Abstract
Transcriptional regulation of gene expression is fundamental to most cellular processes, including determination of cellular fates. Quantitative studies of transcription in cultured cells have led to significant advances in identifying mechanisms underlying transcriptional control. Recent progress allowed implementation of these same quantitative methods in multicellular organisms to ask how transcriptional regulation unfolds both in vivo and at the single molecule level in the context of embryonic development. Here we review some of these advances in early Drosophila development, which bring the embryo on par with its single-celled counterparts. In particular, we discuss progress in methods to measure mRNA and protein distributions in fixed and living embryos, and we highlight some initial applications that lead to fundamental new insights about molecular transcription processes. We end with an outlook on how to further exploit the unique advantages that come with investigating transcriptional control in the developmental context of the embryo.
Keywords: gene regulatory networks, embryogenesis, quantitative biology, single molecule FISH, live imaging
1) Transcription in the early embryo
All biological systems require the appropriately timed expression of gene products in amounts sufficient to carry out cellular activities [1]. Nowhere is this maxim better illustrated than in the patterning of the early Drosophila embryo. In this system, maternally supplied patterning cues direct the establishment of distinct gene expression programs with exquisite precision and reproducibility. During the first three hours following egg fertilization, cells receive patterning inputs in the form of transcription factors whose nuclear concentration differs by less than 10% between cells at a given position along the anterior-posterior (AP) axis. This leads to the establishment of spatial identities along the long axis of the egg that are reproducible from embryo to embryo to within less than the linear dimension of a single cell, i.e. less than 1% egg length [2–8]. These features of early fly embryogenesis have provided researchers with unprecedented opportunities for assessing general properties of transcriptional regulation of gene expression (Box 1). Many central concepts of metazoan transcription, such as enhancer modularity, the combinatorial activities of transcription factors (both cooperative and competitive in nature), long range interactions of enhancers with promoters, and the phenomenon of polymerase pausing, have emerged from over three decades of research in Drosophila embryos [9–11]. These studies have led to first generation quantitative measurements exploring the interplay between maternal signals and zygotically expressed patterning factors generates diverse gene expression patterns [12, 13].
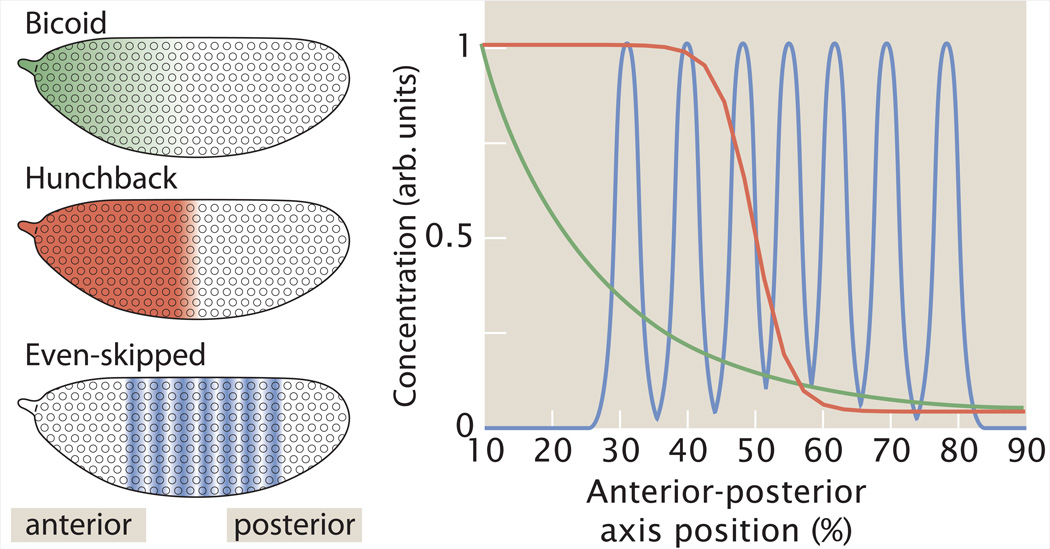
Box 1: Segmentation by transcription.
During the first three hours of its development, the 500µm long Drosophila embryo transitions from a single cell to a differentiated multicellular structure with a single layer of ~6000 regularly arranged cells just below its surface. These cells express differential combinations and amounts of gene products specifying cell types, thus laying out a spatial blueprint for the structures in the future adult organism. This blueprint originates during the construction of the egg, when localization processes place symmetry breaking gene products at the poles along the axes of the egg. The signaling cascades that initiate patterning are triggered upon fertilization at time zero, establishing maternal activity gradients that spread along the anterior-posterior (AP) axis. One such maternal factor is Bicoid, a transcription factor required to determine anterior fates. Maternal gradients are established during the first 1–2 hours following fertilization, when nuclei undergo 13 rounds of mitotic division without cytokinesis. The absence of membranes between nuclei permits the free diffusion of molecules within the embryonic syncytium. Bicoid and other maternal factors activate the zygotic patterning genes in specific spatial domains, generating the aforementioned blueprint. The extended 14th interphase takes place during the third hour of development, when zygotic gene products accumulate to high levels and membranes are deposited between nuclei forming individual cells. The patterning genes compose a hierarchical transcription network with three layers (Figure I): maternal genes, such as Bicoid (green); gap genes, whose expression domains demarcate large territories spanning many cell diameters (Hunchback, red); and pairrule genes that form an iterative pattern of 7 stripes (Even-skipped, blue), presaging the formation of the larval body segments. Three maternal AP patterning systems regulate the expression of >12 gap genes whose combined activity regulates 7 pair-rule genes. These genes encode a network of transcription factors, the interactions of which determine the positions of gene expression boundaries.
Figure I: Hierarchy of patterning genes in the early Drosophila embryo. The maternal factor Bicoid activates hunchback and various other gap genes in broad domains which all work together to regulate the activity of pair-rule genes such as even-skipped.
Gene expression patterns, cell fate determination, and morphogenesis events unfold in a characteristic and extremely reproducible manner across all wild-type embryos. The process of development, or the “experiment”, takes place without the intervention of the experimenter. By merely observing the underlying molecular processes, the experimenter can discern how embryo-wide patterns of gene expression emerge from discrete molecular events, such as the association and dissociation of transcription factors with promoter and enhancer elements of individual gene loci–effectively using the embryo as a laboratory. Subsequently, it becomes straightforward to construct mathematical models from these observations, and thereafter test their predictions using genetic methods to alter transcription factor activity and transgenesis to test the effects of modified DNA regulatory elements.
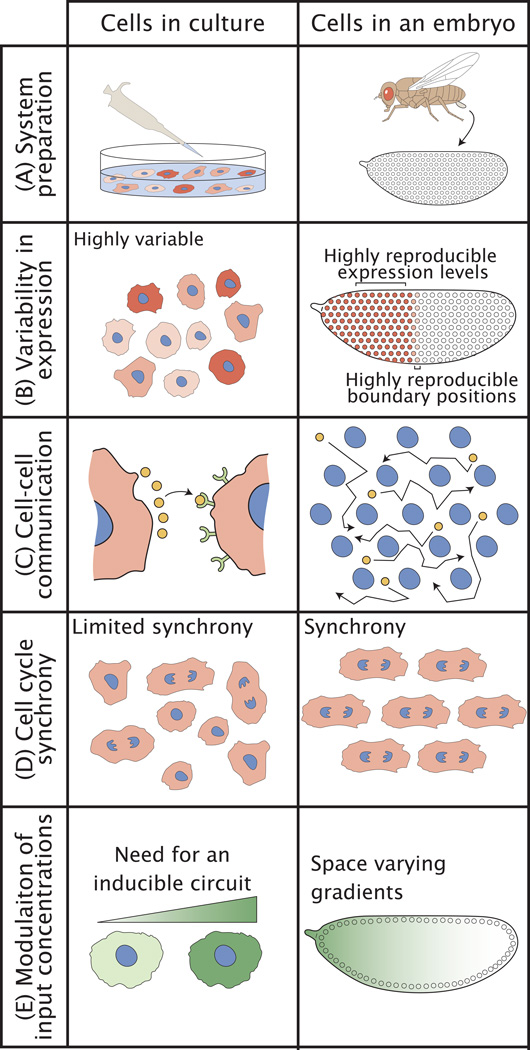
These approaches are greatly aided by several advantages of early embryos. First, developmental reproducibility under a given set of environmental conditions is well documented at both the molecular and morphologic levels [14, 15]. Spatially, extremely low variability in gene expression between embryos results in the unique specification of the fates of individual rows of cells along the anterior-posterior axis [2, 3]. Temporally, observations of the formation of morphological features [16] indicate that morphogenetic events occur with differences between embryos of no more than one to two minutes [3]. Because early patterning events are driven by maternally supplied factors, this reproducibility suggests that every individual instance of the experiment, i.e. each embryo, possesses very similar starting conditions. For example, mRNA and protein amounts of the maternally supplied transcription factor Bicoid (Bcd) are highly reproducible between embryos [2, 17]. Thus, available evidence suggests that embryos themselves represent nearly identical iterations of the same set of processes (Figure 1A,B).
