Abstract
Background & Aims
Alcohol abuse is a major cause of liver injury. The pathologic features of alcoholic liver disease develop over prolonged periods, yet the cellular defense mechanisms against the detrimental effects of alcohol are not well understood. We investigated whether macroautophagy, an evolutionarily conserved cellular mechanism that is commonly activated in response to stress, could protect liver cells from ethanol toxicity.
Methods
Mice were acutely given ethanol by gavage. The effects of ethanol on primary hepatocytes and hepatic cell lines were studied in vitro.
Results
Ethanol-induced macroautophagy in the livers of mice and cultured cells required ethanol metabolism, generated reactive oxygen species, and inhibited mTOR signaling. Suppression of macroautophagy with pharmacological agents or small interfering RNAs significantly increased hepatocyte apoptosis and liver injury; macroautophagy therefore protected cells from the toxic effects of ethanol. Macroautophagy induced by ethanol seemed to be selective for cells with damaged mitochondria and accumulated lipid droplets, but not long-lived proteins, which could account for its protective effects. Increasing macroautophagy pharmacologically reduced hepatotoxicity and steatosis associated with acute ethanol exposure.
Conclusions
Macroautophagy protects against ethanol-induced toxicity in livers of mice. Reagents that modify macroautophagy might be developed as therapeutics for patients with alcoholic liver disease.
Keywords: GFP-LC3, bodipy staining, autophagy flux, long-lived protein degradation
Introduction
Alcohol is widely consumed around the world. While a moderate use of alcohol may be beneficial to human health, excessive drinking, whether in a binge or chronic fashion, can lead to considerable health concerns. Binge drinking can cause acute glycogen depletion, hypoglycemia and acidosis in addition to behavioral and psychological changes1. At the cellular level, binge drinking results in mitochondria damage, generation of free radicals, inhibition of insulin signaling, and steatosis, although cell death is usually minor unless other susceptibility factors exist2–7. In chronic ethanol abuse, the histological manifestation of the liver injury can progress from steatosis to focal inflammation, focal necrosis, fibrosis and cirrhosis, resulting in alcoholic liver disease1, 2, 5.
Ethanol pathogenesis is contributed by multiple factors1, 2, 5, 7, 8. Early events such as mitochondrial damage, ROS generation and steatosis seem to be the direct outcome of ethanol metabolism, which are common features of acute and chronic alcohol exposure. While greater efforts have been spent on the understanding of these mechanisms, little is known about the cellular protective mechanism against the detrimental effects of ethanol. We hypothesized that a cellular protective response to ethanol exposure is critical to the control of ethanol-induced liver pathology.
Among the potential protective mechanisms, we investigated whether macroautophagy could play a role in limiting ethanol-induced liver injury. Macroautophagy (hereafter referred to as autophagy) is a bulk intracellular degradation system responsible for the degradation of macromolecules and subcellular organelles9. Autophagy involves the formation of double-membraned autophagosomes, which envelop the substrates and fuse with the lysosomes for degradation. More than thirty Atg genes have been defined in yeast that participate in autophagy, many of which have mammalian homologues9. The functions of Atg8/LC3, Atg7, and Atg6/Beclin 1 are among the best characterized in mammalian cells. The Atg6/Beclin 1 forms a complex with the Class III phosphoinositide 3-kinase (PI3K) complex, VPS34 and VPS15, which leads to autophagy specific generation of phosphatidylinositol-3-phosphate (PtdIns3P). PtdIns3P is required for a series of events that ultimately contributes to the conjugation of Atg8/LC3 to phosphatidylethanolamine (PE) at the autophagosomal membranes, which also requires Atg7. PE-conjugated Atg8/LC3, also known as LC3-II (in contrast to the unlipidated LC3-I), is important in autophagosome formation9.
Autophagy is now known to be widely involved in the pathogenesis of many diseases and is activated under a variety of stress conditions. Autophagy seems to constitute an effective cellular defense system against multiple pathological insults. Thus, we investigated whether autophagy is activated in response to ethanol exposure and whether it plays a key role in mitigating ethanol-induced pathology. Using an ethanol binge model and primary hepatocyte culture in which the ethanol-induced early pathological events are well defined, we determined that ethanol could induce a selective autophagy process in primary hepatocytes dependent on its metabolism, ROS production and mTOR inhibition. Furthermore, autophagy protects hepatocytes against the detrimental effects of ethanol likely by removing damaged mitochondria and accumulated lipid droplets.
Materials and Methods
Animals experiments
Wild type C57BL/6 mice and GFP-LC3 transgenic mice10 were used in this study. All animals received humane care. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Pittsburgh.
Ethanol binge was conducted as previously described11. This model was designed to achieve blood alcohol levels, behavioral effects, and physiological effects comparable to human binge drinking. After 6 hours of fasting, male mice were given 33% (v/v) ethanol at a total accumulative dosage of 4.5 g/kg body weight by four equally divided gavages in 20-minute intervals. Control mice received the same volume of water. For autophagy inhibition or induction, chloroquine (CQ, 60 mg/kg) or rapamycin (2 mg/kg) were given (i.p.) to the mice 30 minutes before the administration of ethanol. Mice were analyzed 16 hours later. For inhibition of Atg7 expression in murine livers, siRNA (see supplemental information) was injected (0.7 nmol/g) through the tail vein using the hydrodynamic technique. Forty-eight hours later, the mice were subjected to the ethanol treatment.
Cell culture
Murine hepatocytes were isolated and cultured as previously described12. After initial culture in William’s medium E with 10% FCS for attachment, cells were cultured in serum-free William’s medium E overnight before treatment. HepG2 and VL-17A cells13 were maintained in DMEM with 10% FCS and other standard supplements. Ethanol and the following chemicals were applied as indicated in the figure legends: 3-MA (10 mM), CQ (10 µM), rapamycin (10 µM), 4-MP (4 mM), MnTBAP (1 mM) and NAC (20 mM).
Microscopic analysis
Cells (2 × 105/well in 12-well plate) were infected with adenoviral GFP-LC3 overnight or loaded with MitoTracker Red (50 nM) for 15 minutes before other treatments. Apoptotic cell death was determined by nuclear staining with Hoechst 33342 or TUNEL staining. Cryosection of GFP-LC3 transgenic livers were performed as previously described10. Sections were stained with Bodipy 581/591-C11 (1 µM) or Bodipy 493/503 (0.1 µM) for 15 minutes before analysis. All fluorescence images were digitally acquired with an Olympus Fluoview 1000 confocal microscope. Electron microscopy was conducted with standard procedures using a JEM 1016CX electron microscope.
Biochemical analysis
Serum ALT level was determined using a kit from Biotron Diagnostics (Hemet, CA). Hepatic triglyceride was determined as in reference14. Caspase-3 activity was determined using Ac-DEVD-AFC as the substrate12. For inhibition of Beclin 1 expression in VL-17A cells, siRNA (see supplemental information) were transfected into VL-17A cells at the concentration of 0.12 µM per 1×106 cells using Oligofectamine. Cells were subjected to other treatment 36 hours later. Densitometry of LC3-II and p62 bands was conducted and the values were normalized to the loading control (β-actin or LAMP1) and then converted to relative units to the untreated control. Autophagy flux was conducted as described in Table S1. Long-lived proteins degradation assay was performed as described previously9, 15.
Statistical analysis
Quantitative data are subjected to one way ANOVA with Holm-Sidak analysis, Student’s t test or z test where appropriate using SigmaStat 3.5 (Systat Software, Inc., San Jose, CA),
Results
Acute ethanol exposure induced autophagy
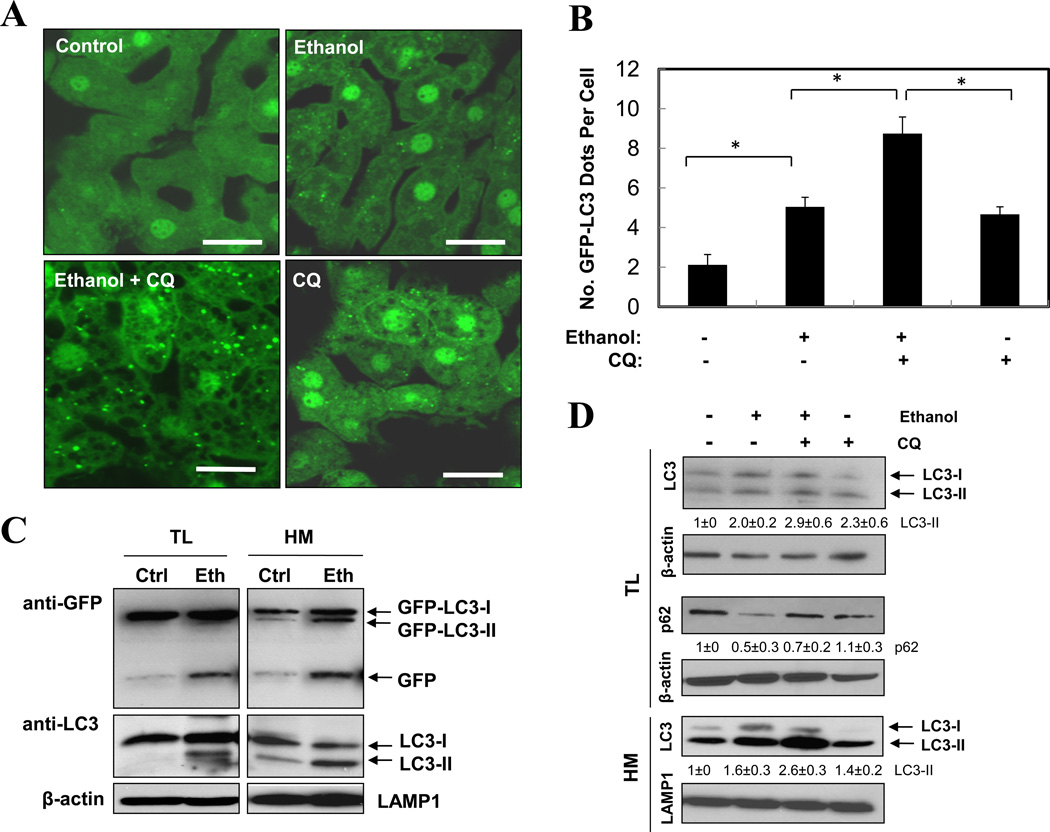
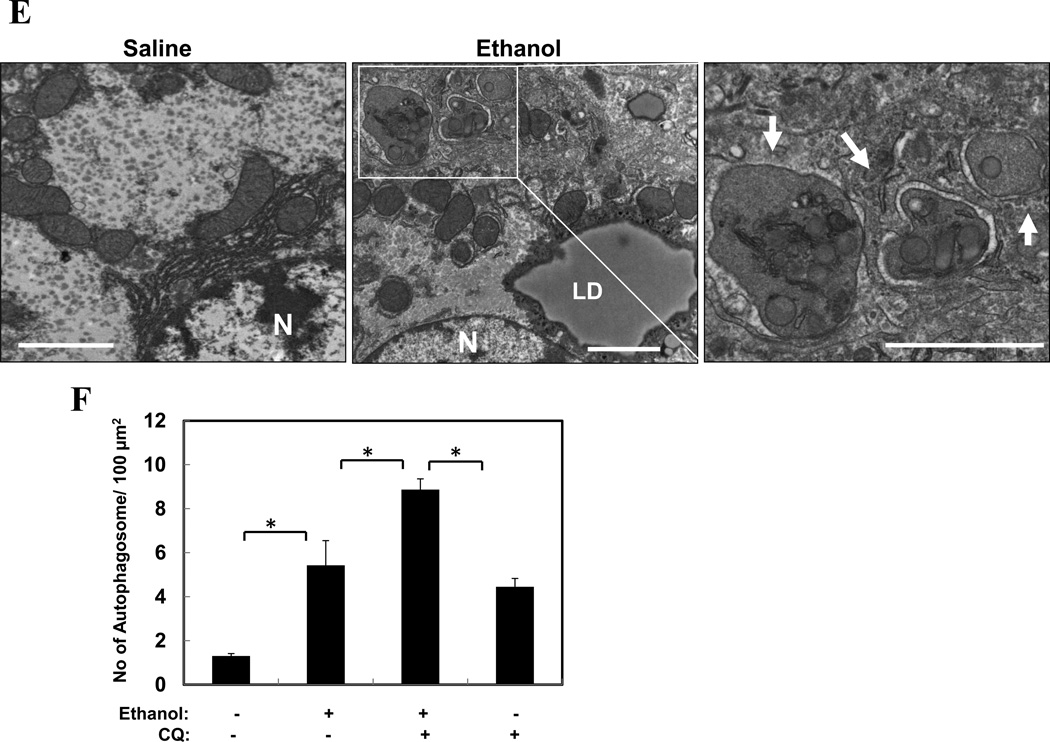
Activation of autophagy by ethanol was first examined in GFP-LC3 transgenic mice10. Binge ethanol treatment induced a significant elevation of GFP-LC3 puncta in the liver (Fig. 1A–B), which represented autophagosomes. Immunoblot analysis confirmed the increase of the membrane-associated PE-conjugated form of GFP-LC3 (GFP-LC3-II) and the endogenous LC3 (LC3-II) in ethanol-treated livers (Fig. 1C–D). The lipidated LC3 was particularly enriched in the LAMP1-positive heavy membrane fraction, where the lysosome was located, reflecting its destination as the autophagosome fused with the lysosome to form the degradative autolysosomes. Importantly, electron microscopic (EM) analysis indicated an apparent accumulation of autophagosomes following ethanol treatment (Fig. 1E–F), which could be suppressed by siRNA-mediated knockdown of Atg7 (see below).
