expression pattern are available, isolate genomic DNA (Section 3.6) from some of the extra embryos for identification of insertion site via LMPCR. Otherwise, genomic DNA can be isolated from tailfin biopsies at 6 weeks of age for identification of the insert site via LMPCR.
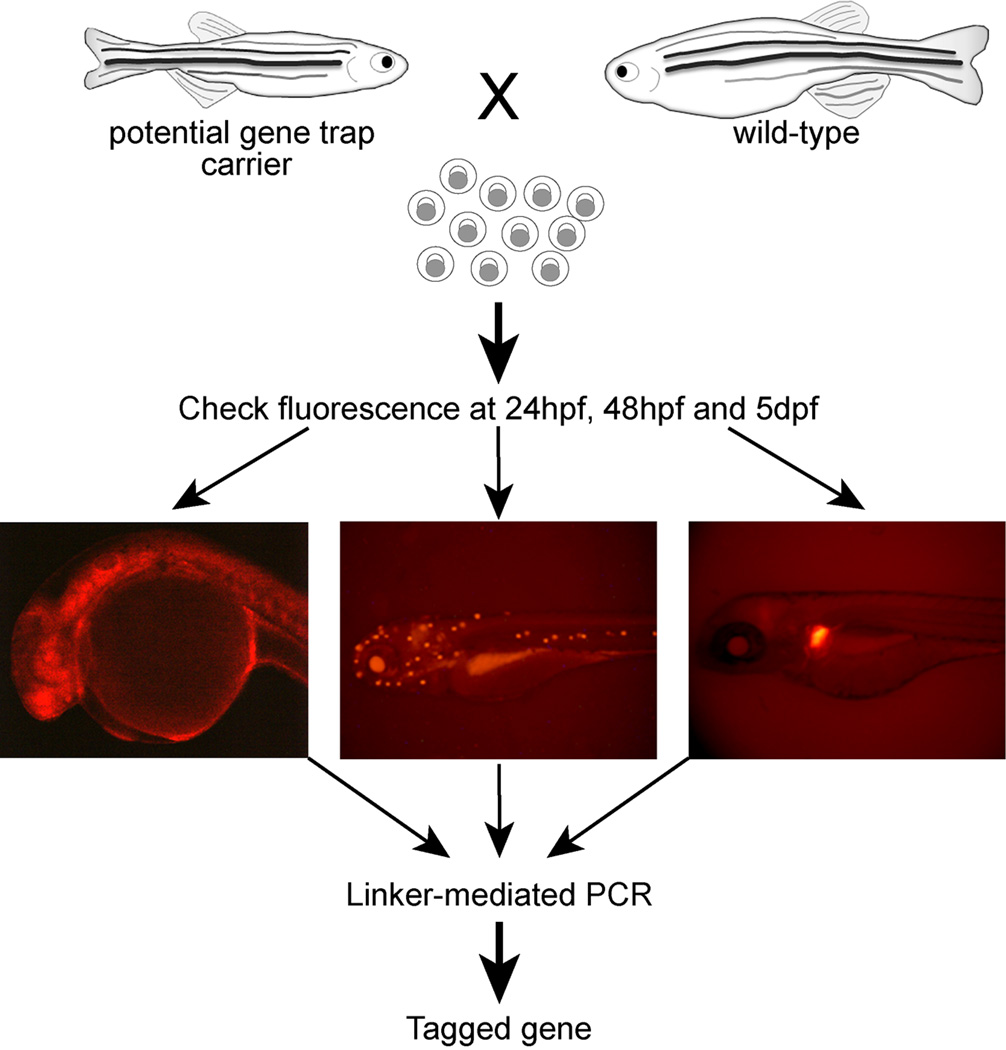
Figure 3. Screening for gene-trapping events.
An adult fish that had been injected with the gene-trap cassette as an embryo is mated with a wild type fish and embryos collected. At multiple time points, the embryos should be examined for mCherry fluorescence. The examples demonstrate that mCherry may be visible at different developmental stages and can be widely expressed or in specific cell and tissue types. These embryos will then be used to determine the location of the gene-trap insertion.
3.3 Insert identification via linker-mediated PCR
The genomic location of the insertion can be easily identified via linker-mediated PCR (see Note 6). This can be accomplished using genomic DNA from mCherry-expressing fish. Detailed information on the method has been presented elsewhere (26). It should be considered that many fish will carry more than one insertion. One of these insertions should result in mCherry expression, but others may not lead to mCherry expression due to wrong reading frame or orientation. Some of the latter insertions may still be useful to determine gene function.