Figure 1. Cells in different contexts: culture vs. embryo.
(A) Cells in culture require careful preparation to assure reproducibility across experiments. Female flies naturally prepare embryos in a high-throughput and reproducible manner. (B) Culturing allows examination of gene regulatory network behavior across a range of conditions in a synthetic environment. As such, it is not a priori evident which part of observed cell-to-cell variability (which can range between 50% and 500%) is due to sample preparation versus inherent fluctuations in the system. Studies of embryos reveal the degree of natural variability in gene expression that is tolerated by an intact system. As a system, the embryo is designed to generate a specific gene expression pattern within tight temporal and spatial constraints. This tight control results in levels of gene expression that can be reproducible within 10% and boundary positions with a 1% egg length reproducibility. (C) The absence of membranes between nuclei in the early embryo allows for the mixing of regulatory factors by diffusion. (D) Synchrony of the cell cycle is hardwired in the early fly developmental program. (E) Titrating input concentrations of transcription factors in cell culture requires the implementation of synthetic inducible circuits in multiple experiments. By contrast, transcription factor gradients in the early embryo span the natural range of patterning dynamics required for proper development.
Second, the newly fertilized embryo undergoes 13 rounds of rapid mitosis without cytokinesis, resulting in a hexagonal 2D array of syncytial nuclei, nearly all of which are found just beneath the egg cortex inhabiting a shared cytoplasm. The positioning of the nuclear layer facilitates imaging by minimizing specimen thickness in a geometrically simple arrangement. The shared cytoplasm simplifies inter-nuclear communication and minimizes variability between nuclei in the numbers of vital molecules such as RNA polymerases or transcription factors (Figure 1C). Therefore the extent of so-called “extrinsic” or environment-driven variability that is typically observed in bacterial cultures [18, 19] is greatly reduced, unmasking “intrinsic” or process-specific events that determine the magnitude and variability of expression dynamics.
Third, this array of about 6000 nuclei is synchronized by the mitotic cycle, all of which simultaneously undertake gene expression decisions (Figure 1D). Each nucleus acts as an independent unit responding to the natural gradients of patterning activity, wherein the position of a nucleus in the embryo correlates with the amount (or concentration) of input activity that the nucleus observes. Thus, transcriptional responses can be observed simultaneously across a physiologically relevant range of signaling input levels. This minimizes potential challenges encountered in cultured cells to ensure synchronization, and in which probing the effects of different input conditions requires multiple experiments (Figure 1E). Thus, the design features of early embryos present unique advantages over cells prepared in culture.
Despite these advantages of the developing Drosophila embryo, it has proven challenging to assess transcriptional events with the quantitative rigor achieved in organisms grown in laboratory culture. For cultured organisms, recently developed technologies allow the visualization of absolute numbers of biomolecules in living cells on a cell-by-cell basis [20, 21]. The measurement of gene expression in absolute units has generated exciting new insights into the regulatory mechanisms that produce mRNA and protein molecules [22, 23], and enabled the formulation and testing of thermodynamic and stochastic quantitative models of gene expression [24–28]. Among the more striking findings has been that, at least for cells grown in culture, gene expression is exceedingly variable: genetically identical cells almost always possess large differences in their absolute mRNA and protein content [29–33], in stark contrast to observations from fly embryos [2, 3, 34]. Many questions thus arise regarding how embryos achieve precise gene expression. In particular, are the observations from cultured cells relevant to understanding patterning in a multicellular organism? Or, do embryos employ specialized mechanisms to ensure precision? We currently have a very limited grasp of how the magnitude of gene expression, i.e. the rate of transcription of target genes, is controlled by maternal patterning inputs and/or interactions between zygotic gene products at a given position in the embryo. Moreover, we possess limited understanding regarding the underlying mechanisms that result in highly variable stochastic gene expression [35]. In general, to resolve any of these problems–be it in single cells or in whole embryos–we need to know how discrete transcription factor binding events at specific DNA elements lead to the control of RNA polymerase II (Pol II) activity.
Recent advances now endow the study of Drosophila embryos with the same quantitative rigor achieved in studies of cultured cells. Combined with an array of genetic and molecular tools, fly embryos are set to lead to profound new molecular insights regarding transcriptional regulation. Here, we highlight some of the advantages of examining gene expression in Drosophila and briefly review some recent examples of quantitative studies in the early embryo, with particular emphasis on the patterning along the anterior-posterior axis. Our goal is to transmit the current excitement for investigating metazoan transcription with new quantitative approaches, bringing real opportunities for novel mechanistic insights into the fundamental processes governing transcription across phyla.
2) Quantitative methods to measure transcription in the embryo
Studies of fly patterning over the past few decades have revealed how maternally supplied patterning gradients subdivide the anterior-posterior axis of the early embryo into broad territories of zygotic gene expression [36], and how these events are necessary to generate the familiar iterated pattern of segmented gene expression stripes observed during the third hour following fertilization [37] (Box 1). We now possess an essentially complete list of the critical maternal inputs, the vital zygotically expressed factors that ensure correct spatial modulation of transcription, and we know how zygotic gene products regulate each other’s expression [12, 38]. Yet many questions remain unaddressed. Broadly speaking, we would like to relate the discrete transcription factor binding events at promoters and enhancers to the dynamics of Pol II activity, how these dynamics give rise to mRNA and protein expression rates and variability, and how the resulting gene products interact at DNA elements and refine transcriptional behavior to ultimately give rise to the precise placement of gene expression boundaries.
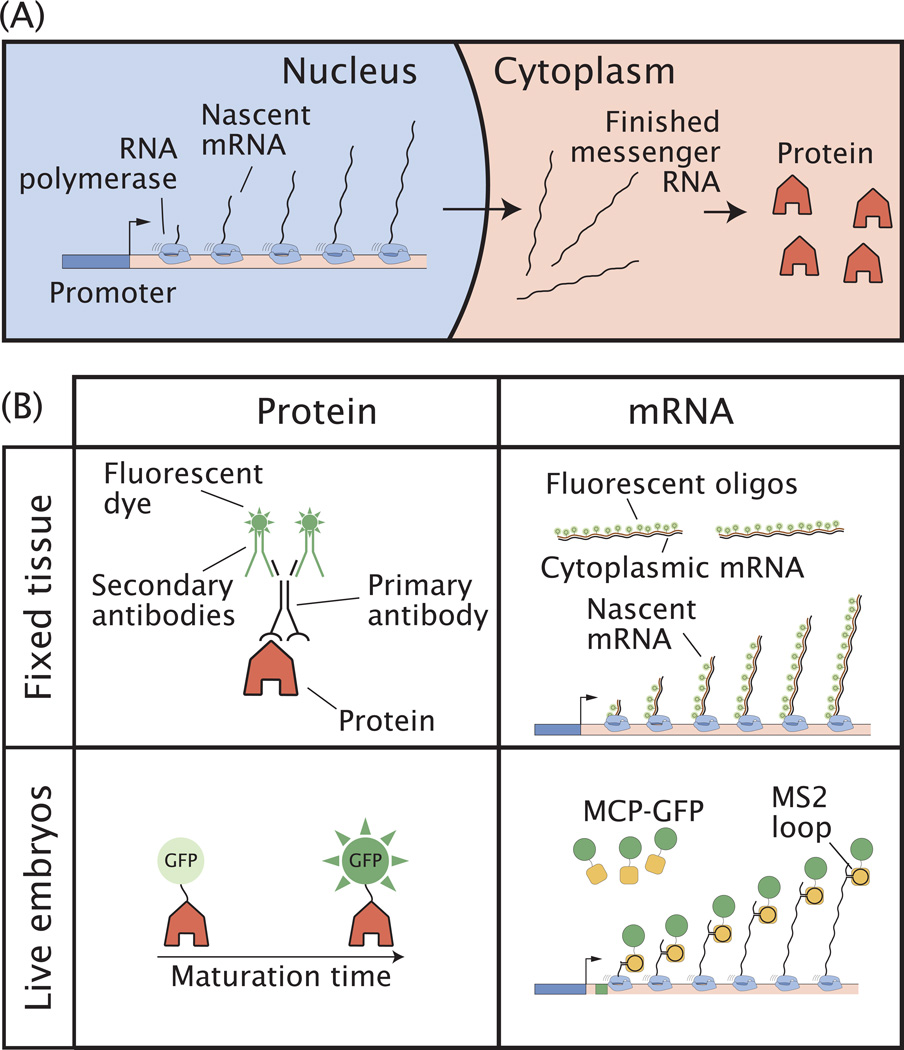
Advancing our understanding of complex systems such as the fruit fly segmentation network requires direct quantitative access to the spatiotemporal evolution of network components. Ideally, we would like to measure at all times and all locations in the embryo the production rates of any mRNA and protein of interest (Figure 2A). For each molecular species, two complementary methods exist: one based on snapshots of fixed embryos, the other based on live imaging. In the first method, proteins or mRNA molecules are labeled in chemically fixed embryos to provide a very accurate snapshot of the intact system. Although it requires halting the developmental process, this approach has the advantage that it can be readily applied to any genetic background as long as labeling reagents are available. The second method allows the direct observation of dynamics by labeling with genetically encoded fluorescent reporters. In these cases embryos can be observed in real time as development unfolds. However, much care must be taken to determine whether such labeling alters the characteristics of the molecules in question and of the developmental process in general, which might be perturbed by the addition of synthetic reporters. Here we outline applications of both fixed and live measurement techniques of proteins and mRNA molecules (Figure 2B):
Figure 2. Visualizing the central dogma in the embryo.
