Abstract
A new method is presented for the identification of oligosaccharides obtained by enzymatic digestion of hyaluronan (HA) with bacterial hyaluronidase (E.C. 4.2.2.1, from Streptomyces hyalurolyticus) using matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOFMS). Mixtures containing HA oligosaccharides of tetrasaccharide (4-mer)–34-mer were analyzed using this method. The carboxyl groups of the glucuronate residues in the prepared HA oligomers, were modified as the acidic form (—COOH), sodium salts (—COONa), organic ammonium salts, or methylesters before MALDI-TOFMS measurement. Among these samples, the methylester form of glucuronate residues in HA oligosaccharides, prepared by methylation using trimethylsilyl diazomethane, afforded high sensitivity for spectra. This simple modification method for carboxyl group methylation of acidic polysaccharides [Hirano et al., Carbohydr. Res., 340, (2005) 2297–2304] provides samples suitable for MALDI-TOF mass spectrometric analysis throughout a significantly enhanced range of masses.
Keywords: Hyaluronan oligosaccharide, Matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOFMS), Methylester
1. Introduction
Hyaluronan (HA) is comprised of long, unbranched chains of variable length that consists of a disaccharide, glucuronic acid-β1,3-N-acetylglucosamine-β1,4-, repeated several thousand times. Generally, HA is found in the extracellular matrix and its role for the essential maintenance of tissue architecture is well known [1,2]. HA is also involved in the modulation of cellular adhesion, signaling, and motility in mammals [3]. HA has also been discovered to assist in the control of morphogenesis [4–7], tissue regeneration [8], wound healing [9], inflammation [10] and cancer metastasis [11–14]. The biological, physiological and pathophysiological functions of HA have not been fully elucidated. Interestingly, there is evidence that some of the functions mentioned above might be related to the molecular size of HA. For example, high molecular weight HA enhances cell growth in culture [15,16]. While high molecular weight HA can inhibit angiogenesis [17], low molecular weight HA shows strong activation of angiogenesis [18] and there have been published several papers on hyaluronidase inhibitors to prevent angiogenesis and tumor cell growth [19–21]. Thus, the effect of HA size on the regulation of angiogenesis is still unclear.
The preparation of homogenous, structurally characterized HA oligosaccharides is essential for the investigation of the physiological and biochemical functions of HA. A complex mixture of HA oligosaccharides is obtained by hyaluronan lyase and hyaluronidase catalyzed digestion of HA [22–24]. It is difficult to apply structurally undefined oligosaccharides to establish structure activity relationships (SAR) for HA. The average molecular weight and molecular weight distribution of HA chains depend on the extent of enzymatic digestion. Therefore, the first step of any SAR studies must be the precise evaluation of the chain length distribution in such oligosaccharide mixtures. MALDI-TOFMS offers a particularly powerful method to determine molecular weight and dispersity. While there are several reports on analysis of HA oligosaccharides by MALDI-TOFMS and electrospray ionization mass spectrometry (ESI-MS), there are few quantitative approaches for the analysis of HA oligosaccharides, due to the different ionization efficiency of HA oligosaccharides of various sizes [25].
In this study, we have developed a useful method using trimethylsilyl diazomethane for methyl esterification of HA [26] to facilitate a more quantitative approach on MALDI-TOFMS detection for the measurement of HA. This method should have value in the analysis of biological preparations and pharmaceutical formulations of HA.
2. Materials and methods
2.1. Chemicals and instruments
Hyaluronan sodium salt (M.W. 20,000) from Streptococcus zooepidemicus was purchased from Kibun Food Chemipha Co., Tokyo, Japan. Hyaluronidase from Streptomyces hyalurolyticus (lyase, E.C.4.2.2.1) for HA oligosaccharide preparation was purchased from Seikagaku Kogyo Co., Tokyo, Japan. A column (4.4 cm I.D. × 1 m) for low pressure gel permeation chromatography was purchased from Millipore Japan Co., (Tokyo). Sephadex G-50 (superfine) resin and a Hi-Trap desalting column were purchased from Pharmacia Biotech, Tokyo, Japan. Dialysis tubing (MWCO 500) was purchased from Wako Pure Chemicals Ltd., Osaka, Japan. All other chemicals were analytical reagent grade. Capillary electrophoresis (CE)was performed on a Beckman capillary electrophoresis system (P/ACE 5010) equipped with a UV detector and an operation system using version 0.4 P/ACE station on an IBM-compatible PC, from Beckman Coulter (Fullerton, CA, USA). JEOL GSX500A and ECP600 NMR instruments (Tokyo, Japan), equipped with a 5 mm field gradient tunable probe with standard JEOL software, were used for 1H NMR experiments at 30 °C on 500 µL each sample.