3.4 Analysis of global gene function and conditionality
Before utilizing a conditional mutation to analyze tissue specific function, the phenotypic consequence of global inactivation of the gene should be examined. If a lethal or complex phenotype is observed, tissue-specific inactivation of the tagged gene is justified. Figure 4 demonstrates one such mutation with additional analysis of tissue specific mutation of the gene in Figure 5. Detailed information on this and other mutations is available elsewhere (27). Outlined below is a general protocol for using conditional mutations, with an emphasis on the logistics of the fish crosses.
- Global gene inactivation
- Cross blocking insertion carriers to each other and collect eggs from successful matings. Embryos should be carefully examined daily for any abnormalities through at least 7 days since not all gene mutations will result in an early phenotype. It is expected that 25% of the clutch should have the same phenotype if the blocking insertion causes a morphological phenotype. If less than 5% of the clutch has an abnormal phenotype or if the phenotype is inconsistent, then it is a likely result of non-specific factors and not the blocking insertion.
- If there is not a clear phenotype, embryos can be randomly selected for analysis of gene expression and genotype. If a phenotype is evident, both phenotypic and non-phenotypic embryos should be selected for analysis of expression and genotype.
- RT-PCR
- Isolate RNA and genomic DNA from individual embryos using TRIzol® per manufacturer instructions. Sufficient RNA and DNA can be obtained from single embryos but care must be taken not to lose any material. Resuspend the RNA in 10 µL RNase free water and use the entire volume to generate cDNA.
- Generate cDNA using oligo dT as the primer using MMLV reverse transcriptase. Incubate at 37°C for 1 hour, followed by 95°C for 10 min to stop the reaction and chill to 4°C. Dilute the cDNA 1:5 with sterile water and store unused cDNA at −20°C.
1. For a single reaction Embryo RNA 10.0 µL 5× MMLV reaction buffer 4.0 µL 10mM dNTP 1.0 µL 50 uM oligo dT 0.5 µL RNase inhibitor (20U/µL) 1.0 µL MMLV RT (200U/µL) 1.0 µL RNase free water 8.0 µL - For amplification of the target (see Note 7), use 1.0 µL of the diluted cDNA and the gene-specific primers. It is recommended that all primer pairs span an intron to distinguish cDNA from potentially contaminating genomic DNA. A control gene, such as β-actin, should be amplified in a separate reaction to ensure that the yield of RNA and cDNA were sufficient. It is simpler to prepare a PCR reaction cocktail sufficient for the number of samples. For amplification use 25–35 cycles with annealing temperature appropriate for the primer pairs (generally 60–65°C) with extension at 72°C for 1 minute per kb of amplicon size. The number of cycles should be determined empirically so that it is semi-quantitative.
For a single reaction Diluted cDNA 1.0 µL 5× GoTaq Buffer (Promega) 5.0 µL Sterile Water 14.8 µL 25 mM MgCl2 2.5 µL 10 mM dNTP 0.5 µL 10 µM gene-specific primer 1 0.5 µL 10 µM gene-specific primer 2 0.5 µL Taq DNA Pol (5U/µL) 0.2 µL
- To determine the genotype of individual embryos to correlate genotype with gene expression, a 3-primer PCR genotyping protocol is used (see Notes 8 and 9). Two of the three primers target the genomic DNA flanking the insertion and will produce an amplicon from the wildtype allele. The third primer is specific to the insertion and will produce an amplicon with one of the other two primers when the insertion is present. The TIR primers in 5’ TIR1, 5’TIR2, 3’ TIR1 and 3’TIR2 (Table 1) can be used as the gene-trap primers. However, these TIR-specific primers are not ideal to distinguish the orientation of the gene trap. To determine the orientation of the gene trap, use either one of two primers, GT1 and GT2, that are internal to the FlEx modules (Table 1). Design the gene-specific primers for genotyping so that amplicon size is less than 500 bp and the bands for the wild-type and insertion allele are sufficiently different to allow clear separation on a gel.
- Prepare a PCR reaction cocktail sufficient for the number of samples. For each reaction use 1 µL of the genomic DNA isolated that corresponds to the sample analyzed by RT-PCR. For amplification use 35 cycles with annealing temperature of 65°C and with extension at 72°C for 1minute per kb of amplicon size. Run 15 µL of each sample on a 1 to 2% agarose gel depending on amplicon size (see Note 10).