(A) Developmental decisions can be assessed at multiple stages of the central dogma: from nascent mRNA transcript formation (and thus the number of active RNA polymerases loaded on the promoter), to cytoplasmic mRNA and protein distributions. (B) Methods to measure mRNA and protein distributions during development in fixed and living embryos (see text).
i. Relative protein level measurements in fixed tissue
Immunohistochemistry has been used historically to discern protein localization and relative levels of expression [39]. Many regulatory relationships between early expressed genes have been inferred by antibody labeling of zygotically expressed transcription factors and subsequent testing by use of mutants [40, 41] [42]. Such studies have found that activating cues are provided by maternal inputs [43–47], and that the majority of zygotically expressed transcription factors inhibit transcription, mutually repressing each other, thus establishing expression boundaries that define broad pattern territories [48–51]. More recently, much effort has been put toward assembling a full time course of spatial protein expression profiles by preparing embryos at varying ages and carefully quantifying expression levels [3, 52, 53]. These methods rely on a strong ability to estimate embryo ages from fixed tissue, which can be done with varying accuracy. Prior to the 14th interphase, the mitotic cycles allow staging of embryos with an accuracy of 2 to 4 minutes [54, 55]. During the third hour of development, when most of the gene expression dynamics of the segmentation network occurs, membranes are deposited from the surface and invaginate, thus dividing the nuclei in the syncytium and giving rise to individual cells. The timing of membrane deposition provides a timer that correlates precisely with age and allows for staging accuracy of 1 to 2 minutes [3, 16]. However, analysis of fixed tissue data can only indirectly assess dynamics, and absolute protein levels are accessible only in combination with biochemistry on bulk samples and are thus associated with excessive measurement error. Overall, relative protein concentration measurements have led to insights into pattern formation, but direct access to gene expression rates has been limited.
ii. Live protein concentration measurements with fluorescent fusion proteins
The advent of green fluorescent protein (GFP) and the creation of fluorescent protein fusions have granted us direct access to transcription factor dynamics using live microscopy. The technique has worked exceedingly well for examining many aspects of gene expression dynamics in the fly. For example in early embryos, fluorescent tags to maternally provided gene products have revealed the dynamics of mitotic divisions through fusions to histones [56], intricate cell mechanics during gastrulation [57], and cell membrane formation through protein traps [58]. Dynamics of the maternal transcription factors Bcd and Dorsal have led to a number of important quantitative insights regarding their activities [2, 59–63]. For Bcd in particular, it is notable that the gradient of nuclear localization is quite similar between nuclear cycles despite the geometric increase in nuclear number [60]. Indeed, various functional protein fusions to Bcd have allowed the measurements of absolute protein concentrations, production rates, diffusion constants and degradation times [2, 64–67], all of which are attuned to the requirements of the system. In combination with the antibody-based observations mentioned above, these studies have determined that Bcd and its downstream targets exhibit very similar degrees of precision [2, 3]. These findings have led to a model in which tightly controlled transcriptional inputs are processed by a highly reliable, precise set of processes to result in the correct degree of gene expression precision with low variability (see the section “From transcription to patterns” below).
But also beyond the early embryo, during larval development, fusions of fluorescent proteins to the secreted factors Decapentaplegic (Dpp) and Wingless (Wg) have allowed the estimation of biophysical parameters that give rise to the observed gradients in the wing imaginal disc, the larval precursor of the adult wing [68]. These studies have demonstrated, for example, the scaling of the Dpp gradient with tissue size [69], although for both Dpp and Wg, many questions remain regarding the mechanism(s) underlying gradient formation [70]. Indeed, for Wg, a gradient arising from extracellular protein transport is dispensable [71]. Dpp and Wg dynamics using fluorescent protein fusions will no doubt prove invaluable to addressing how the graded distributions of these factors contribute to patterning and growth.
Whereas the study of maternally supplied molecular activities has benefitted greatly from fluorescent fusions, the study of early zygotic gene expression using fluorescent tags remains challenging. Ideally, one would like to observe the interactions of transcription factors with DNA, requiring single molecule measurements that are just becoming possible in single cells [72]. Significant technological improvements will be necessary to increase the sensitivity of these imaging techniques for use in embryos. Another challenge is the fact that the dynamics of gene expression occur at time scales comparable to the time required for newly synthesized reporter proteins to mature and acquire fluorescence [73]. The time lag between fluorescent protein translation and the moment when it actually becomes fluorescent complicates the interpretation of dynamic observations, as revealed by differences between live and fixed measurements [3, 8, 67, 74, 75]. Moreover, an engineered fusion protein should ideally possess wild-type functionality when expressed at or near the levels of the endogenous protein, a requirement that is not always easy to assess. For many of these requirements new advances at the level of the chemistry of fluorescent protein engineering will be necessary, and it will take another round of interdisciplinary interactions to solve the challenge of a new generation of fluorescent proteins.
iii. Fixed tissue mRNA counting
Because patterns emerge from the modulation of transcription in space and time, full understanding of the segmentation network can only be achieved through examining transcriptional activity directly. Historically, the use of in situ hybridization has been instrumental in determining the spatial positions of gene expression domains [76, 77] and in the analysis of artificial reporter constructs in which critical DNA elements are altered or missing [78–81]. Such studies have determined, for example, how striped patterns of pair-rule genes emerge from combinatorial regulation by transcription factors [82] and the combined activity of multiple enhancer elements [83]. Overall, such studies have revealed many qualitative insights about the spatial modulation of gene expression levels, but these have been limited in part by non-linearities introduced by the prevalent use of enzymatic amplification to label mRNA molecules.
Recent advances have enabled the visualization of individual transcripts through the use of nucleic acid probes complementary to a gene of interest, termed single molecule fluorescence in situ hybridization (smFISH). These probes are either covalently linked to a fluorophore [34] or contain an epitope that is subsequently recognized by labeling with fluorescently tagged antibody [5, 84]. The resulting signal-to-noise ratio is substantially higher than observed for previous approaches that rely on enzymatic reactions to produce either fluorescence or solid precipitate. Such high contrast allows for the resolution of individual mRNA molecules, which has now been demonstrated in a variety of contexts [33, 85–88]. This method permits simultaneous visualization of transcript levels of multiple genes. Studies employing FISH have shown that the transcripts of several early expressed genes accumulate at very similar rates with high precision in the syncytial embryo (discussed below [34]), whereas at late times, following the formation of cells in the embryo, transcript content can vary substantially between cells [5].
In many cases, studies employing FISH focus on measuring cytoplasmic mRNA distributions in order to infer the microscopic mechanisms behind transcriptional regulation [5, 84]. Cytoplasmic RNA counts necessarily result from many processes: the production of mRNAs in the nucleus, the processing of pre-mRNAs to produce mature transcripts, export to the cytoplasm, and transcript degradation. Thus, mRNA counts do not provide the most direct access to the processes that direct Pol II holoenzyme assembly and entry into productive elongation. Nevertheless, FISH allows assessment of the process of transcriptional initiation through the careful examination of fluorescence intensities at sites of nascent transcript formation inside nuclei [89, 90]. Such foci have traditionally been used to determine whether a given nucleus is engaged in transcription [89, 91–93]. However, these foci can provide additional information: their fluorescence intensities are a measure of the amount of nascent mRNA, and therefore an estimate of the number of bound Pol II complexes, present at a transcription site at the moment of embryo fixation [34]. Thus, it becomes possible to use fluorescent intensities to gauge either relative transcriptional activity [94], or to determine activity in absolute units by normalizing transcription site intensities to the mean intensity of labeled cytoplasmic transcripts [34].
iv. Live mRNA measurements and polymerase counting
While the single molecule precision of smFISH can provide indirect insights about the temporal evolution of transcriptional decisions, the observations of transcript production in real time is now commonplace in the context of single cells [95–98]. Such dynamical measurements are achieved with the use of an mRNA tagging system in which a reporter transgene is tagged with multiple repeats of a stem loop sequence recognized by a cognate binding protein fused to GFP. One example is the MS2 bacteriophage stem loop and the MS2 coat protein (MCP) tagged with GFP [99, 100]. This system has been implemented to study maternal mRNA transport in fly oocytes [101] and has recently been applied to the study of transcription [102, 103]. In this context, the MCP-GFP fusion is provided maternally, so that all GFP molecules are fluorescent prior to fertilization. As a result, once Pol II transcribes the stem loops, the nascent mRNA becomes fluorescently tagged. Thus, unlike the dynamics of fluorescent reporters of zygotic gene activity, mRNA dynamics can be accessed on time scales that are limited only by the diffusion and binding of MCP-GFP to its target mRNA. In embryos, the detection threshold of this method is around a few hundred fluorescent proteins [2, 102], unlike in cultured cells where single molecule detection is commonplace [104]. However, live fluorescence signals in embryos can be converted to absolute units of transcribing Pol II molecules and numbers of produced mRNA molecules by calibrating MS2 signals and smFISH measurements [102], thereby producing dynamic descriptions of gene expression in absolute terms.