2.2. Preparation of HA oligosaccharides
Hyaluronan (HA) was depolymerized using bacterial hyaluronidase (hyaluronan lyase, E.C.4.2.2.1) to obtain an oligosaccharide mixture containing chain sizes from a tetrasaccharide (a 4-mer) to a 34-mer. The oligosaccharide mixture was neutralized with 0.1 M sodium hydroxide to make sodium salts of the HA oligosaccharides. Although each oligosaccharide derived by this enzymatic treatment contains unusual double bond at the nonreducing end uronate. This functionality represents an advantage as it facilitates the detection of these HA oligosaccharides in purification procedures based on its absorbance in the UV at 230 nm. The hyaluronidase digestion was performed for different periods of time to obtain different quantities of various sized HA oligosaccharides. Digestions of 20, 40 and 60% completion ([absorbance at 230 nm after partial digestion/absorbance at 230 nm after complete digestion] × 100) were prepared.
2.3. MALDI-TOFMS
MALDI-TOF mass spectra were collected as follows: Mass analysis was carried out in negative/positive linear and reflectron mode using an Axima™ (Shimadzu Kratos Inc., Kyoto, Japan) equipped with a 337 nm nitrogen laser. The acceleration voltage was set to 19 kV and the delay time was 450 ns. A total of 200 mass spectra were acquired and summed for each sample spot. All data were collected by searching an adequate spot on the target sample plate manually using “Raster mode”. Mass calibrations were performed over several m/z ranges, using commercially available protein and peptide standards. For the sample preparation, several matrices were tested and optimized (see Section 3). Briefly, 1 mg of sample was mixed with 100 µL of solvent mixture (acetonitrile/0.1% trifluoroacetic acid, 1:2, v/v). One micrliter of sample solution was mixed with 10 µL of a 10 mg mL−1 solution of CHCA (α-cyano-4-hydroxycinnamic acid) in TA buffer (30% acetonitrile containing 0.1% trifluoroacetic acid). This preparation (0.2 µL) was placed onto a MALDI-sample plate and spectra were collected by raster irradiation on the sample surface. The results shown in the text were obtained by using HA oligosaccharide sample prepared under the conditions described above.
2.4. Capillary electrophoresis (CE)
CE was performed using a system with advanced computer interface, equipped with high voltage power supply capable of constant or gradient voltage control using a fused silica capillary from GL Science, Tokyo, Japan. The compositional analysis of HA oligosaccharide mixture was confirmed by CE under normal polarity mode using a mixture of 40 mM disodium phosphate/40 mM sodium dodecylsulfate/10 mM tetraborate adjusted to pH 9.0 with 1.0 M hydrochloride as described previously [27]. The fused silica capillary (75 µm I.D. × 375 µm O.D., 67 cm long) was automatically washed before use with 0.1 M sodium hydroxide, followed nitrogen gas pressure injection (5 s) at a constant current 15 kV. The samples (0.1 mg mL−1) were dissolved in water and loaded (7 nL) with nitrogen gas pressure injection.
2.5. Sample preparation for MALDI-TOFMS experiments
To convert the sodium salts of HA oligosaccharides to the acidic form and organic ammonium salts, dried sample (~10 mg)was dissolved in 0.5 mL water and applied to a Dowex 50×8 cation exchange column (7.5 mm I.D. × 87 mm, H+ form). The acidic form of the HA oligosaccharide fraction was collected manually by checking pH of the eluate and the fraction was immediately freeze-dried to prepare acidic form of the HA oligosaccharide fraction. In contrast, the acidic fraction from the cation exchange column was immediately neutralized with an aqueous solution of organic amine such as tetraethyl ammonium (TEA) at 0 °C, and then freeze-dried. To obtain HA oligosaccharide permethylester, the dried acidic (—COOH) form of the HA oligosaccharides (~10 mg) was suspended in a mixture of 1 mL dry dimethylsulfoxide and 100 µL methanol and 30 µL trimethylsilyl diazomethane was added to the sample solution under stirring vigorously at room temperature for 2 h after which 1 mL of water was added to quench the reaction and the sample was freeze-dried three times (1 mL of water was added after each freeze-drying step).