Figure 1. Acute ethanol exposure induces autophagy in the liver.
(A–C). GFP-LC3 transgenic mice (n=3–5) were treated as indicated and the liver sections were analyzed by confocal microscopy (A). The number of GFP-LC3 dots (mean+SEM) were quantified from each animal (B). The total lysates (TL) and the heavy membrane fraction (HM) of the liver were analyzed by immunoblot assay (C). (D) Wild type mice were treated as indicated (n=3) and the liver fractions were analyzed by immunoblot assay. Densitometry analysis of the LC3-II and p62 was performed for each sample (mean+SEM). (E). Wild type liver samples were processed for EM. Arrows denote autophagosomes. N: nucleus, LD: lipid droplet. (F). The number of autophagosomes per given area (mean+SEM) was determined (n=3–4). Scale bar: 20 µm (A), 2 µm (E). *: p<0.05.
Pre-treatment of mice with the lysosome-inhibitor chloroquine (CQ) further increased the levels of GFP-LC3 puncta (Fig. 1A–B), the LC3-II form (Fig. 1D), and the number of autophagosomes (Fig. 1F), suggesting that at least a portion of autophagosomes resulted from this elevation was degraded in the lysosome. The appearance of the single GFP moiety (Fig. 1C), due to the breakdown of GFP-LC3, and the reduction of p62/SQSTM1 (Fig. 1D), a selective autophagy adaptor molecule that brings ubiquitinated autophagy substrates to autophagosomes for degradation together9, 16, in the ethanol-treated livers would also support an enhanced autophagic degradation by ethanol. Indeed, detailed flux analysis (Table S1) indicated that there was not only an apparent increase in autophagosome synthesis, but also an increased autophagic degradation based on changes in GFP-LC3 dots (Fig. 1B), autophagosome number (Fig. 1F), and LC3-II level in the heavy membranes (Fig. 1D). Such analysis with LC3-II in the total lysates still indicated an increase in synthesis, but not in degradation, perhaps due to a lower enrichment of LC3-II in the total lysates than in the heavy membranes as mentioned above, which reduced the sensitivity of the assay.
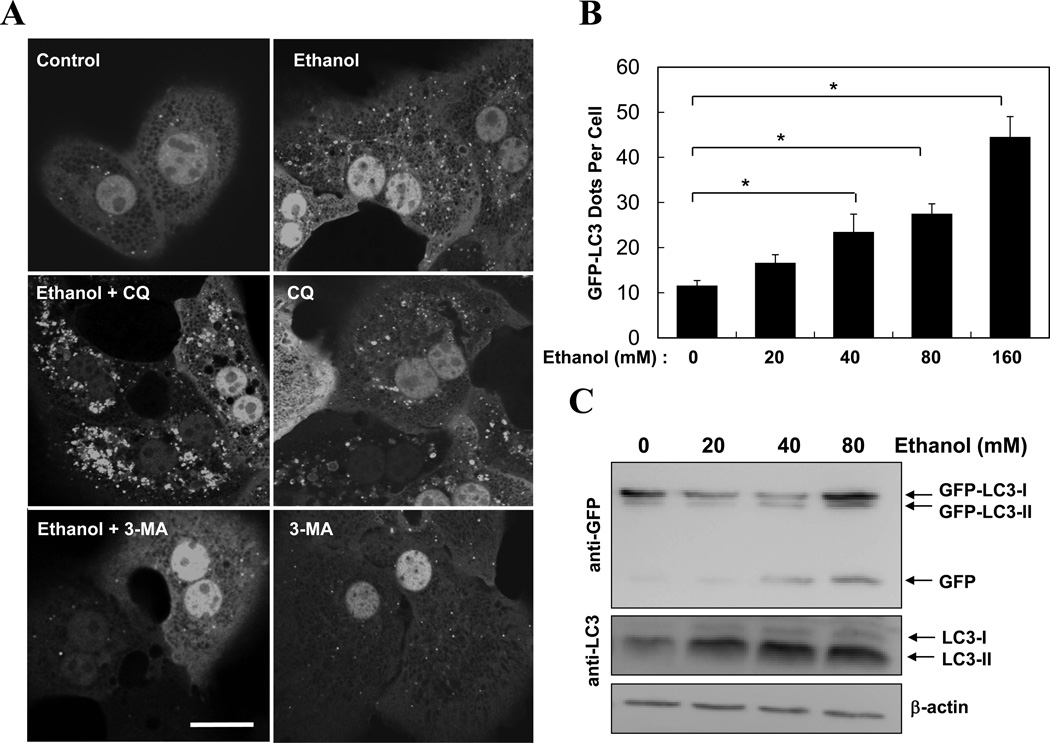
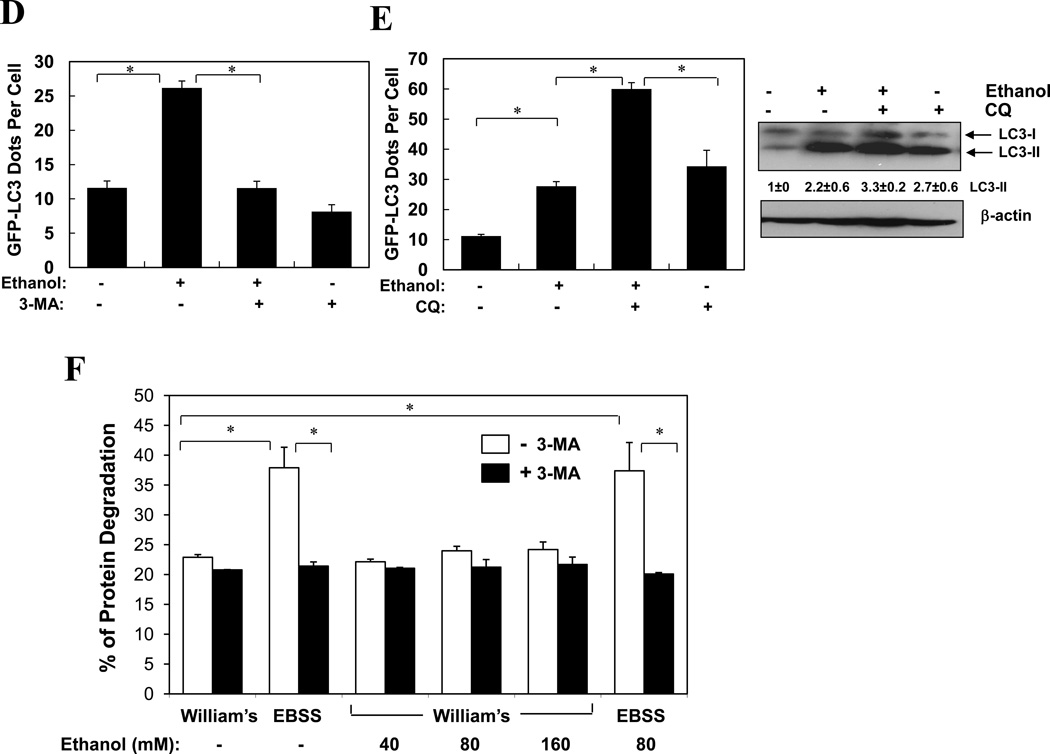
We next examined the effect of ethanol on cultured primary hepatocytes. Consistent with the in vivo observations, ethanol treatment in vitro also caused a dose-dependent increase in GFP-LC3 puncta (Fig. 2A–B), LC3-II formation (Fig. 2C), and the formation of more and larger autophagosomes (Fig. S1). LC3 punctation was suppressed by 3-MA (Fig. 2A, D), an inhibitor of the Class III PI3K, VPS34. Conversely, these changes could be enhanced by the co-treatment with CQ (Fig. 2A, E). Flux analysis (Table S1) indicated an increased autophagosome synthesis based on both GFP-LC3 puncta and LC3-II levels, although the increased degradation was only evident based on the level of GFP-LC3 puncta, but not based on the LC3-II level in the total lysate, likely due to the same reason discussed above.