3) From transcription to patterns
The tools described in the previous section allow for the quantitative study of transcriptional regulation in a multicellular organism and its role in the formation of patterns and subsequent specification of biological structures. Already, these tools have provided key new insights into developmental decision-making that challenge the picture of transcriptional regulation stemming from classic studies performed in cell culture.
i. Transcription noise vs. developmental precision
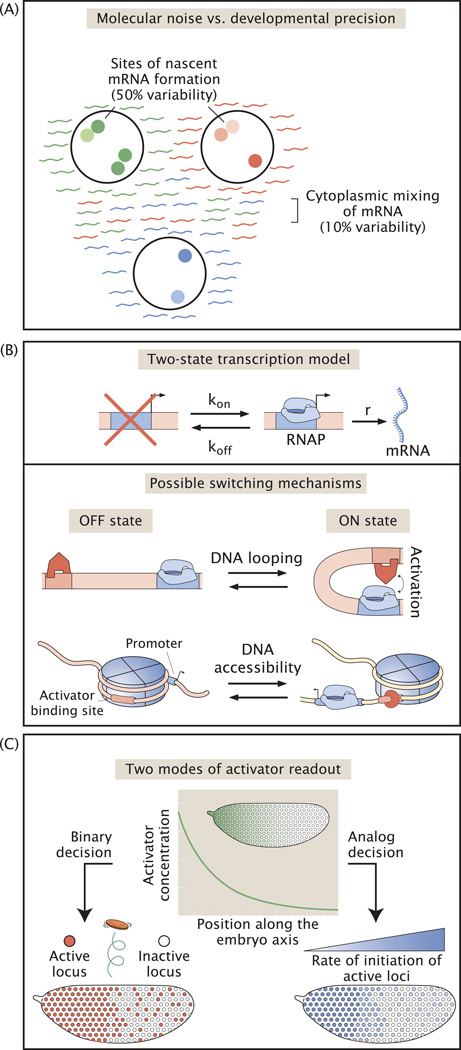
The inherent variability in transcription processes is a widely appreciated phenomenon. In particular, a multitude of experiments performed in single cells have shown that variability in transcriptional output –calculated as the ratio of the standard deviation over the mean number of mRNAs of a given gene per cell– can be as high as 500% [33, 86, 105], leading to a view that transcription is not precisely controlled. If transcriptional activity is inherently variable, how then are precise developmental outcomes achieved? Recent work has found that the instantaneous transcriptional activity in the early fly embryo can vary up to 50% despite the fact that the resulting cytoplasmic mRNA and protein distributions vary by less than 10% [34]. In the fly embryo this high transcriptional variability is buffered by the syncytial cytoplasm: in the absence of membranes, variable mRNA production in an individual nucleus is averaged both in space and time between neighbors. Compared to the mRNAs of cultured cells, the lifetime of which may be on the order of hours [106], transcripts of the early embryo are relatively short lived, especially during the highly dynamic 14th interphase [76, 107]. However, prior to this time point the mRNA lifetime is sufficiently long (on the order of an hour) to permit rapid accumulation [28]. By sharing long-lived transcripts, the embryo reduces the variability in the cytoplasmic distribution of both mRNA and protein molecules [34] (Figure 3A). Straightforward spatial averaging thus reduces the requirement for more complicated feedback mechanisms that are usually proposed to ensure precision. As spatial averaging tends to smoothen patterns, additional regulatory interactions may be required to ensure “sharp” borders [93, 108]. Furthermore, spatial averaging can only occur in the absence of membranes, thus begging the question of how transcriptional variability is buffered during later developmental stages. In order to reveal the mechanisms of variability buffering in later stages, staining and imaging protocols will have to be adapted to access the more complex 3D structures that form after embryo gastrulation. It is tempting to speculate that the precision in mRNA counts at later times would be aided by rapid transcript production: compared to cultured cells where transcripts may be produced at only 1 mRNA per hour [109], fly embryos engage in extremely rapid transcription with up to ~5 mRNAs per minute [28]. If mRNAs are relatively stable and allowed to accumulate, high production rates reduce fluctuations in potentially variable processes, such as Pol II binding and progression, by integrating production over time, thus allowing each cell in a given collection a greater chance of achieving the mean level of gene expression and minimizing variability.
Figure 3. Signatures for the transcriptional basis of pattern formation in the fly embryo.
(A) The variability in the instantaneous amount of mRNA being produced at sites of nascent transcript formation is as high as 50%. However, the mRNA molecules produced in different nuclei are averaged in the common cytoplasm of the syncytium, effectively reducing variability in the distribution of cytoplasmic mRNA molecules to 10%. (B) The variability in mRNA production observed in nuclei in (A) suggests a mechanism of transcription where the promoter switches between an ON and an OFF state. The molecular nature of this mechanism could be associated with enhancer-promoter looping or transient changes in chromatin accessibility, for example. (C) Live mRNA production monitoring reveals that patterns are formed by two serial steps of transcriptional regulation in single cells. First, a locus of a given gene makes a random binary decision whether to turn ON or not, with the local concentration of activator biasing this decision. Loci that are OFF will remain so for the whole interphase.By contrast, loci that turn ON will produce mRNA at a rate that in modulated by the local concentration of activator in an analog fashion. It is the combination of these two regulatory strategies that leads to macroscopic patterns of expression throughout the embryo.
ii. Variability in transcription and the two-state model
Over the last few years the use of variability (or “noise”) in gene expression as a lens through which to view the in vivo molecular mechanisms of transcription has gained traction [27, 110]. As discussed in Box 2, the simplest model of transcription states that the distribution of Pol II molecules on a given gene will be given by Poisson statistics, with variance equal to the mean. However, experiments performed both in single-celled and multicellular organisms have yielded values for the variance that are usually several times higher than the mean, arguing against a simple model of gene expression and suggesting the presence of additional rate-limiting step(s).
Box 2: Variability as a window into molecular mechanisms.
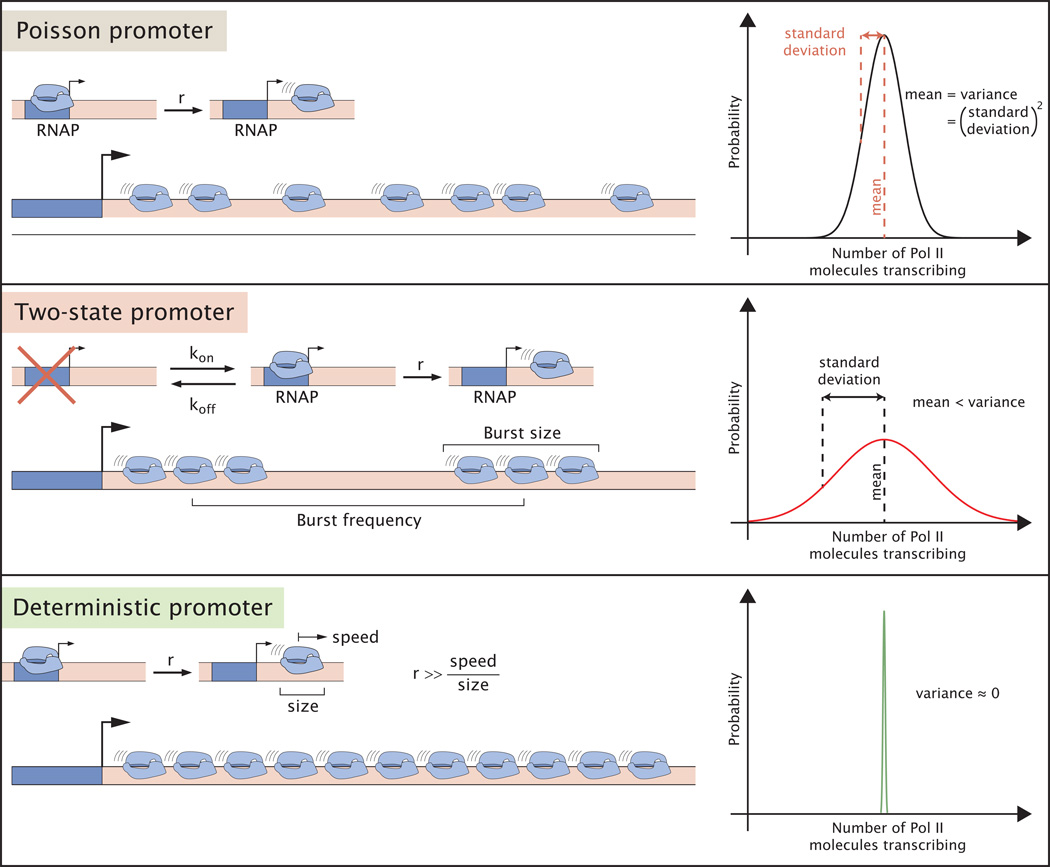
A straightforward model of transcription, the Poisson promoter, proposes that Pol II initiates transcription at a constant rate (Figure I). This rate is then modulated by the relative abundance of activators and repressors [145]. Because of the inherent stochasticity of biochemical reactions, a constant rate of transcriptional initiation leads to a Poisson distribution of transcribing Pol II molecules along the gene, with variance equal to the mean number of Pol II molecules. A variance higher than the mean number of actively transcribing Pol II molecules indicates the presence of extra regulatory steps along the transcriptional cascade. The simplest model consistent with increased variance consists of a promoter that switches between transcriptionally permissive (“ON”) and restrictive (“OFF”) states, a two-state promoter (Figure I). When switching occurs more slowly than the rate of transcriptional initiation, mRNA molecules are produced in “bursts,” increasing the variability in Pol II loading. Cartoons such as shown in Figure 2A, where all Pol II molecules are uniformly loaded on the DNA, represent a special case. Pol II molecules have a physical footprint such that there is a minimum spacing between contiguous molecules on a gene. When the rate at which Pol II can attempt initiation is higher than the time required for the previously loaded Pol II to traverse the length of its footprint, the rate of Pol II clearance from the promoter becomes limiting. As a result of this “traffic jam” of Pol II molecules, the variance in Pol II loading will be smaller than its mean, resulting in a sub-Poissonian distribution [146] i.e. a deterministic promoter (Figure I). Finally, it is important to note that the simple reaction schemes discussed here represent only a small subset of all possible mechanisms of transcription initiation. More complex regulatory behavior, such as the regulation of transcriptional elongation [147], can lead to an increase or decrease of the variance with respect to the mean [23, 146]. As a result, making claims about regulatory mechanisms from the magnitude of the variance with respect to the mean are generally insufficient. Uncovering the intricacies of transcriptional cascades in vivo requires experimentation wherein regulatory parameters are varied systematically and their effect on the variance is measured [110].