5× GoTaq Buffer (Promega) 5.0 µL Sterile Water 14.3 µL 25 mM MgCl2 2.5 µL 10mM dNTP 0.5 µL 10 µM gene-specific primer 1 0.5 µL 10 µM gene-specific primer 2 0.5 µL 10 µM transposon-specific primer 0.5 µL Taq DNA Pol (5U/µL) 0.2 µL
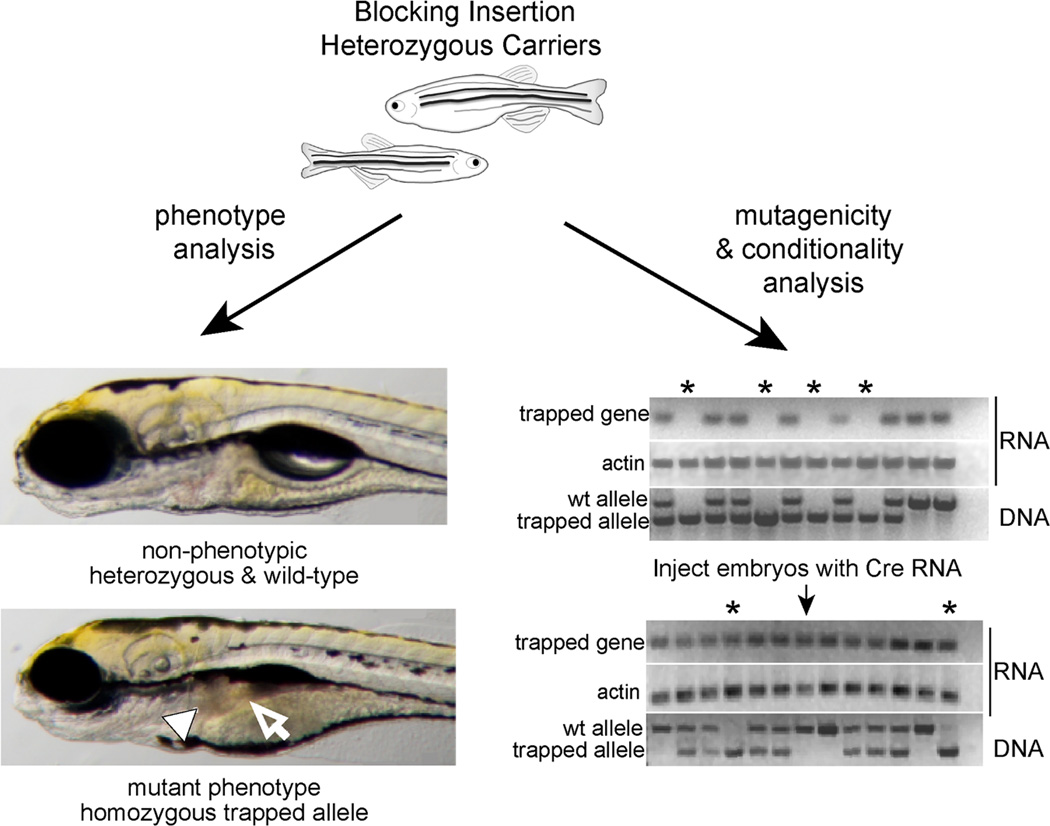
Figure 4. Mutagenicity and conditionality of the gene-trap cassette.
Once an insertion of interest has been identified, the consequence of a global knockout can be observed from an incross of two carrier fish. In this example, in the embryos homozygous for the trapped allele, several phenotypes are evident including small eyes, abnormal liver (arrowhead) and intestine (arrow). The embryos can be used for transcript analysis by RTPCR and corresponding genotype analysis by 3-primer PCR. The example shows little to no transcript of the tagged gene in homozygous embryos (asterisk) and that injection of Cre RNA in these embryos can invert the gene-trap to the neutral orientation and restore gene transcription.