Figure 2. Ethanol induces autophagy in hepatocytes in vitro.
(A–C). Ad-GFP-LC3 infected hepatocytes were treated as indicated (80 mM of ethanol in A) for 6 hours and examined by confocal microscopy (A) or immunoblot assay (C). Scale bar: 20 µm. GFP-LC3 dots (mean+SEM) were quantified for each experiment (n=3). (D–E). Ad-GFP-LC3 infected hepatocytes were treated with ethanol (80 mM, D, 40 mM, E–F) with or without 3-MA (D) or CQ (E, F) for 6 hours. GFP-LC3 dots (mean+SEM) were quantified for each experiment (n=3). Total lysates were subjected to immunoblot assay and densitometry analysis (mean+SEM) of the LC3-II was performed (n=3). (F). Hepatocytes were cultured in William’s medium/10% serum or in EBSS without or with ethanol for 15 hr and long-lived protein degradation was determined (mean+SEM, n=2–3). *: p<0.001.
To further look into the ability of ethanol to promote autophagy degradation, we examined whether it could affect the clearance of long-lived proteins, commonly associated with the general non-selective autophagy9. Markedly, ethanol did not inhibit long-lived protein degradation, nor did it promote such a degradation in hepatocytes, at either basal level or under starvation (Fig. 2F). This suggested that ethanol-induced elevation in autophagosomal markers and autophagosomes were not due to the suppression of a general autophagic degradation. On the other hand, the lack of enhanced long-lived protein degradation, coupled with a significant reduction of p62/SQSTM1 (Fig. 1D), led us to suspect that ethanol might induce mainly a selective autophagy process that targeted to organelles rather than long-lived proteins (see below).
Ethanol induced autophagy via metabolic intermediates, ROS and mTOR inhibition
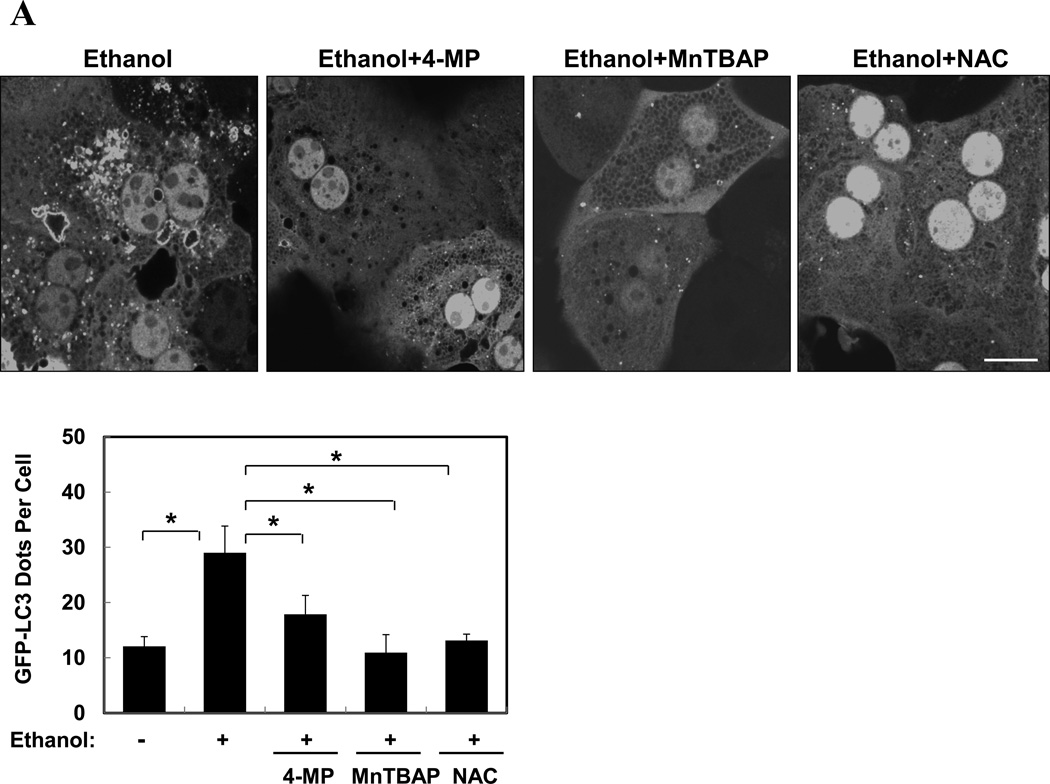
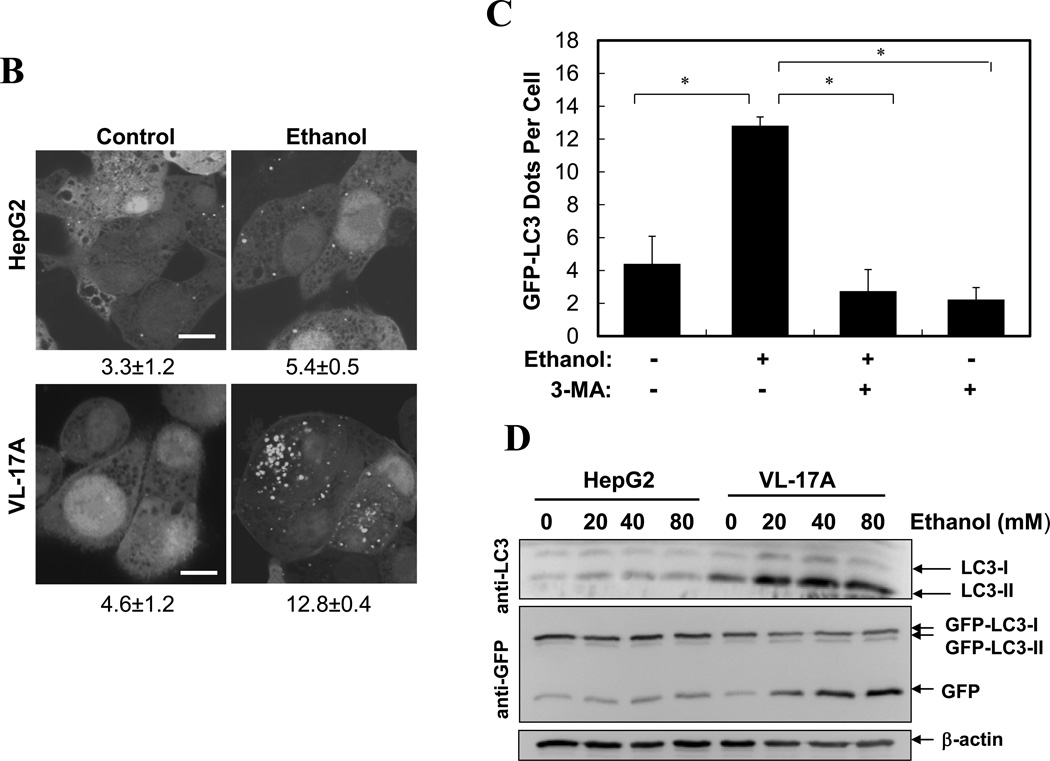
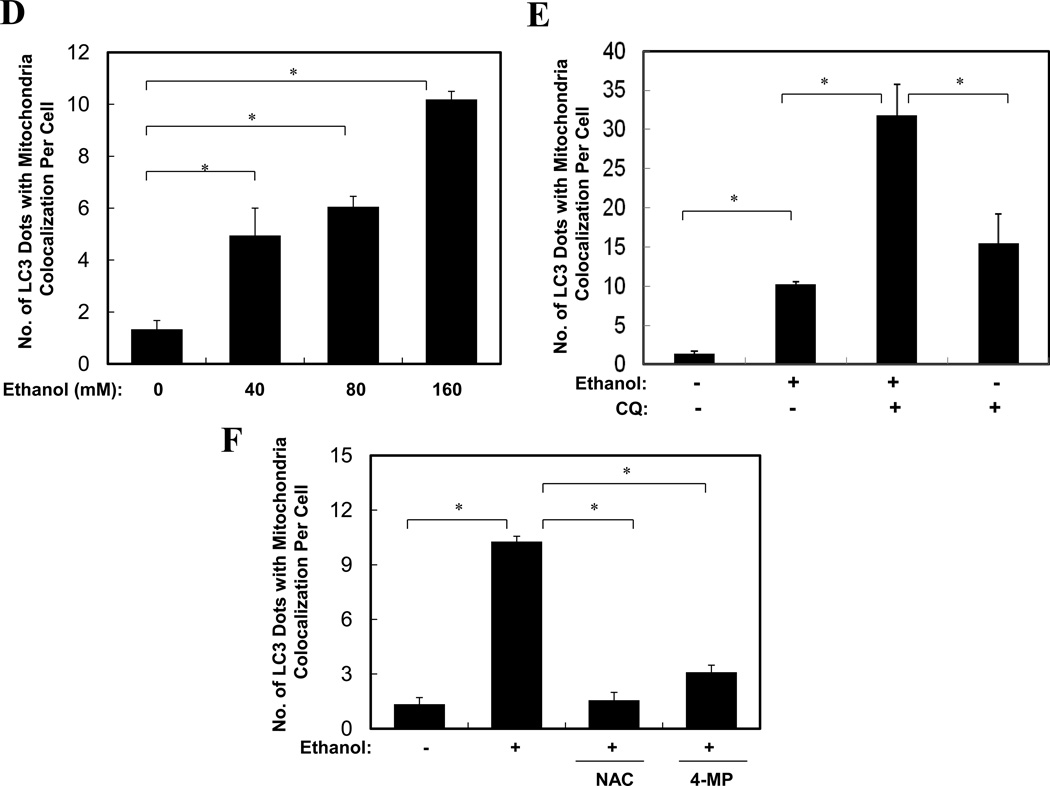
Most ethanol-mediated effects depend on its metabolism. The first step of ethanol oxidization is catalyzed mainly by alcohol dehydrogenase (ADH), and to a lesser extent, by cytochrome P450 2E1 (CYP2E1) and catalase. Ethanol oxidation results in the generation of ROS, particularly by the later two enzymatic reactions. The production of ROS is considered a major factor in ethanol-induced pathologies1, 3, 7. We found that ethanol-induced GFP-LC3 puncta was significantly diminished in the presence of 4-methyl pyrazole (4-MP), an inhibitor of both ADH and CYP2E1, or MnTBAP or N-acetylcysteine (NAC), two antioxidants (Fig. 3A), indicating that ethanol-induced autophagy required ethanol metabolism and ROS production. To further confirm this requirement, we used HepG2, a human hepatoma cell line that has a very weak capability of metabolizing ethanol, and VL-17A, a cell line derived from HepG2, which over-expresses both ADH and CYP2E1 and efficiently metabolizes ethanol13. We found that ethanol treatment significantly increased the number of GFP-LC3 puncta in VL-17A cells, but not in HepG2 cells (Fig. 3B). This effect could be suppressed by 3-MA (Fig. 3C), but further enhanced in the presence of lysosomal inhibitors (data not shown). In addition, the levels of endogenous LC3-II and GFP-LC3-II, as well as the level of free GFP moiety, were all increased in VL-17A cells, but not in HepG2 cells (Fig. 3D). Thus, ethanol-induced autophagy required ethanol metabolism and ROS production in hepatocytes.