Figure I. Variability of Pol II gene loading and molecular mechanisms of transcriptional initiation. Different mechanisms of promoter clearance by Pol II and transcriptional initiation lead to qualitatively different distributions of Pol II molecules along the gene.In the Poisson promoter Pol II molecules escape the promoter at a constant rate. In the two-state promoter the promoter switches between an ON and and OFF state. Only when the promoter is in the ON state do Pol II molecules initiate transcription. In the deterministic promoter the rate of Pol II loading is higher than the time it takes for the previous Pol II to escape the promoter leading to a chain of closely spaced Pol II molecules along the gene.
One simple mechanism to invoke this increased variance is that of a promoter that can switch between an ON and an OFF state (Box 2). This model predicts that mRNA molecules are produced in “bursts,” increasing the variability in the mRNA output, which is consistent with observations made for single-cell organisms [96]. In this model, repressor molecules can switch the promoter to the OFF state, and the action of activators increases the probability of finding the promoter in the ON state. Further, a saturating concentration of activator effectively fixes the promoter in the ON state. This reverts expression to the simple case of a promoter producing mRNA at a constant rate with Poissonian fluctuations.
This simple prediction has been tested and falsified in the context of the early embryo: even at saturating concentrations of activating factor(s), where gene expression output is maximal, the observed variance of several early expressed genes is more than two times higher than the mean [34]. This argues for the presence of further “switching” steps, even for fully activated genes, that both limit the maximum achievable magnitude of gene expression and constrain the degree of attainable precision. The molecular identity of these mechanisms is unclear, but it could be related to nucleosome remodeling, changes in chromatin state, Pol II promoter proximal pausing or stochastic processes of enhancer-promoter looping [111–114] (Figure 3B). Recent theoretical work demonstrates that some molecular mechanisms can reduce variability instead of increase it [23]. As a result, even variability consistent with a Poisson mechanism can result from the interplay between noise-increasing and noise-decreasing mechanisms. Though more theoretical and experimental work is required, this example demonstrates that the embryo is now poised to reveal fundamental mechanisms behind transcriptional regulation with a degree of quantitative rigor previously reserved for its single-celled counterparts.
iii. Dynamic strategies for pattern formation
Most of our knowledge of transcriptional decisions in development stems from examining fixed tissue. Extracting dynamic parameters from such snapshots depends on the adoption of a specific model. The model of promoter ON/OFF switching put forth above has been explicitly tested in some cases in single cells [35, 96, 105, 115, 116], but not yet in the context of the development of a multicellular organism. Recent work based on fixed tissue has also suggested that promoters enriched for paused Pol II can turn on in a more synchronous fashion than those promoters that do not exhibit Pol II pausing [91, 114]. Additionally, using similar techniques the existence of “memory” has been proposed in the regulation of transcription throughout the developmental cascade in the early embryo [92]. All these claims are of a dynamical nature and can be best tested by accessing transcriptional dynamics in a living embryo.
The genetically encoded MS2 system in combination with live imaging, as described above, should provide such access to the dynamics of developmental decisions. For example, this method has shown that the final structure of gene expression patterns arises from multiple independent transcriptional decisions [102, 103]. A given gene locus of a hunchback reporter first determines whether to enter the ON or OFF state in a random manner, biased by the local concentration of activator. The ON/OFF decision is subsequently maintained throughout interphase, so that an inactive locus remains inactive continuously. Loci that enter the ON state modulate their rate of transcription initiation by, once again, reading out the local concentration of the activator. Thus, the resulting pattern of gene expression emerges from the combination of decisions of whether to express a gene and, if so, at what level of activity (Figure 3C). Such a finding cannot be derived from fixed analysis alone since only living embryos reveal whether and how often switching occurs between active and inactive states.
The molecular origin of the random decision to turn a gene locus ON or OFF remains elusive. Interestingly, in previous nuclear cycles all loci of this hunchback reporter are active (or in the ON state), indicating that this random decision is only made in the third hour of development. This temporal change in the behavior of nuclear activity suggests the presence of a new regulatory landscape as a result of, for example, the action of newly expressed zygotic repressors or change in local chromatin modification [117–120]. A new set of mutant screens for the involved molecular players will be necessary and these efforts will present the opportunity to shed light on the role of chromatin conformation in transcriptional regulation in single cells, a process that had been previously mostly assessed in bulk experiments [26, 121, 122]
4) From Genome to Form
By taking advantage of the methods described above, the examples we have presented make it clear that fly embryos present new opportunities for investigating metazoan transcription. As such we are undoubtedly at the brink of a new set of mechanistic insights into the processes that ensure the presence of the necessary amount of gene product at the correct place and time in the developing embryo and beyond. The study of transcription in an intact developing organism opens new possibilities that simply cannot be addressed in unicellular systems. For example, one striking aspect of development is the generation of diverse morphologies from a limited number of network components and signaling systems [38, 123, 124]. Can any aspect of form be predicted by the linear string of nucleotides [125]? This question can now be addressed quantitatively by asking how expression rates change and expression boundaries shift as a function of sequence alterations to enhancer and promoter elements. This effort becomes all the more tractable given recent technological developments such as BAC transgenesis [126] and CRISPR-mediated genome modification [127] that allow such alterations to be made at endogenous loci. In addition, the insertion of stem-loop sequences into endogenous genes for live imaging bypasses the need to construct reporter transgenes, minimizing position effects that are often observed in transgenesis experiments [128]. Such experiments will quantitatively address, for example, the role of “shadow” enhancers (secondary enhancers that work together with a primary enhancer, which typically has been discovered first) functioning as redundant systems that ensure correct gene expression patterns [129–131].
Additionally, the last few years have resulted in an accumulated body of evidence for the role of nucleosome positioning, chromatin conformation, and modifications in modulating transcriptional activity in a plethora of developmental contexts [10, 132–135]. Current technology, however, has only allowed for the observation of these processes in bulk, often leading to the loss of the spatiotemporal information behind them. The ability to access transcriptional decisions in space and time with high precision in the context of the embryo laboratory provides the exciting prospect of the development of new technologies aimed at visualizing enhancer-promoter looping [136] and its coupling to DNA accessibility and chromatin interactions [137] in real time [138].
Finally, an exciting possibility is that of simultaneously labeling multiple network components to assess regulatory relationships and correlations in expression dynamics. For example, a live mRNA labeling system based on the PP7 bacteriophage can be combined with MS2 to observe the simultaneous expression of two genes using different colored coat protein fusions [139]. Prior observations of co-expressed gene products have relied on fixed material [3, 34]. Data from such experiments have given rise to models of cross-regulatory relationships between pairs of genes expressed in spatially overlapping domains. Such models generally propose that reproducible positioning of expression boundaries emerges largely from interactions across expression borders [7, 63, 140–144]. These models thus provide predictions (either qualitative or quantitative) about how expression rates will change as zygotic gene products accumulate at different locations in the embryo. The ability to monitor transcription in real time will provide the first rigorous quantitative test of these models. Because expression patterns emerge from numerous interactions of many activators and repressors at gene loci, experiments of this type will at long last allow researchers to ascertain the contribution of such interactions to boundary formation. By combining the above methods to measure expression rates with genetic manipulations of transcription factor activity, it will now become possible to determine how each activating or repressing factor contributes to expression rates, and thus to the dynamics of pattern formation, for any target gene of interest.