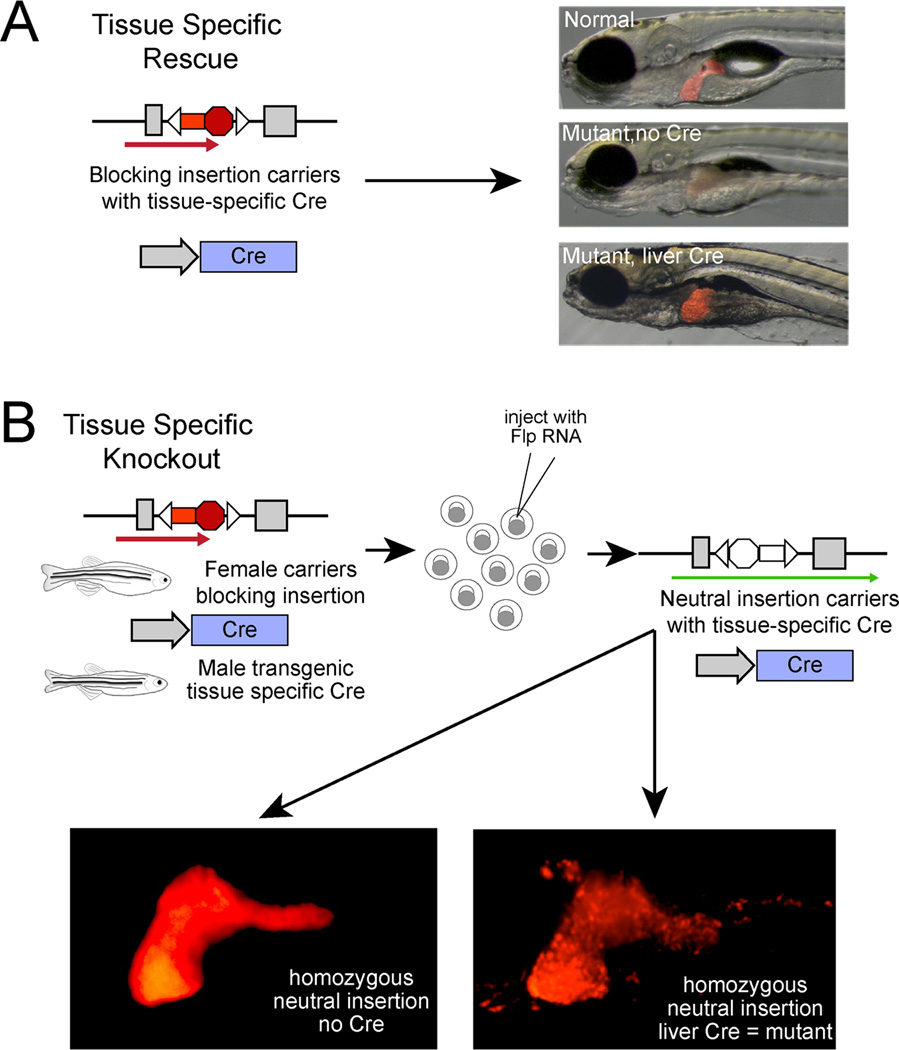
Figure 5. tissue specific analysis.
A – An example of tissue specific rescue where fish with the blocking insertion and a liver (hepatocyte) specific Cre. The liver is highlighted using a transgenic line (31). In the absence of Cre, the liver of embryos homozygous for the insertion is nearly absent. When Cre is present in the hepatocytes, the liver defect in homozygotes is rescued but the other affected tissues are still abnormal. B – An example of tissue specific knockout. The same insertion as in A was rendered neutral by injection of Flp RNA. This was done in embryos that carry both the insertion and the transgene for the liver specific Cre, allowing double carriers to be obtained in a single generation. In the liver, re-inversion of the neutral allele into a blocking insertion, mediated by Cre, resulted in detachment of hepatocytes and the eventual death of the larvae.
3.5 Analysis of tissue specific function
Once the genomic location and orientation of the insertions have been determined, the orientation of the gene trap can be manipulated using Cre or Flp transgenic lines or by RNA injection. Due to the greater availability of transgenic Cre lines in zebrafish, Flp recombinase is ideal for germline inversion. Although transgenic Flp lines can be used, injection of Flp RNA into the 1-cell embryos can result in 100% germline inversion. By injecting the Flp RNA into progeny of a gene trap carrier and a Cre transgenic fish, both germline inversion and generation of Cre-expressing carriers of conditional alleles can be accomplished in one generation. Although the initial screening for gene trap events is based on mCherry expression, it is likely that multiple insertions can be identified even in these families. Select progeny with a single insertion for further analysis. Any orientation of the insertion can be manipulated to determine tissue specific functions (Figure 1). As Cre become a more frequently used reagent in zebrafish, an increasing number of transgenic lines with tissue specific expression of Cre, as well as ligand inducible forms of Cre such as CreER (3; 4), will be available to allow for both tissue specific and temporal analysis of gene function. Here we will focus on using insertions that are in the blocking orientation but will also briefly address using insertions that are in the neutral orientation.