Figure 3. Ethanol-induced autophagy requires ethanol metabolism, ROS and mTOR inhibition.
(A). Ad-GFP-LC3-infected hepatocytes were treated with ethanol (80 mM) with or without 4-MP, MnTBAP or NAC for 6 hours. GFP-LC3 dots (mean+SEM) were quantified from each experiment (n=3). (B–D). Ad-GFP-LC3 infected HepG2 (B, D) and VL-17A (B, C, D) cells were treated with ethanol (40 mM unless indicated) and other agents for 24 hours. GFP-LC3 dots per cell (mean+SEM) were quantified from each experiment (B, C, n=3) and immunoblot analysis conducted (D). (E–F). Primary hepatocytes were treated as indicated for 6 hours and total lysate were subjected to immunoblot analysis. Scale bar: 10 µm (A–B). *: p<0.05.
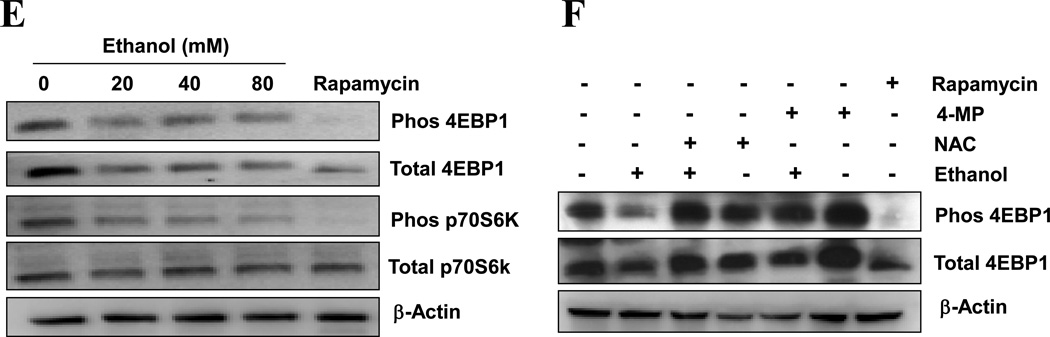
The mTOR pathway is a major regulatory pathway suppressing the activation of autophagy. We found that in primary hepatocytes ethanol caused a dose-dependent inhibition of mTOR, as manifested by the reduction of both phosphorylated p70 S6 kinase and 4E-BP1, two major downstream targets of mTOR (Fig. 3E). Interestingly, the total levels of these molecules were also reduced, perhaps reflecting an outcome of mTOR inhibition in protein synthesis. Suppression of mTOR was also observed in the livers of ethanol-treated mice (data not shown). Notably, the presence of 4-MP or NAC could reverse the inhibition of mTOR by ethanol (Fig. 3F). Taken together, these findings supported the notion that ethanol could induce autophagy by inhibiting mTOR activity via ROS.
Inhibition of autophagy enhanced ethanol-induced hepatocyte apoptosis and liver injury
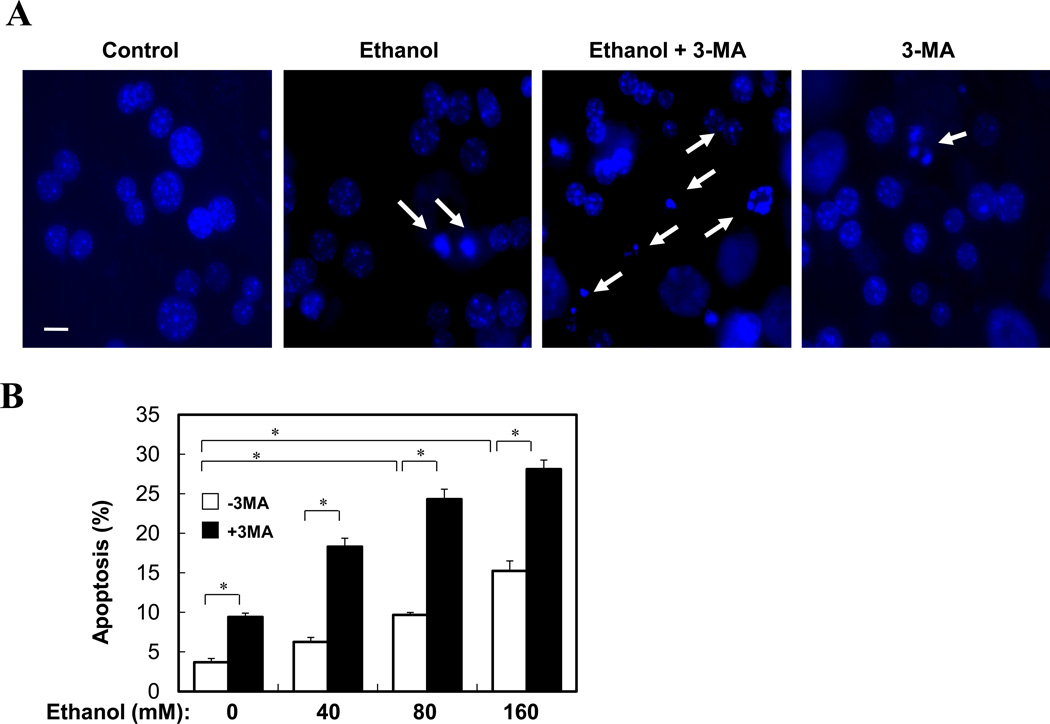
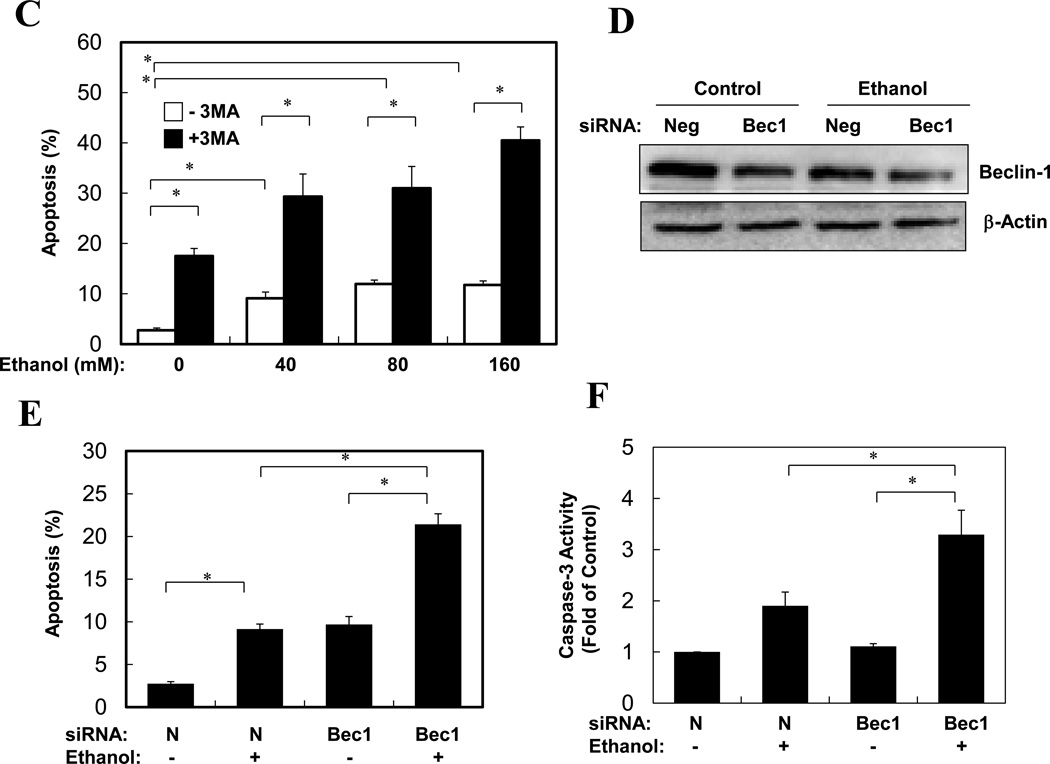
Ethanol is able to induce hepatocyte apoptosis and liver injury, which, however, is minor17, 18, 19. Autophagy could be a major protective mechanism limiting ethanol toxicity. Indeed, while only about 15% of hepatocytes were apoptotic when treated with ethanol alone, 3-MA co-treatment almost doubled the percentage of cells undergoing apoptosis (Fig. 4A–B). An even stronger enhancement of apoptosis and caspase activation by 3-MA or by a Beclin 1-specifiic siRNA could be observed in ethanol-treated VL-17A cells (Fig. 4C–F, S2). On the other hand, ethanol did not seem to noticeably promote necrosis in this model, although autophagy could have an independent effect (Fig. S2).
Figure 4. Inhibition of autophagy promotes apoptosis in ethanol-treated primary hepatocytes and VL-17 cells.
(A–B). Primary hepatocytes were treated as indicated (ethanol at 80 mM in A) and stained with Hoechst 33342. Cells with fragmented or condensed nuclei (arrows in A) (mean+SEM) were quantified for each experiment (n=2). Scale bar: 10 µm. (C). VL-17A cells were treated as indicated for 24 hours. Apoptosis (mean+SEM) was determined as in (A)(n=2). (D–F). VL-17A cells were treated as indicated (ethanol at 80 mM) for 24 hours. Beclin 1 expression was analyzed by immunoblot assay (D). Apoptosis level (E) and caspase activity (F) was determined (mean+SEM, n=2). *: p<0.05.