These are early days for the quantitative study of transcription in the context of developmental programs. A revolution in our understanding of the microscopic processes leading to cellular decision-making has been spearheaded by studies in cultured cells over the last 15 years. The novel experimental methods to quantify and manipulate transcription in embryos combined with theoretical models aimed at predicting regulatory behavior have the potential of furthering this revolution and bringing it to the forefront of metazoan evolution.
Highlights.
-
-
Early Drosophila embryos possess many features advantageous to studying transcription
-
-
New methods to measure gene expression developed in cultured organisms now allow quantification in early fly embryos
-
-
Protein and mRNA expression, transcription rates, and RNA polymerase dynamics can be measured in terms of absolute molecular numbers
-
-
Recent studies reveal features of transcriptional regulation specific to the fly and universal between organisms
Acknowledgements
We thank J. Bothma, H. Grabmayr, W. Li, and A. Sgro for comments on the manuscript. This review does not aim to be a comprehensive survey of the literature. Rather, it is a reflection of some of our favorite examples, which we believe to be illustrative of this new wave of quantitative dissection of developmental decisions in the early fly embryo. We apologize to the community if we omitted some pertinent references. This work was supported by National Institutes of Health Grant P50 GM071508 and R01 GM097275, and by Searle Scholar Award 10-SSP-274 to T.G. H.G.G. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund and a Princeton Dicke Fellowship.
Glossary
- Zygotic genes
Genes whose expression occurs in the embryo (i.e. the zygote), not during oogenesis in the female
- Oogenesis
Process of egg formation in the female (lasts 2–3 days in Drosophila melanogaster)
- Segmentation
Process of generating body segments
- Syncytium
Cell with many nuclei that are not separated by cell membranes
- Maternal gradients
Protein concentration gradients in the embryo that are set up by the female during oogenesis
- Gene expression
Act of generating gene products, such as mRNA and protein; often stands for the amount of mRNA or protein present in a particular cell
- Gap genes
Class of genes that orchestrate anterior-posterior patterning in the early Drosophila embryo (see Box 1); typical expression pattern has 2–3 broad domains during nuclear cycle 14
- Pair-rule genes
Class of genes that orchestrate anterior-posterior patterning in the early Drosophila embryo (see Box 1); typical expression pattern is 7 stripes during mid-nuclear cycle 14
- Expression noise
Stochastic fluctuations in mRNA or protein expression levels; typically reported as a coefficient of variation, i.e. the ratio between standard deviation and mean expression level
- Gene regulatory network
Gene products that can regulate the activity of other genes; genes are nodes and interactions between genes are links of the network
- Intrinsic noise
Gene-intrinsic fluctuations in gene expression levels, attributed to the stochastic nature of the biochemical processes in transcription such as Pol II binding to the DNA
- Extrinsic noise
Gene-independent fluctuations in gene expression levels, attributed to environmental fluctuations such as the number of available Pol II molecules per nucleus
- smFISH
Single molecule fluorescence in situ hybridization is an mRNA labeling technique that uses fluorescently-tagged complementary DNA probes that bind mRNA molecules in fixed tissue. These DNA probes can be bound to a fluorophore for direct detection or to a molecule that is then detected using antibodies
- MS2-system
RNA stem loops that are bound by complementary coat proteins
- Antibody staining
Technique to label proteins in fixed tissue with fluorescently tagged antibodies
- Transcription factors
Proteins that bind to promoters and enhancers to control gene expression
- Enhancers
Regulatory DNA regions that control the expression of nearby genes
- Polymerase (Pol) II
Molecular complex that binds promoters on the DNA and transcribes DNA into mRNA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Balazsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144(6):910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gregor T, et al. Probing the limits to positional information. Cell. 2007;130(1):153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubuis JO, Samanta R, Gregor T. Accurate measurements of dynamics and reproducibility in small genetic networks. Mol Syst Biol. 2013;9:639. doi: 10.1038/msb.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crauk O, Dostatni N. Bicoid determines sharp and precise target gene expression in the Drosophila embryo. Curr Biol. 2005;15(21):1888–1898. doi: 10.1016/j.cub.2005.09.046. [DOI] [PubMed] [Google Scholar]
- 5.Pare A, et al. Visualization of individual Scr mRNAs during Drosophila embryogenesis yields evidence for transcriptional bursting. Curr Biol. 2009;19(23):2037–2042. doi: 10.1016/j.cub.2009.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaeger J, et al. Dynamic control of positional information in the early Drosophila embryo. Nature. 2004;430(6997):368–371. doi: 10.1038/nature02678. [DOI] [PubMed] [Google Scholar]
- 7.Manu, et al. Canalization of gene expression in the Drosophila blastoderm by gap gene cross regulation. PLoS Biol. 2009;7(3):e1000049. doi: 10.1371/journal.pbio.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu F, Morrison AH, Gregor T. Dynamic interpretation of maternal inputs by the Drosophila segmentation gene network. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1220912110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20(17):R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li LM, Arnosti DN. Long- and short-range transcriptional repressors induce distinct chromatin states on repressed genes. Curr Biol. 2011;21(5):406–412. doi: 10.1016/j.cub.2011.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lagha M, Bothma JP, Levine M. Mechanisms of transcriptional precision in animal development. Trends Genet. 2012;28(8):409–416. doi: 10.1016/j.tig.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaeger J, Manu, Reinitz J. Drosophila blastoderm patterning. Curr Opin Genet Dev. 2012;22(6):533–541. doi: 10.1016/j.gde.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Knowles DW, Biggin MD. Building quantitative, three-dimensional atlases of gene expression and morphology at cellular resolution. Wiley Interdiscip Rev Dev Biol. 2013;2(6):767–779. doi: 10.1002/wdev.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gergen JP, Coulter D, Wieschaus E. Segmental pattern and blastoderm cell identities. In: Gall JG, editor. Gametogenesis and the Early Embryo. New York: Liss Inc; 1986. pp. 195–220. [Google Scholar]
- 15.Kornberg TB, Tabata T. Segmentation of the Drosophila embryo. Curr Opin Genet Dev. 1993;3(4):585–594. doi: 10.1016/0959-437x(93)90094-6. [DOI] [PubMed] [Google Scholar]
- 16.Lecuit T, Samanta R, Wieschaus E. slam encodes a developmental regulator of polarized membrane growth during cleavage of the Drosophila embryo. Dev Cell. 2002;2(4):425–436. doi: 10.1016/s1534-5807(02)00141-7. [DOI] [PubMed] [Google Scholar]
- 17.Petkova MD, et al. Maternal origins of developmental reproducibility. Curr Biol. 2014;24(11):1283–1288. doi: 10.1016/j.cub.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenfeld N, et al. Gene regulation at the single-cell level. Science. 2005;307(5717):1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 19.Hensel Z, et al. Stochastic expression dynamics of a transcription factor revealed by single-molecule noise analysis. Nat Struct Mol Biol. 2012;19(8):797–802. doi: 10.1038/nsmb.2336. [DOI] [PubMed] [Google Scholar]
- 20.Geiler-Samerotte KA, et al. The details in the distributions: why and how to study phenotypic variability. Curr Opin Biotechnol. 2013;24(4):752–759. doi: 10.1016/j.copbio.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia T, Li N, Fang X. Single-molecule fluorescence imaging in living cells. Annu Rev Phys Chem. 2013;64:459–480. doi: 10.1146/annurev-physchem-040412-110127. [DOI] [PubMed] [Google Scholar]
- 22.Li GW, Xie XS. Central dogma at the single-molecule level in living cells. Nature. 2011;475(7356):308–315. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez A, Choubey S, Kondev J. Regulation of noise in gene expression. Annu Rev Biophys. 2013;42:469–491. doi: 10.1146/annurev-biophys-083012-130401. [DOI] [PubMed] [Google Scholar]
- 24.Bintu L, et al. Transcriptional regulation by the numbers: models. Curr Opin Genet Dev. 2005;15(2):116–124. doi: 10.1016/j.gde.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bintu L, et al. Transcriptional regulation by the numbers: applications. Curr Opin Genet Dev. 2005;15(2):125–135. doi: 10.1016/j.gde.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segal E, Widom J. From DNA sequence to transcriptional behaviour: a quantitative approach. Nat Rev Genet. 2009;10(7):443–456. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munsky B, Neuert G, van Oudenaarden A. Using gene expression noise to understand gene regulation. Science. 2012;336(6078):183–187. doi: 10.1126/science.1216379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coulon A, et al. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat Rev Genet. 2013;14(8):572–584. doi: 10.1038/nrg3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elowitz MB, et al. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 30.Raser JM, O'Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309(5743):2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441(7095):840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 32.Cohen AA, et al. Protein dynamics in individual human cells: experiment and theory. PLoS One. 2009;4(4):e4901. doi: 10.1371/journal.pone.0004901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329(5991):533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Little SC, Tikhonov M, Gregor T. Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell. 2013;154(4):789–800. doi: 10.1016/j.cell.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez A, Golding I. Genetic determinants and cellular constraints in noisy gene expression. Science. 2013;342(6163):1188–1193. doi: 10.1126/science.1242975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaeger J. The gap gene network. Cell Mol Life Sci. 2011;68(2):243–274. doi: 10.1007/s00018-010-0536-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papatsenko D. Stripe formation in the early fly embryo: principles, models, and networks. Bioessays. 2009;31(11):1172–1180. doi: 10.1002/bies.200900096. [DOI] [PubMed] [Google Scholar]