- To determine the contribution of the tissue of interest in a complex phenotype, a tissue specific rescue experiment can be carried out using a Cre transgenic line of choice (Figure 5A).
- Cross the insertion carriers to Cre transgenic fish, collect embryos and raise to maturity. Identify double carrier progeny using genomic DNA isolated from the tailfin (outlined in Section 3.6) and previously outlined PCR protocols (see Note 11).
- To obtain embryos that have a homozygous blocking insertion and tissue specific Cre expression, cross double carriers (carries both the Cre transgene and the blocking insertion) to a fish that only carries the blocking insertion. It is expected that half of the embryos will have the Cre transgene and of this half, one quarter (or 12.5% of the clutch) will be a tissue specific rescue. Global knockout mutants should also be present at 12.5% in the same clutch and confirms the genotype of the parent fish.
- For tissue specific knockout (Figure 5B), the appropriate genetic modifications can be generated in a single generation. However, it is important that a male Cre transgenic be used to avoid potential complications of undesired inversion in the germline (see Note 12). If this is not a viable option, the desired double carriers can be generated over two generations.
- To invert the blocking insertion to a neutral insertion (Figure 1), injection of Flp RNA will be used. Cross female blocking insertion carriers to male Cre transgenic fish and inject 50–100pg Flp RNA into the one-cell stage embryos as outlined in Section 3.1. Remove any abnormal embryos and raise the remaining fish to maturity.
- Identify injected fish with high levels of germline inversion by crossing the Flp injected fish to a wildtype fish. Select 48 embryos randomly and isolate genomic DNA (Section 3.6). Determine the frequency of inversion using primers that are specifically designed for this purpose and not simply the primers for detection of the insertion (Section 3.4.3).
- To generate the tissue specific knockout, cross the fish that have high level of germline inversion (>90%) and carry the Cre transgene to another fish with high levels of germline inversion but no Cre transgene. Observe progeny for a phenotype in the target tissue which is expected in >10% of the clutch (see Note 13). The phenotype may be different than that observed in the global knockout mutant and may appear much later. Use assays appropriate to the tissue of interest for phenotype analysis.
- For tissue specific knockout starting from a neutral insertion, cross the insertion carriers to the Cre transgenic fish. Again, here it is advised to use male Cre transgenic carriers and female insertion carriers. Once the appropriate genotypes are generated, the procedure is similar to that in section 3.5.2c.
- For global gene inactivation starting with a neutral insertion. Cross the neutral insertion carrier to wild-type fish. Inject 100pg Flp RNA in one-cell stage embryos as outlined in Section 3.1. Similar to Section 3.5.2b, it is recommended to determine the frequency of inversion. Incross fish with high levels of germline inversion, ideally close to 100%. Observe the progeny for phenotypes, with the expectation that 25% of the embryos will be homozygous for the now blocking insertion. Subsequent analysis should be done as outlined in Section 3.4.
3.6 Genotyping adult fish and embryos
There are multiple stages where it is likely necessary to genotype adult fish to determine if they are heterozygous or homozygous insertion carrier. Furthermore, the orientation of the insertion may need to be determined or confirmed. The specific protocols and primers to use for these different purposes are outlined in the above sections. This section outlines a general protocol for rapid isolation of genomic DNA for PCR-based genotyping..
For DNA isolation from adult fish, anaesthetize the fish in 0.05% MESAB and cut the tail posterior to the caudal peduncle. Transfer the tail to a 1.5 mL microfuge tube containing 250 µL of 50 mM NaOH. Place fish in individual plastic cups containing 350 ml of system water until PCR analysis is completed.
For DNA isolation from embryos, place individual embryos in a tube containing 100 µL of 50 mM NaOH.
To isolate the genomic DNA from either group, incubate the samples at 95°C for 20 minutes and chill to 4°C. Vortex well until homogenous. Add 1/5 volume of 1M Tris.HCl (pH approximately 3) to neutralize the samples (50 µL for tailfin, 20 µL for embryos).
Footnotes
Plasmids are ideally prepared by midi or maxi prep so they will be free of any contaminants. For the purpose of DNA injection, if the gene-trap plasmid has been prepared by midi or maxi prep, no additional purification is necessary. However, for DNA injection if only a plasmid miniprep is available, it is recommended for the plasmid to be additionally purified using GeneClean.