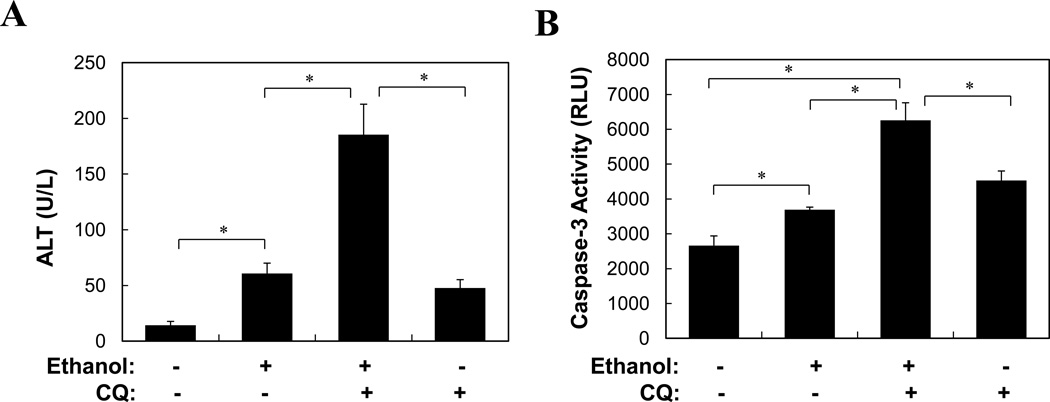
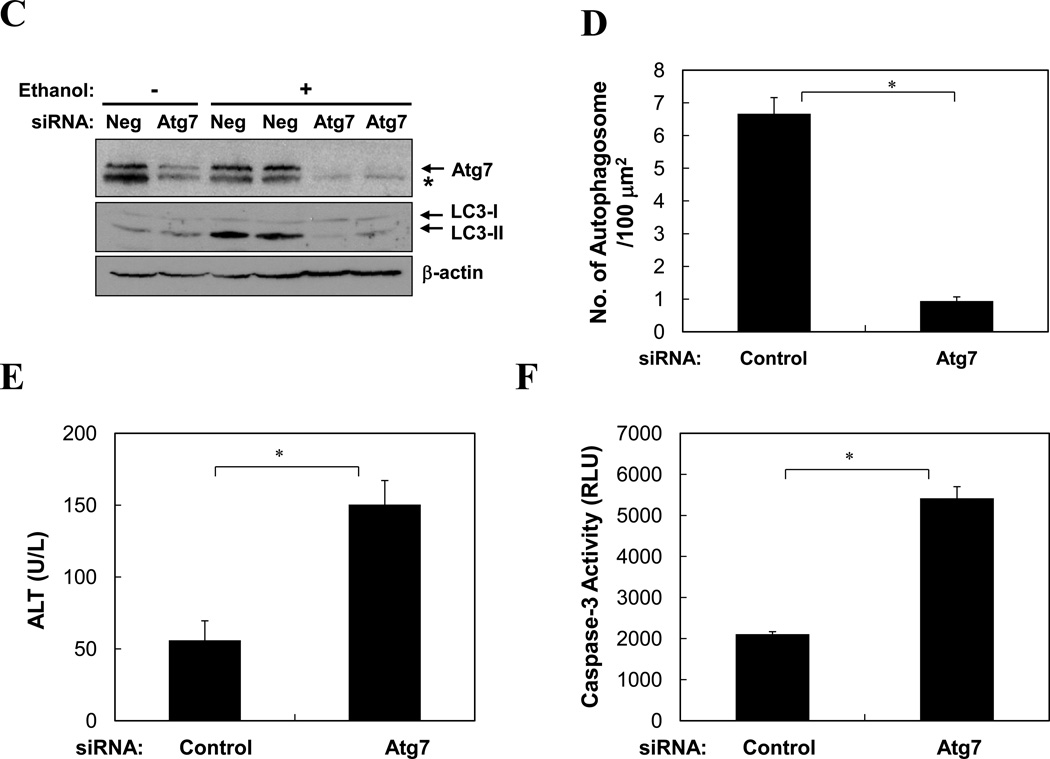
Furthermore, pretreatment of mice with CQ significantly enhanced ethanol-induced liver injury as measured by blood ALT level, hepatic caspase-3 activity and TUNEL staining (Fig. 5A–B, S3). To confirm the specificity of CQ-enhanced liver injury related to autophagy, we pretreated mice with a specific siRNA against Atg7, which effectively knocked down the liver Atg7 expression (Fig. 5C). Subsequent ethanol binge exposure resulted in a reduction in LC3-II level and autophagosome formation (Fig. 5C–D), enhanced elevation of blood ALT and caspase activation in the liver (Fig. 5E–F). Overall, while ethanol alone caused about a 3–5-fold increase in blood ALT levels, suppression of autophagy during ethanol treatment led to an additional 3-fold or more increase in blood ALT levels (Fig. 5A, E), despite that gross parenchymal alterations an cell death were minimal (Figs. S4–S5). These findings thus indicated that autophagy played an active role in limiting ethanol-induced cellular injury.
Figure 5. Inhibition of autophagy enhances ethanol-induced liver injury.
(A–B). Wild type mice (n=4–6) were treated as indicated and analyzed for blood ALT level (A) and hepatic caspase-3 activity (B)(mean+SEM). (C–F). Wild type mice were given control or Atg7-specific siRNA for 48 hours before treatment with ethanol for another 16 hours. Livers were subjected to immunoblot assay (C), EM (D) and caspase-3 activity assay (F), while blood was analyzed for ALT level (E). Data (mean+SEM) were determined from each mouse (n=3). In C, each lane represented one sample and asterisk indicates a non-specific band. *: p<0.05.
Ethanol-induced autophagy was selective for the mitochondria and lipid droplets
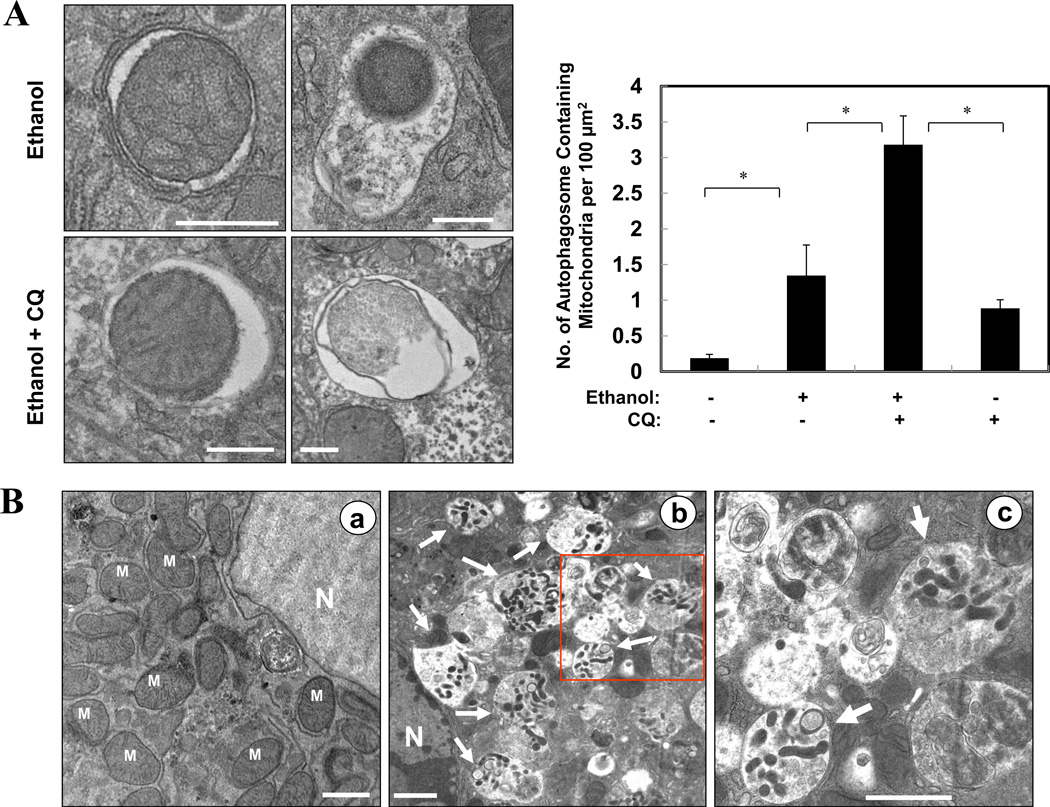
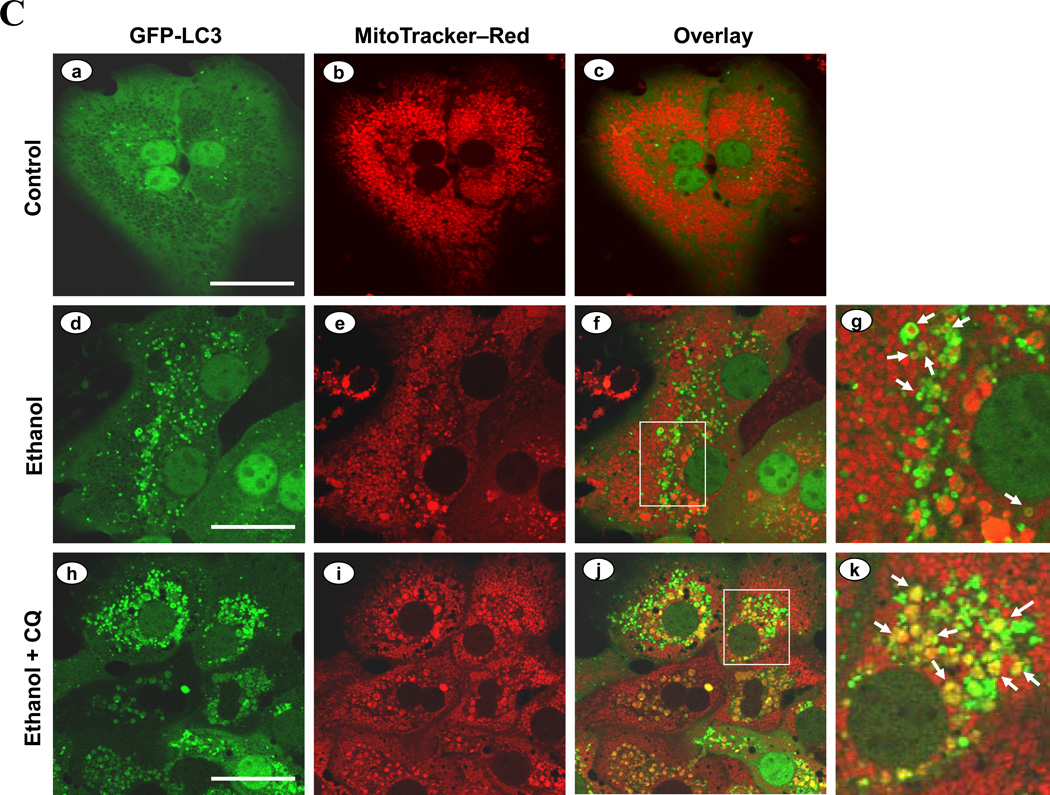
To understand the mechanisms by which autophagy protected against ethanol-induced hepatocyte injury, we examined the nature of the autophagy induced by ethanol. Studies presented above had suggested that ethanol could mainly induce selective autophagy (Fig. 1D, 2F). We observed by EM that a notable fraction of ethanol-induced autophagosomes in the liver contained mitochondria, which was further increased by pre-treatment with CQ (Fig. 6A). These mitophagic vesicles were particularly obvious in primary hepatocytes cultured in the presence of ethanol (Fig. 6B). The engulfed mitochondria seemed to be damaged (Fig. 6A), and appeared to be much smaller than the nonengulfed mitochondria (Fig. 6B), suggesting that they could have been fragmented.