- 38.Rosenberg MI, Lynch JA, Desplan C. Heads and tails: evolution of anteroposterior patterning in insects. Biochim Biophys Acta. 2009;1789(4):333–342. doi: 10.1016/j.bbagrm.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warn R, Bullard B, Maleki S. Myosin as a constituent of the Drosophila egg cortex. Nature. 1979;278(5705):651–653. doi: 10.1038/278651a0. [DOI] [PubMed] [Google Scholar]
- 40.Carroll SB, Scott MP. Zygotically Active Genes That Affect the Spatial Expression of the Fushi-Tarazu Segmentation Gene during Early Drosophila Embryogenesis. Cell. 1986;45(1):113–126. doi: 10.1016/0092-8674(86)90543-x. [DOI] [PubMed] [Google Scholar]
- 41.Frasch M, Levine M. Complementary patterns of even-skipped and fushi tarazu expression involve their differential regulation by a common set of segmentation genes in Drosophila. Genes Dev. 1987;1(9):981–995. doi: 10.1101/gad.1.9.981. [DOI] [PubMed] [Google Scholar]
- 42.Stanojevic D, Hoey T, Levine M. Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Kruppel in Drosophila. Nature. 1989;341(6240):331–335. doi: 10.1038/341331a0. [DOI] [PubMed] [Google Scholar]
- 43.Driever W, Nusslein-Volhard C. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature. 1989;337(6203):138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- 44.Rivera-Pomar R, et al. Activation of posterior gap gene expression in the Drosophila blastoderm. Nature. 1995;376(6537):253–256. doi: 10.1038/376253a0. [DOI] [PubMed] [Google Scholar]
- 45.Hou XS, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84(3):411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- 46.Yan R, et al. Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84(3):421–430. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- 47.Liang HL, et al. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456(7220):400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carroll SB, Vavra SH. The zygotic control of Drosophila pair-rule gene expression. II. Spatial repression by gap and pair-rule gene products. Development. 1989;107(3):673–683. doi: 10.1242/dev.107.3.673. [DOI] [PubMed] [Google Scholar]
- 49.Hulskamp M, Pfeifle C, Tautz D. A morphogenetic gradient of hunchback protein organizes the expression of the gap genes Kruppel and knirps in the early Drosophila embryo. Nature. 1990;346(6284):577–580. doi: 10.1038/346577a0. [DOI] [PubMed] [Google Scholar]
- 50.Kraut R, Levine M. Mutually repressive interactions between the gap genes giant and Kruppel define middle body regions of the Drosophila embryo. Development. 1991;111(2):611–621. doi: 10.1242/dev.111.2.611. [DOI] [PubMed] [Google Scholar]
- 51.Steingrimsson E, et al. Dual Role of the Drosophila Pattern Gene Tailless in Embryonic Termini. Science. 1991;254(5030):418–421. doi: 10.1126/science.1925599. [DOI] [PubMed] [Google Scholar]
- 52.Fowlkes CC, et al. A quantitative spatiotemporal atlas of gene expression in the Drosophila blastoderm. Cell. 2008;133(2):364–374. doi: 10.1016/j.cell.2008.01.053. [DOI] [PubMed] [Google Scholar]
- 53.Surkova S, et al. Quantitative dynamics and increased variability of segmentation gene expression in the Drosophila Kruppel and knirps mutants. Dev Biol. 2013;376(1):99–112. doi: 10.1016/j.ydbio.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 54.Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- 55.Di Talia S, et al. Posttranslational control of Cdc25 degradation terminates Drosophila's early cell-cycle program. Curr Biol. 2013;23(2):127–132. doi: 10.1016/j.cub.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clarkson M, Saint R. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 1999;18(6):457–462. doi: 10.1089/104454999315178. [DOI] [PubMed] [Google Scholar]
- 57.Mason FM, Tworoger M, Martin AC. Apical domain polarization localizes actin-myosin activity to drive ratchet-like apical constriction. Nat Cell Biol. 2013;15(8):926–936. doi: 10.1038/ncb2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morin X, et al. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci U S A. 2001;98(26):15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeLotto R, et al. Nucleocytoplasmic shuttling mediates the dynamic maintenance of nuclear Dorsal levels during Drosophila embryogenesis. Development. 2007;134(23):4233–4241. doi: 10.1242/dev.010934. [DOI] [PubMed] [Google Scholar]
- 60.Gregor T, et al. Stability and nuclear dynamics of the bicoid morphogen gradient. Cell. 2007;130(1):141–152. doi: 10.1016/j.cell.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gregor T, McGregor AP, Wieschaus EF. Shape and function of the Bicoid morphogen gradient in dipteran species with different sized embryos. Dev Biol. 2008;316(2):350–358. doi: 10.1016/j.ydbio.2008.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kanodia JS, et al. Dynamics of the Dorsal morphogen gradient. Proc Natl Acad Sci U S A. 2009;106(51):21707–21712. doi: 10.1073/pnas.0912395106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reeves GT, et al. Dorsal-ventral gene expression in the Drosophila embryo reflects the dynamics and precision of the dorsal nuclear gradient. Dev Cell. 2012;22(3):544–557. doi: 10.1016/j.devcel.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abu-Arish A, et al. High mobility of bicoid captured by fluorescence correlation spectroscopy: implication for the rapid establishment of its gradient. Biophys J. 2010;99(4):L33–L35. doi: 10.1016/j.bpj.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grimm O, Wieschaus E. The Bicoid gradient is shaped independently of nuclei. Development. 2010;137(17):2857–2862. doi: 10.1242/dev.052589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drocco JA, et al. Measurement and perturbation of morphogen lifetime: effects on gradient shape. Biophys J. 2011;101(8):1807–1815. doi: 10.1016/j.bpj.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Little SC, et al. The formation of the Bicoid morphogen gradient requires protein movement from anteriorly localized mRNA. PLoS Biol. 2011;9(3):e1000596. doi: 10.1371/journal.pbio.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kicheva A, et al. Investigating the principles of morphogen gradient formation: from tissues to cells. Curr Opin Genet Dev. 2012;22(6):527–532. doi: 10.1016/j.gde.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 69.Wartlick O, et al. Dynamics of Dpp signaling and proliferation control. Science. 2011;331(6021):1154–1159. doi: 10.1126/science.1200037. [DOI] [PubMed] [Google Scholar]
- 70.Kornberg TB. The imperatives of context and contour for morphogen dispersion. Biophys J. 2012;103(11):2252–2256. doi: 10.1016/j.bpj.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alexandre C, Baena-Lopez A, Vincent JP. Patterning and growth control by membrane-tethered Wingless. Nature. 2014;505(7482):180–185. doi: 10.1038/nature12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hammar P, et al. The lac repressor displays facilitated diffusion in living cells. Science. 2012;336(6088):1595–1598. doi: 10.1126/science.1221648. [DOI] [PubMed] [Google Scholar]
- 73.Cubitt AB, et al. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20(11):448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 74.Ludwig MZ, et al. Consequences of eukaryotic enhancer architecture for gene expression dynamics, development, and fitness. PLoS Genet. 2011;7(11):e1002364. doi: 10.1371/journal.pgen.1002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berezhkovskii AM, Shvartsman SY. On the GFP-Based Analysis of Dynamic Concentration Profiles. Biophys J. 2014;106(3):L13–L15. doi: 10.1016/j.bpj.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Edgar BA, et al. Repression and turnover pattern fushi tarazu RNA in the early Drosophila embryo. Cell. 1986;47(5):747–754. doi: 10.1016/0092-8674(86)90517-9. [DOI] [PubMed] [Google Scholar]
- 77.Jackle H, et al. Cross-regulatory interactions among the gap genes of Drosophila. Nature. 1986;324(18):668–670. [Google Scholar]
- 78.Driever W, Thoma G, Nusslein-Volhard C. Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity of binding sites for the bicoid morphogen. Nature. 1989;340(6232):363–367. doi: 10.1038/340363a0. [DOI] [PubMed] [Google Scholar]
- 79.Schroder C, et al. Differential regulation of the two transcripts from the Drosophila gap segmentation gene hunchback. Embo J. 1988;7(9):2881–2887. doi: 10.1002/j.1460-2075.1988.tb03145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiang J, et al. The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes Dev. 1991;5(10):1881–1891. doi: 10.1101/gad.5.10.1881. [DOI] [PubMed] [Google Scholar]
- 81.Pankratz MJ, et al. Spatial control of the gap gene knirps in the Drosophila embryo by posterior morphogen system. Science. 1992;255(5047):986–989. doi: 10.1126/science.1546296. [DOI] [PubMed] [Google Scholar]
- 82.Stanojevic D, Small S, Levine M. Regulation of a Segmentation Stripe by Overlapping Activators and Repressors in the Drosophila Embryo. Science. 1991;254(5036):1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- 83.Clyde DE, et al. A self-organizing system of repressor gradients establishes segmental complexity in Drosophila. Nature. 2003;426(6968):849–853. doi: 10.1038/nature02189. [DOI] [PubMed] [Google Scholar]