For PCR, any Taq polymerase buffer can be used. We prefer to use the GoTaq Green buffer from Promega since a gel can be loaded directly from the PCR without the need to add gel loading buffer.
When calibrating the liquid drop for injection, if the drop is too large with a minimum pulse duration, the needle opening is too large and a new needle will have to be prepared. If the bubble is too small with maximum pulse duration, carefully trim the needle so the opening is slightly larger.
One can continue to inject into the two-cell stage, but if division has proceeded beyond that new one-cell stage embryos should be used. To ensure that there is a continuous supply of the appropriate stage embryo, do not remove the dividers of the mating tanks all at once.
There are some limitations of this version of the gene-trap cassette. With mCherry expression only visible if the insertion is in a gene in a proper orientation and dependent on the frame of the fusion between the tagged gene and mCherry, some intronic insertions may be missed. With the wide availability of high throughput sequencing technologies, it is feasible to use LMPCR with bar coded linkers as a primary screen to identify all insertions regardless of mCherry expression. If it is desirable to reveal expression pattern of genes tagged with an out of frame insertion, one may design a pair of TALENs that target the coding sequence upstream of mCherry. Small indels from imperfect repair may correct the out of frame fusion.
During LMPCR, it is very likely that insertions in addition to the one responsible for fluorescence expression will be identified. It is important that careful analysis of all of the identified insertions be carried out, even if mCherry expression can not be used as trace for gene expression. Because of potential recombinase-catalyzed chromosomal rearrangement, it is recommended to use single insertion carriers for phenotype analysis.
When designing primers for gene expression analysis, the location of the insertion should be considered and primers that lie downstream or flanking the insertion should be used. Since the gene-trap should result in premature termination of transcription, there should be no read-through past the insertion but sequence upstream of the insertion may still be transcribed.
Optimization of the 3-primer PCR protocol will be necessary. Some of the insertion primers will be more efficient than others depending on the genomic locus. Altering primer concentration can often solve problems with biased amplification. If the initial set of primers chosen have less than optimal results, other primer sets should be tried.
When designing the gene specific primers, it is important to make sure that the size of the amplicon from the wild-type allele is sufficiently different from the amplicon from the mutant allele to easily distinguish them in a conventional agarose gel.
If genomic DNA of sufficient quality is difficult to isolate using TRIzol®, an alternative is to use a scalpel or razor blade divide the embryo into two sections. One section can then be used to isolate RNA for RT-PCR analysis and the other section to isolate genomic DNA directly.
We and others have found that using a fluorescent marker as an indicator of transgenic carriers can streamline genotyping and analysis of embryos. For example, the Tg(−1.5ins:Cre, −.58cryaa:Venus) transgenic line has expression of a yellow fluorescent protein in the lens (30). Therefore, carriers of this transgene can be easily determined using a fluorescent microscope.
In generating both inversion of a trapping insertion and a carrier for Cre expression, it is ideal to use a male Cre carrier. We have found that even using previously characterized tissue specific promoters with no maternal activity, germline inversion may still occur. Therefore, using a male Cre carrier avoids the complication of germline inversion and the desired outcome is easily achieved.
For the tissue specific knockout, after the initial inversion by Flp RNA, it is useful to obtain fish that carry two alleles of the neutral insertion. This will allow for an increased percentage of the clutch resulting in the genotype for a tissue specific knockout.