Figure 6. Ethanol induces mitophagy.
(A). Wild type mice (n=3–4) were treated as indicated. Representative liver EM images of autophagosomes containing mitochondria were shown and data (mean+SEM) were quantified. (B). Primary hepatocytes were treated with vehicle control (a) or ethanol (b) for 6 hours and examined by EM. Panel c was enlarged from the boxed area in panel b. Arrows denote autophagosomes containing fragmented mitochondria. M: mitochondria; N: nuclei. (C–F). Ad-GFP-LC3-infected primary hepatocytes were treated as indicated for 6 hours and imaged (C). Panels C-g and C-k were enlarged from the boxed areas in Panels C-f and C-j, respectively. Arrows denote selected GFP-LC3 ring structures that contained the mitochondria, which were quantified (mean+SEM)(from each experiment (n=3) D–F). Scale bar: 0.5 µm (A), 1 µm (B), 20 µm (C). *: p<0.05. Ethanol was given at 80 mM unless indicated.
In untreated primary hepatocytes infected with adenoviral-GFP-LC3 and loaded with MitoTracker Red (MTR)20, there were very few GFP-LC3 punctated structures and the mitochondria exhibited normal spherical morphology (Fig. 6C, panels a–c). Ethanol treatment greatly increased GFP-LC3 positive punctated structures, and some of them were co-localized with the MTR signals (Fig. 6C). More importantly, mitochondria engulfed by the GFP-LC3 rings appeared smaller than the nonengulfed mitochondria, implying that they might be fragmented, consistent with the EM analysis (Fig. 6B). Quantitative analysis indicated that the number of GFP-LC3 puncta colocalized with MTR in ethanol-treated hepatocytes was increased in a dose-dependent manner (Fig. 6D), and could be further enhanced by the presence of CQ (Fig. 6C, E). Indeed, flux analysis indicated that both the synthesis and the degradation of the mitochondria-containing autophagosomes were significantly increased by ethanol treatment (Fig. 6A, 6E, Table S1). Together these observations indicated that ethanol promoted autophagic degradation of mitochondria, or mitophagy. Furthermore, this process could be suppressed by 4-MP or NAC (Fig. 6F), supporting the role of ethanol metabolism and ROS in promoting mitophagy.
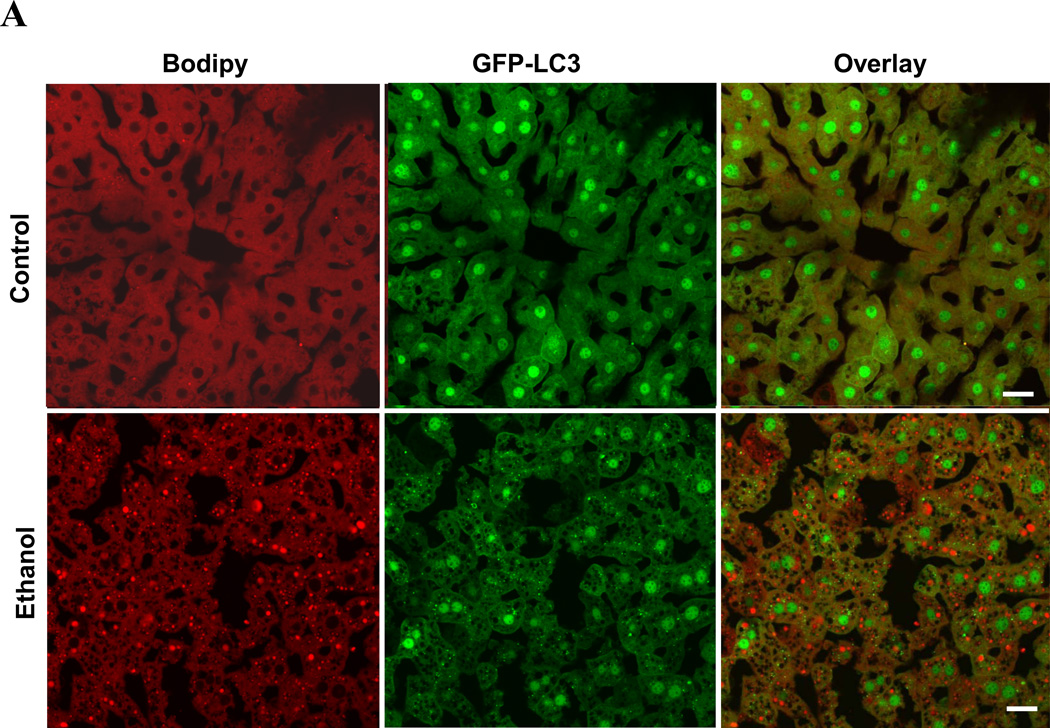
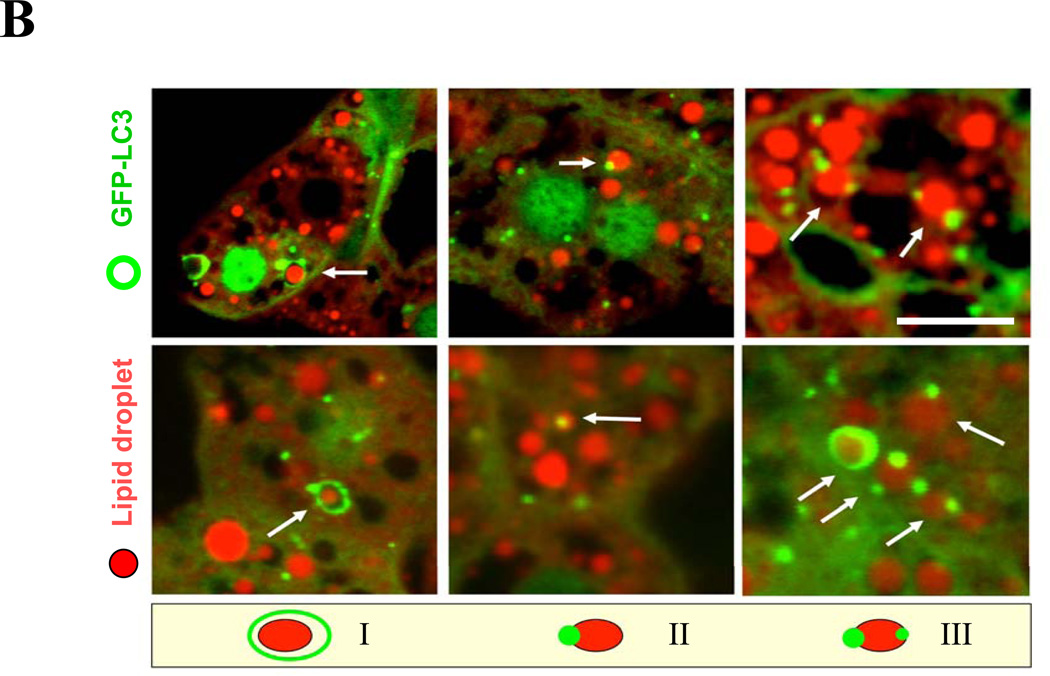
Another significant feature of ethanol-induced liver pathology is microvesicular steatosis1, 2, 5, which is considered pathogenic. By using the lipid-binding compound, Bodipy 581/591-C11 (Fig. 7A) or Bodipy 493/503 (Fig. S6), we confirmed the accumulation of lipids in the livers of mice given ethanol binge treatment. In addition, GFP-LC3 puncta or ring-like structures were found to colocalize with Bodipy-positive lipid droplets (LDs) (Fig. 7A), suggesting the activation of lipophagy, or autophagic removal of lipid droplets. While LDs that were completely encircled by GFP-LC3 rings (Pattern I) were rare, LDs that were in contact with one (Pattern II) or more (Pattern III) GFP-LC3 dots were much more common (Fig. 7B).
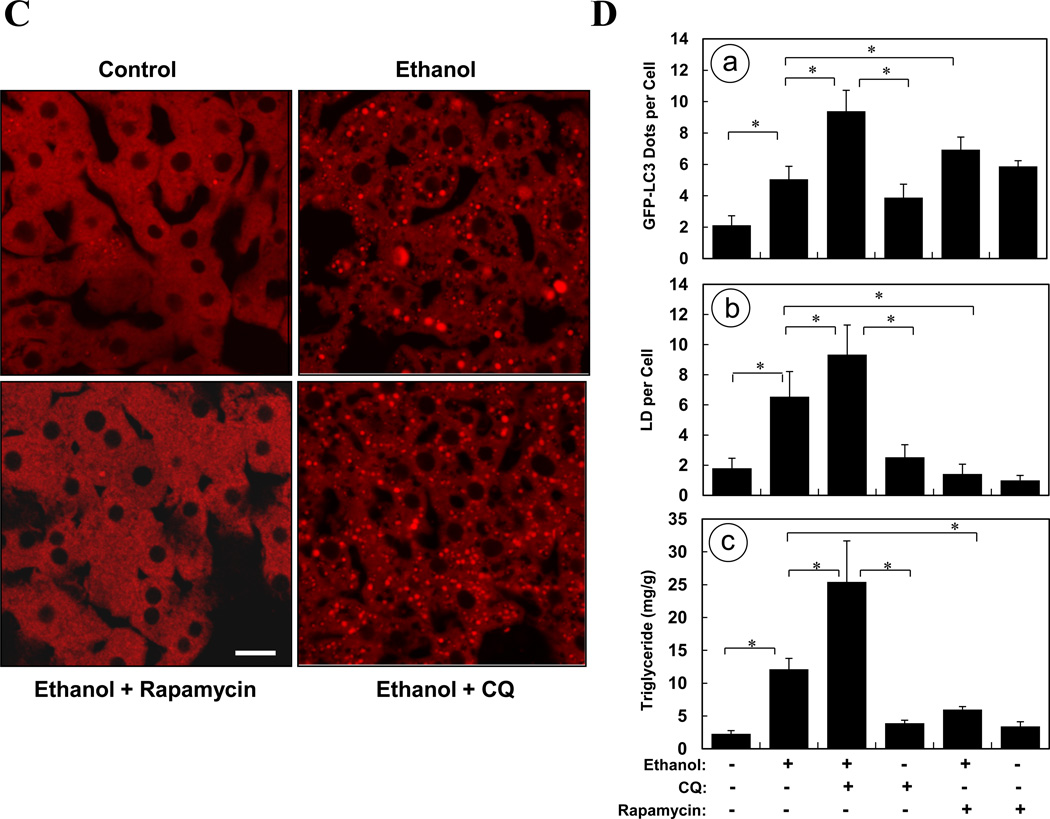
Figure 7. Modulating autophagy affects ethanol-induced hepatic steatosis.