- 84.Boettiger AN, Levine M. Rapid Transcription Fosters Coordinate snail Expression in the Drosophila Embryo. Cell Rep. 2013;3(1):8–15. doi: 10.1016/j.celrep.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Femino AM, et al. Visualization of single RNA transcripts in situ. Science. 1998;280(5363):585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 86.Raj A, et al. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4(10):e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15(12):1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Raj A, et al. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463(7283):913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shermoen AW, O'Farrell PH. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67(2):303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wilkie GS, et al. Transcribed genes are localized according to chromosomal position within polarized Drosophila embryonic nuclei. Curr Biol. 1999;9(21):1263–1266. doi: 10.1016/s0960-9822(99)80509-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325(5939):471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Porcher A, et al. The time to measure positional information: maternal hunchback is required for the synchrony of the Bicoid transcriptional response at the onset of zygotic transcription. Development. 2010;137(16):2795–2804. doi: 10.1242/dev.051300. [DOI] [PubMed] [Google Scholar]
- 93.Perry MW, et al. Precision of hunchback expression in the Drosophila embryo. Curr Biol. 2012;22(23):2247–2252. doi: 10.1016/j.cub.2012.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McHale P, et al. Gene length may contribute to graded transcriptional responses in the Drosophila embryo. Dev Biol. 2011;360(1):230–240. doi: 10.1016/j.ydbio.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fusco D, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13(2):161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Golding I, et al. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123(6):1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 97.Larson DR, et al. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332(6028):475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lionnet T, et al. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods. 2011;8(2):165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bertrand E, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2(4):437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 100.Urbinati CR, Long RM. Techniques for following the movement of single RNAs in living cells. Wiley Interdiscip Rev RNA. 2011;2(4):601–609. doi: 10.1002/wrna.83. [DOI] [PubMed] [Google Scholar]
- 101.Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13(14):1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 102.Garcia HG, et al. Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr Biol. 2013;23(21):2140–2145. doi: 10.1016/j.cub.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lucas T, et al. Live imaging of bicoid-dependent transcription in Drosophila embryos. Curr Biol. 2013;23(21):2135–2139. doi: 10.1016/j.cub.2013.08.053. [DOI] [PubMed] [Google Scholar]
- 104.Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316(5828):1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.So LH, et al. General properties of transcriptional time series in Escherichia coli. Nat Genet. 2011;43(6):554–560. doi: 10.1038/ng.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Perez-Ortin JE, et al. Eukaryotic mRNA decay: methodologies, pathways, and links to other stages of gene expression. J Mol Biol. 2013;425(20):3750–3775. doi: 10.1016/j.jmb.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 107.Edgar BA, Datar SA. Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila's early cell cycle program. Genes Dev. 1996;10(15):1966–1977. doi: 10.1101/gad.10.15.1966. [DOI] [PubMed] [Google Scholar]
- 108.Perry MW, Boettiger AN, Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc Natl Acad Sci U S A. 2011;108(33):13570–13575. doi: 10.1073/pnas.1109873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sun M, et al. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res. 2012;22(7):1350–1359. doi: 10.1101/gr.130161.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sanchez A, et al. Effect of promoter architecture on the cell-to-cell variability in gene expression. PLoS Comput Biol. 2011;7(3):e1001100. doi: 10.1371/journal.pcbi.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Buecker C, Wysocka J. Enhancers as information integration hubs in development: lessons from genomics. Trends Genet. 2012;28(6):276–284. doi: 10.1016/j.tig.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Krivega I, Dean A. Enhancer and promoter interactions-long distance calls. Curr Opin Genet Dev. 2012;22(2):79–85. doi: 10.1016/j.gde.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Petesch SJ, Lis JT. Overcoming the nucleosome barrier during transcript elongation. Trends Genet. 2012;28(6):285–294. doi: 10.1016/j.tig.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lagha M, et al. Paused Pol II Coordinates Tissue Morphogenesis in the Drosophila Embryo. Cell. 2013;153(5):976–987. doi: 10.1016/j.cell.2013.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chubb JR, et al. Transcriptional pulsing of a developmental gene. Curr Biol. 2006;16(10):1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yunger S, et al. Single-allele analysis of transcription kinetics in living mammalian cells. Nat Methods. 2010;7(8):631–633. doi: 10.1038/nmeth.1482. [DOI] [PubMed] [Google Scholar]
- 117.Ahmad K, Henikoff S. Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell. 2001;104(6):839–847. doi: 10.1016/s0092-8674(01)00281-1. [DOI] [PubMed] [Google Scholar]
- 118.Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136(18):3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- 119.Bai L, Ondracka A, Cross FR. Multiple sequence-specific factors generate the nucleosome-depleted region on CLN2 promoter. Mol Cell. 2011;42(4):465–476. doi: 10.1016/j.molcel.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen H, et al. A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes. Cell. 2012;149(3):618–629. doi: 10.1016/j.cell.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Naumova N, Dekker J. Integrating one-dimensional and three-dimensional maps of genomes. J Cell Sci. 2010;123(Pt 12):1979–1988. doi: 10.1242/jcs.051631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2014;15(2):69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu PZ, Kaufman TC. Short and long germ segmentation: unanswered questions in the evolution of a developmental mode. Evol Dev. 2005;7(6):629–646. doi: 10.1111/j.1525-142X.2005.05066.x. [DOI] [PubMed] [Google Scholar]
- 124.Perry MW, et al. Evolution of insect dorsoventral patterning mechanisms. Cold Spring Harb Symp Quant Biol. 2009;74:275–279. doi: 10.1101/sqb.2009.74.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Stern DL, Orgogozo V. The loci of evolution: how predictable is genetic evolution? Evolution. 2008;62(9):2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Venken KJ, et al. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314(5806):1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 127.Bassett AR, et al. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4(1):220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Markstein M, et al. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40(4):476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Frankel N, et al. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466(7305):490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Perry MW, et al. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20(17):1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Barolo S. Shadow enhancers: frequently asked questions about distributed cisregulatory information and enhancer redundancy. Bioessays. 2011;34(2):135–141. doi: 10.1002/bies.201100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463(7280):474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Van Nostrand EL, Kim SK. Seeing elegance in gene regulatory networks of the worm. Curr Opin Genet Dev. 2011;21(6):776–786. doi: 10.1016/j.gde.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vastenhouw NL, Schier AF. Bivalent histone modifications in early embryogenesis. Curr Opin Cell Biol. 2012;24(3):374–386. doi: 10.1016/j.ceb.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.White R. Packaging the fly genome: domains and dynamics. Brief Funct Genomics. 2012;11(5):347–355. doi: 10.1093/bfgp/els020. [DOI] [PubMed] [Google Scholar]
- 136.Ronshaugen M, Levine M. Visualization of trans-homolog enhancer-promoter interactions at the Abd-B Hox locus in the Drosophila embryo. Dev Cell. 2004;7(6):925–932. doi: 10.1016/j.devcel.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 137.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2014;503(7475):290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fisher JK, et al. Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell. 2013;153(4):882–895. doi: 10.1016/j.cell.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hocine S, et al. Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat Methods. 2013;10(2):119–121. doi: 10.1038/nmeth.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jaeger J. Modelling the Drosophila embryo. Mol Biosyst. 2009;5(12):1549–1568. doi: 10.1039/b904722k. [DOI] [PubMed] [Google Scholar]
- 141.Bieler J, Pozzorini C, Naef F. Whole-embryo modeling of early segmentation in Drosophila identifies robust and fragile expression domains. Biophys J. 2011;101(2):287–296. doi: 10.1016/j.bpj.2011.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kicheva A, Cohen M, Briscoe J. Developmental pattern formation: insights from physics and biology. Science. 2012;338(6104):210–212. doi: 10.1126/science.1225182. [DOI] [PubMed] [Google Scholar]
- 143.Kozlov K, et al. Modeling of gap gene expression in Drosophila Kruppel mutants. PLoS Comput Biol. 2012;8(8):e1002635. doi: 10.1371/journal.pcbi.1002635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sokolowski TR, Erdmann T, ten Wolde PR. Mutual repression enhances the steepness and precision of gene expression boundaries. PLoS Comput Biol. 2012;8(8):e1002654. doi: 10.1371/journal.pcbi.1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kaern M, et al. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6(6):451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 146.Boettiger AN. Analytic approaches to stochastic gene expression in multicellular systems. Biophys J. 2013;105(12):2629–2640. doi: 10.1016/j.bpj.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]