References
- 1.Feil R. Conditional somatic mutagenesis in the mouse using site-specific recombinases. Handb Exp Pharmacol. 2007:3–28. doi: 10.1007/978-3-540-35109-2_1. [DOI] [PubMed] [Google Scholar]
- 2.Gu H, Zou YR, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 3.Casanova E, Fehsenfeld S, Lemberger T, et al. ER-based double iCre fusion protein allows partial recombination in forebrain. Genesis. 2002;34:208–214. doi: 10.1002/gene.10153. [DOI] [PubMed] [Google Scholar]
- 4.Indra AK, Warot X, Brocard J, et al. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ERT) and Cre-ERT2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jao LE, Maddison L, Chen W, et al. Using retroviruses as a mutagenesis tool to explore the zebrafish genome. Brief Funct Genomic Proteomic. 2008;7:427–443. doi: 10.1093/bfgp/eln038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moens CB, Donn TM, Wolf-Saxon ER, et al. Reverse genetics in zebrafish by TILLING. Brief Funct Genomic Proteomic. 2008;7:454–459. doi: 10.1093/bfgp/eln046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sood R, English MA, Jones M, et al. Methods for reverse genetic screening in zebrafish by resequencing and TILLING. Methods. 2006;39:220–227. doi: 10.1016/j.ymeth.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Wienholds E, van Eeden F, Kosters M, et al. Efficient target-selected mutagenesis in zebrafish. Genome Res. 2003;13:2700–2707. doi: 10.1101/gr.1725103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doyon Y, McCammon JM, Miller JC, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foley JE, Yeh JR, Maeder ML, et al. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PLoS One. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng X, Noyes MB, Zhu LJ, et al. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cade L, Reyon D, Hwang WY, et al. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40:8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahlem TJ, Hoshijima K, Jurynec MJ, et al. Simple Methods for Generating and Detecting Locus-Specific Mutations Induced with TALENs in the Zebrafish Genome. PLoS Genet. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang P, Xiao A, Zhou M, et al. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 15.Sander JD, Cade L, Khayter C, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawakami K, Noda T. Transposition of the Tol2 element, an Ac-like element from the Japanese medaka fish Oryzias latipes, in mouse embryonic stem cells. Genetics. 2004;166:895–899. doi: 10.1534/genetics.166.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark KJ, Balciunas D, Pogoda HM, et al. In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat Methods. 2011;8:506–515. doi: 10.1038/nmeth.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawakami K, Takeda H, Kawakami N, et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Petzold AM, Balciunas D, Sivasubbu S, et al. Nicotine response genetics in the zebrafish. Proc Natl Acad Sci U S A. 2009;106:18662–18667. doi: 10.1073/pnas.0908247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trinh le A, Hochgreb T, Graham M, et al. A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Genes Dev. 2011;25:2306–2320. doi: 10.1101/gad.174037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnutgen F, De-Zolt S, Van Sloun P, et al. Genomewide production of multipurpose alleles for the functional analysis of the mouse genome. Proc Natl Acad Sci U S A. 2005;102:7221–7226. doi: 10.1073/pnas.0502273102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xin HB, Deng KY, Shui B, et al. Gene trap and gene inversion methods for conditional gene inactivation in the mouse. Nucleic Acids Res. 2005;33:e14. doi: 10.1093/nar/gni016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Floss T, Schnutgen F. Conditional gene trapping using the FLEx system. Methods Mol Biol. 2008;435:127–138. doi: 10.1007/978-1-59745-232-8_9. [DOI] [PubMed] [Google Scholar]
- 24.Schnutgen F, Doerflinger N, Calleja C, et al. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol. 2003;21:562–565. doi: 10.1038/nbt811. [DOI] [PubMed] [Google Scholar]
- 25.Yeo G, Hoon S, Venkatesh B, et al. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc Natl Acad Sci U S A. 2004;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maddison LA, Lu J, Chen W. Generating conditional mutations in zebrafish using gene-trap mutagenesis. Methods Cell Biol. 2012;104:1–22. doi: 10.1016/B978-0-12-374814-0.00001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ni TT, Lu J, Zhu M, et al. Conditional control of gene function by an invertible gene trap in zebrafish. Proc Natl Acad Sci U S A. 2012;109:15389–15394. doi: 10.1073/pnas.1206131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boniface EJ, Lu J, Victoroff T, et al. FlEx-based transgenic reporter lines for visualization of Cre and Flp activity in live zebrafish. Genesis. 2009;47:484–491. doi: 10.1002/dvg.20526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suster ML, Kikuta H, Urasaki A, et al. Transgenesis in zebrafish with the tol2 transposon system. Methods Mol Biol. 2009;561:41–63. doi: 10.1007/978-1-60327-019-9_3. [DOI] [PubMed] [Google Scholar]
- 30.Hesselson D, Anderson RM, Beinat M, et al. Distinct populations of quiescent and proliferative pancreatic beta-cells identified by HOTcre mediated labeling. Proc Natl Acad Sci U S A. 2009;106:14896–14901. doi: 10.1073/pnas.0906348106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korzh S, Pan X, Garcia-Lecea M, et al. Requirement of vasculogenesis and blood circulation in late stages of liver growth in zebrafish. BMC Dev Biol. 2008;8:84. doi: 10.1186/1471-213X-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]