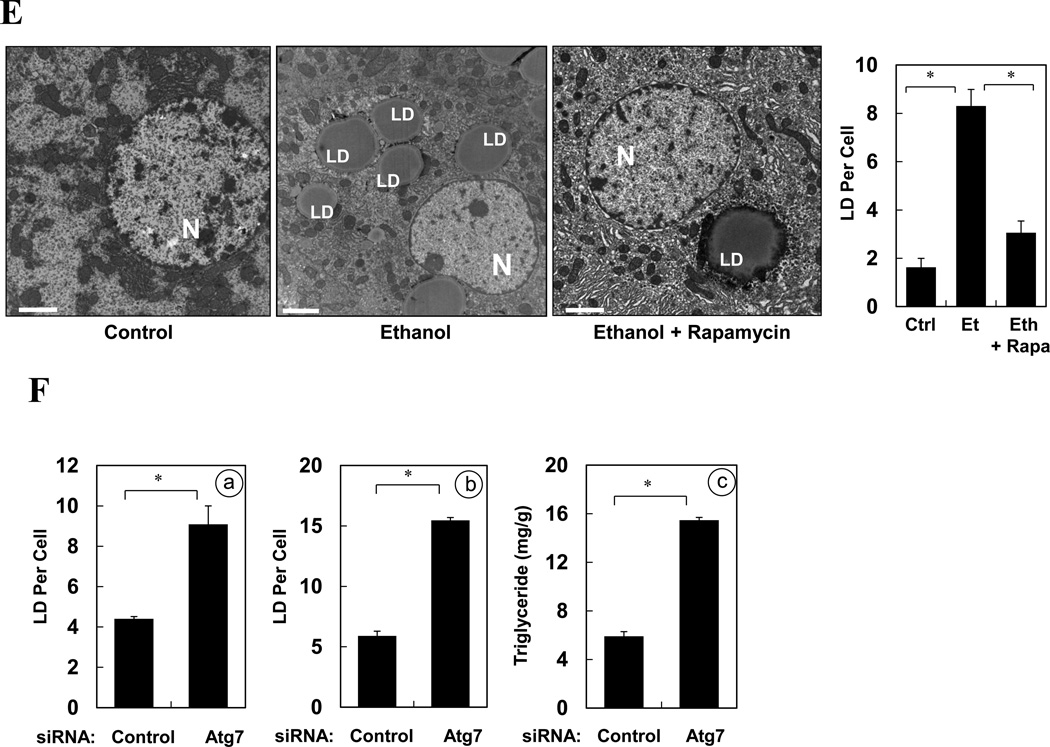
(A). GFP-LC3 transgenic mice were treated as indicated for 16 hours. Cyrosections of livers were stained with Bodipy 581/591-C11. (B). Enlarged images denote the three types of relationship of GFP-LC3 and Bodipy581/591-C11 signals (arrows) as illustrated in the diagram. (C–D). GFP-LC3 transgenic mice (n=3–5) were treated as indicated for 16 hours. Cryosections of livers were stained with Bodipy 581/591-C11 (C). GFP-LC3 dots per cell (a), Bodipy-positive lipid droplets per cell (b) and hepatic triglyceride level (c) (mean+SEM) were quantified (D). (E). Wild type mice (n=3) were treated as indicated. Liver samples were examined by EM and the number of LD per cell (mean+SEM) was quantified. (F). Wild type mice (n=3) were treated with a control or Atg7-specific siRNA for 48 hours and then ethanol for 16 hours. The number of LD per cell in the liver by Bodipy staining (a) or by EM (b) and the hepatic triglyceride level (c) (mean+SEM) were determined. Scale bar: 20 µm (A–C), 2 µm (E). *: p<0.05
To determine whether autophagy was functionally involved in the control of lipid homeostasis in the liver following ethanol treatment, we pre-treated mice with rapamycin and found that this dramatically reduced the number of LDs in the ethanol-treated livers based on Bodipy staining (Fig. 7C–D) and EM analysis (Fig. 7E), and the microvesicular changes in hepatocytes (Fig. S4). Conversely, blockage of lysosomal function with CQ significantly increased the number of LDs (Fig. 7C–D) and the histological alterations (Fig. S4). As the result, the total hepatic triglyceride levels that were elevated due to ethanol treatment were further increased by CQ pre-treatment, but was greatly reduced by rapamycin pre-treatment (Fig. 7D-c). We confirmed the specificity of the effects of these pharmacological modulations in terms of autophagy regulation by knocking down Atg7 in the liver. These mice exhibited a greatly reduced number of autophagosomes (Fig. 5E), a significantly increased number of LDs (Fig. 7F) and increased microvesicular changes (Fig. S5) following ethanol binge treatment. Consistent with this, the levels of hepatic triglycerides were significantly increased (Fig. 7F). These findings indicated that removal of lipid droplets by a selective autophagy process played an important role in mitigating ethanol-induced hepatic steatosis and therefore the injury2, 5, 8.
Discussion
Acute ethanol treatment activates a selective autophagy process
The present study demonstrates that acute ethanol exposure strongly induced autophagosome synthesis and promoted a selective autophagy process to removed damaged mitochondria and hepatic lipids droplets. The evidence for increased synthesis is based on the increase in the autophagosome number and size, and the positive changes in autophagosomal marker, LC3. The increased level of GFP-LC3 puncta was suppressed by 3-MA, and the formation of LC3-II and the increase in autophagosome number were inhibited by Atg7 siRNA, indicating that these events were induced in response to upstream autophagy signals.
Several lines of evidence also supported that ethanol treatment resulted in increased, rather than decreased, autophagic degradation through the lysosome. Ethanol did not inhibit general autophagy at the basal state or under starvation in hepatocytes (Fig 2F) or in VL-17A cells (Fig. S7). Ethanol-promoted formation of LC3-II and LC3 puncta, and the generation of autophagosomes were all further enhanced by CQ co-treatment. Although not without its own limitations, flux analysis (Table S1) confirmed elevated autophagy degradation in the presence of ethanol based on LC3 puncta, LC3-II formation in the heavy membrane fraction and autophagosome number. Notably, when a more specific set of autophagosomes that were associated with mitochondria was analyzed for this purpose, the ethanol-promoted degradation was even more evident.
This last observation suggested that ethanol could particularly activate autophagy targeting to specific substrates, i.e., a selective autophagy process. Consistently, ethanol did not promote long-lived protein degradation, commonly seen in non-selective general autophagy (Fig. 2F, S7). In contrast, ethanol did enhance the degradation of p62/SQSTM1 (Fig. 1D), a key adaptor molecule associated with selective autophagy16. p62/SQSTM1 binds to both LC3 and ubiquitinated autophagic substrates so that they could be taken up by the autophagosome for degradation.
Activation or deactivation of selective autophagy may not affect the degradation of targets of the non-selective autophagy, such as the long-lived proteins21. Subcellular organelles are the major targets of selective autophagy16, which can be activated in response to the changes in these organelles. Notably, a major target of ethanol toxicity is the mitochondria. Functional and structural alternations of mitochondria have been observed in hepatocytes following acute or chronic ethanol treatment3, 22. Damaged mitochondria could trigger, and become targets of, autophagy. Indeed we found that ethanol treatment induced significant autophagic engulfment of damaged and fragmented mitochondria based on both fluorescence and electron microscopy (Fig. 6), indicating the activation of mitophagy.
Another subcellular organelle that would be subjective to selective autophagy is lipid droplets, which are significantly elevated in ethanol-treated livers. Autophagy was recently found to be important in controlling lipid accumulation in hepatocytes subject to abnormal lipid metabolism conditions23. We demonstrated the existence of selective autophagic removal of lipid droplets (lipophagy), primarily based on the double staining of LC3 and Bodipy (Fig. 7A–B), the quantification of lipid droplets and hepatic triglyceride levels (Fig. 7D–F), and the histological observations of the microvesicular changes in the liver (Figs. S4–S5) under a variety of conditions that modulated autophagy, including pharmacological and molecular interventions. Direct observations of lipid droplets being completely engulfed by classical autophagosomes were infrequent with electron microscopy. This may be due to the fact that the large size of lipid droplets could only allow partial engulfment by the autophagosomes23. Indeed, several patterns of autophagosome/lipid droplet interactions could be observed as previously described23. The dynamics of the interaction between lipid droplets and autophagosome membranes will certainly be an interesting area to explore in future studies.
Taken together, our studies provide strong evidence that acute ethanol exposure activates a selective autophagy toward damaged mitochondria and accumulated lipids droplets and promoted their degradation through the lysosome.
Ethanol metabolism and ROS are required for ethanol-induced autophagy
It is generally accepted that ethanol metabolism is required for ethanol-induced pathology1, 2. We found that ethanol-induced autophagy was dependent on ethanol metabolism as it could be suppressed by 4-MP, an inhibitor of ADH and CYP2E1. Consistently, ethanol-induced autophagy was only observed in VL-17A cells, which are reconstituted with ADH and CYP2E1, but not in the parental HepG2 cells that lack these enzymes and are unable to efficiently metabolize ethanol.
ROS generated by ethanol treatment is a major cause of autophagy induction, as autophagy could be inhibited by the antioxidants NAC and MnTBAP (Fig. 3). Ethanol-induced ROS may come from a number of sources, among which ethanol oxidation by CYP2E1 is a major one7. In addition, oxidation of acetaldehyde in the mitochondria results in increases of NADH and its re-oxidation can also contribute to acute ethanol-induced mitochondrial ROS generation3. Furthermore, repeated ethanol binge could result in significant alterations in mitochondrial DNA synthesis, respiration disturbance and ROS generation22. It will be informative to further determine the individual importance of these events in autophagy induction in future studies, since they may constitute additional regulatory checkpoints.
Oxidative stress has been associated with the induction of autophagy in other scenarios, in which the mitochondria were suspected of being the source of ROS24, 25. How ROS trigger autophagy is not entirely clear. One possibility is that ROS impair the Akt/mTOR pathway25. mTOR is a well defined negative regulator of autophagy26. Our findings support that acute ethanol exposure induced autophagy via ROS-dependent inhibition of mTOR activity. An earlier study did find that acute ethanol administration inhibited Akt activation6. Akt is a major upstream activator of mTOR. Thus reduced Akt activity could lead to a reduced mTOR activity, activating autophagy26. Our work, however, does not rule out that ROS can induce autophagy via other mechanisms, which should be explored in the future.
Autophagy could mitigate ethanol-induced hepatotoxicity by removing damaged mitochondria and accumulated fatty acids
Ethanol-induced cell death in vivo is minor, resulting in a modest increase in blood ALT in most models of acute or chronic treatment without obvious gross histological changes unless other susceptibility factors exist (Figs. 5, S4–S5)2, 4, 17–19, 27. Our findings indicated that autophagy establishes an important threshold for ethanol-induced damage because suppressing autophagy significantly increased hepatocyte apoptosis. It seems that functional autophagy in the liver is important to avert the pathological effects of ethanol metabolism and that without this protective mechanism the detrimental effects of alcohol could be significantly amplified. The general molecular mechanisms underlying the pro-survival role of autophagy are not fully understood, but our studies suggest that the autophagy protection during acute ethanol treatment could be mediated by the removal of damaged mitochondria and lipid droplets. Autophagic degradation of mitochondria is well recognized in hepatocytes during nutrient deprivation and glucagon treatment28. However, mitophagy may be more likely involved in constraining detrimental effects associated with the damaged mitochondria28. A critical benefit of mitophagy is the elimination of an important source of ROS, which is associated with the susceptibility of hepatocytes to cellular injury and death2, 3, 5, 22, 28. Thus, it is conceivable that autophagic degradation of damaged mitochondria is a part of the protection mechanism against ethanol toxicity.
Steatosis is a common feature in acute or chronic ethanol treatment1, 2, 5. Although reversible upon ethanol withdrawal, hepatocyte steatosis is now considered important for subsequent pathological changes in ALD, such as cell death, inflammation and fibrosis5, 8. Our study now demonstrated that autophagy had a great impact on ethanol-induced steatosis. We thus propose that lipophagy is an integral part of the autophagy defense system against ethanol-induced liver injury. A key component of the cellular damage caused by ROS is lipid peroxidation, which can damage cellular membranes and produce lipid derivatives of the radicals, such as 4-hydroxynonenal and malondialdehyde, which in turn can mediate secondary insults to promote more ROS production and inflammation1, 2, 5, 7, 8. The level of cellular polyunsaturated fatty acids affects the level of lipid peroxidation and is likely proportional to the level of lipid droplets (containing triglycerides) accumulated in the cell through the equilibrium of hydrolysis of triglycerides and esterification of the free fatty acids with glycerol. Thus, lipophagy may indirectly reduce the level of free fatty acids by removing lipid droplets.
In summary, this study demonstrates that autophagy protects against ethanol-induced hepatocyte apoptosis and liver injury. This protection could be mediated via the removal of damaged mitochondria and accumulated fatty acids, thus concomitantly reducing a major source of ROS and a major target/amplifier of ROS. These findings imply that a proper autophagy capability in the liver may be crucial to reduce the detrimental effects of ethanol consumption and that enhancement of autophagy may be a possible therapeutic strategy to mitigate the pathology associated with alcoholic liver disease.
Supplementary Material
Acknowledgement
The authors would like to thank Dr. N. Mizushima (Tokyo Medical and Dental University, Japan) for the GFP-LC3 transgenic mice and Ms M. Sun (University of Pittsburgh) for technical help in EM.
Grant Support
Supported in part by NIH (X.-M. Y.: R01 CA83817, R01 CA111456; D. L. C.: R01 AA11291, W.-X. D.:R21 AA017421) and Dept. VA (D. L. C).
Abbreviations
- 3-MA
3-methyladenine
- 4-MP
4-methyl pyrazole
- ADH
alcohol dehydrogenase
- CYP2E1
cytochrome P450 2E1
- CQ
chloroquine
- EM
electron microscopy
- LD
lipid droplets
- MTR
MitoTracker Red
- NAC
N-acetylcysteine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contribution of each author
W-XD: study concept and design; data acquisition, analysis and interpretation; manuscript drafting.
ML, XC, H-MN, C-WL, WG, BL, DBS: technical and/or materials support
DLC: material support, manuscript review
X-MY: funding, study concept, design and supervision, data analysis and interpretation, manuscript writing
References
- 1.Zakhari S, Li TK. Determinants of alcohol use and abuse: Impact of quantity and frequency patterns on liver disease. Hepatology. 2007;46:2032–2039. doi: 10.1002/hep.22010. [DOI] [PubMed] [Google Scholar]
- 2.Lumeng L, Crabb DW. Alcoholic liver disease. Curr Opin Gastroenterol. 2000;16:208–218. doi: 10.1097/00001574-200005000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Bailey SM, Cunningham CC. Contribution of mitochondria to oxidative stress associated with alcoholic liver disease. Free Radic Biol Med. 2002;32:11–16. doi: 10.1016/s0891-5849(01)00769-9. [DOI] [PubMed] [Google Scholar]
- 4.Carmiel-Haggai M, Cederbaum AI, Nieto N. Binge ethanol exposure increases liver injury in obese rats. Gastroenterology. 2003;125:1818–1833. doi: 10.1053/j.gastro.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 5.Lieber CS. Alcoholic fatty liver: its pathogenesis and mechanism of progression to inflammation and fibrosis. Alcohol. 2004;34:9–19. doi: 10.1016/j.alcohol.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 6.He J, de la Monte S, Wands JR. Acute ethanol exposure inhibits insulin signaling in the liver. Hepatology. 2007;46:1791–1800. doi: 10.1002/hep.21904. [DOI] [PubMed] [Google Scholar]
- 7.Lu Y, Cederbaum AI. CYP2E1 and oxidative liver injury by alcohol. Free Radic Biol Med. 2008;44:723–738. doi: 10.1016/j.freeradbiomed.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day CP, James OF. Hepatic steatosis: innocent bystander or guilty party? Hepatology. 1998;27:1463–1466. doi: 10.1002/hep.510270601. [DOI] [PubMed] [Google Scholar]
- 9.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carson EJ, Pruett SB. Development and characterization of a binge drinking model in mice for evaluation of the immunological effects of ethanol. Alcohol Clin Exp Res. 1996;20:132–138. doi: 10.1111/j.1530-0277.1996.tb01055.x. [DOI] [PubMed] [Google Scholar]
- 12.Ding WX, Ni HM, DiFrancesca D, Stolz DB, Yin XM. Bid-dependent generation of oxygen radicals promotes death receptor activation-induced apoptosis in murine hepatocytes. Hepatology. 2004;40:403–413. doi: 10.1002/hep.20310. [DOI] [PubMed] [Google Scholar]
- 13.Donohue TM, Osna NA, Clemens DL. Recombinant Hep G2 cells that express alcohol dehydrogenase and cytochrome P450 2E1 as a model of ethanol-elicited cytotoxicity. Int J Biochem Cell Biol. 2006;38:92–101. doi: 10.1016/j.biocel.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Buettner R, Newgard CB, Rhodes CJ, O'Doherty RM. Correction of diet-induced hyperglycemia, hyperinsulinemia, and skeletal muscle insulin resistance by moderate hyperleptinemia. Am J Physiol Endocrinol Metab. 2000;278:E563–E569. doi: 10.1152/ajpendo.2000.278.3.E563. [DOI] [PubMed] [Google Scholar]
- 15.Mareninova OA, Hermann K, French SW, O'Konski MS, Pandol SJ, Webster P, Erickson AH, Katunuma N, Gorelick FS, Gukovsky I, Gukovskaya AS. Impaired autophagic flux mediates acinar cell vacuole formation and trypsinogen activation in rodent models of acute pancreatitis. J Clin Invest. 2009;119:3340–3355. doi: 10.1172/JCI38674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Z, Sun X, Kang YJ. Ethanol-induced apoptosis in mouse liver: Fas- and cytochrome c-mediated caspase-3 activation pathway. Am J Pathol. 2001;159:329–338. doi: 10.1016/S0002-9440(10)61699-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higuchi H, Adachi M, Miura S, Gores GJ, Ishii H. The mitochondrial permeability transition contributes to acute ethanol-induced apoptosis in rat hepatocytes. Hepatology. 2001;34:320–328. doi: 10.1053/jhep.2001.26380. [DOI] [PubMed] [Google Scholar]
- 19.Venugopal SK, Chen J, Zhang Y, Clemens D, Follenzi A, Zern MA. Role of MAPK phosphatase-1 in sustained activation of JNK during ethanol-induced apoptosis in hepatocyte-like VL-17A cells. J Biol Chem. 2007;282:31900–31908. doi: 10.1074/jbc.M703729200. [DOI] [PubMed] [Google Scholar]
- 20.Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. Faseb J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 21.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demeilliers C, Maisonneuve C, Grodet A, Mansouri A, Nguyen R, Tinel M, Letteron P, Degott C, Feldmann G, Pessayre D, Fromenty B. Impaired adaptive resynthesis and prolonged depletion of hepatic mitochondrial DNA after repeated alcohol binges in mice. Gastroenterology. 2002;123:1278–1290. doi: 10.1053/gast.2002.35952. [DOI] [PubMed] [Google Scholar]
- 23.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Gibson SB. Is mitochondrial generation of reactive oxygen species a trigger for autophagy? Autophagy. 2008;4:246–248. doi: 10.4161/auto.5432. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Kong X, Kang J, Su J, Li Y, Zhong J, Sun L. Oxidative stress induces parallel autophagy and mitochondria dysfunction in human glioma U251 cells. Toxicol Sci. 2009;110:376–388. doi: 10.1093/toxsci/kfp101. [DOI] [PubMed] [Google Scholar]
- 26.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarvelainen HA, Lindros KO. Animla models of ethanol-induced liver injury. In: Sherman DIN, Preedy V, Watson RR, editors. Ethanol and the liver. London and New York: Taylor & Francis, Inc; 2002. pp. 358–386. [Google Scholar]
- 28.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.