2.6. NMR spectroscopy
One-dimensional (1D) and two-dimensional (2D) 1H NMR experiments were performed under previously described conditions [28,29]. Briefly, each sample was weighed (~2.0 mg) and was dissolved in 0.5 mL of 2H2O (99.9%) and freeze-dried repeatedly to remove exchangeable protons. The sample was kept in a desiccator over phosphorous pentoxide in vacuo overnight at room temperature. The thoroughly dried sample was the dissolved in 500 µL of 2H2O (99.96%) and passed through 0.45 µm syringe filter and transferred to an NMR tube (5.0 mm O.D. × 25 cm). The HO2H signal was suppressed by presaturation during 3 and 1.5 s for 1D-NMR and 2D-NMR experiments, respectively. To obtain 2D spectra, 1024 × 512 data matrix for a spectral width of 2000 Hz were measured, and the time domain data were multiplied after zero-filling (data matrix size 1 K × 1 K) with a shifted sine-bell window function.
3. Results and discussion
3.1. Preparation of HA oligosaccharide fragments
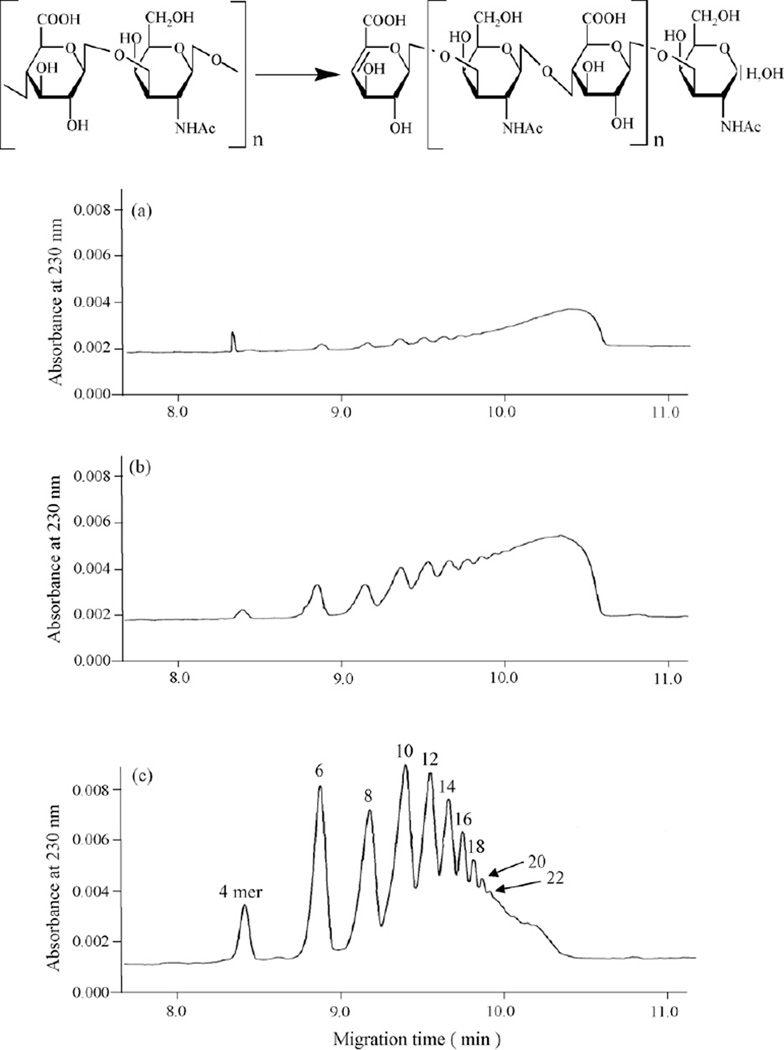
Intact HA (average molecular weight 20,000) was incubated with Streptomyces hyaluronidase for a range of time periods, and the resulting samples were analyzed by capillary electrophoresis. A series of well-separated and symmetrical peaks were detected on CE profiles (Fig. 1). These samples were analyzed by MALDI-TOFMS (as described in detail later) and found to correspond to HA oligosaccharides of increasing length and, thus, of increasing numbers of carboxylic groups of glucuronate residues. The sample corresponding to a hyaluronidase digestion of 18 h (Fig. 1c) resulted in HA oligosaccharides ranging from tetrasaccharides (4-mer) to oligosaccharides greater than 22-mer. As expected, when the digestion time increased, the average size of the oligosaccharides decreased, with a larger proportion of low molecular weight species produced.
Fig. 1.
Capillary electrophoresis of hyaluronan oligosaccharides by partially enzymatic digestion. The structure of hyaluronan and hyaluronan oligosaccharides prepared by partial digestion with a lyase are shown. The structure of a tetrasaccharide (4-mer) is shown where n = 1, a hexasaccharide (6-mer), where n = 2, etc. Hyaluronan degraded with hyaluronidase from Streptmyces hyalurolyticus for 3 h (a), 7 h (b) and 18 h (c).
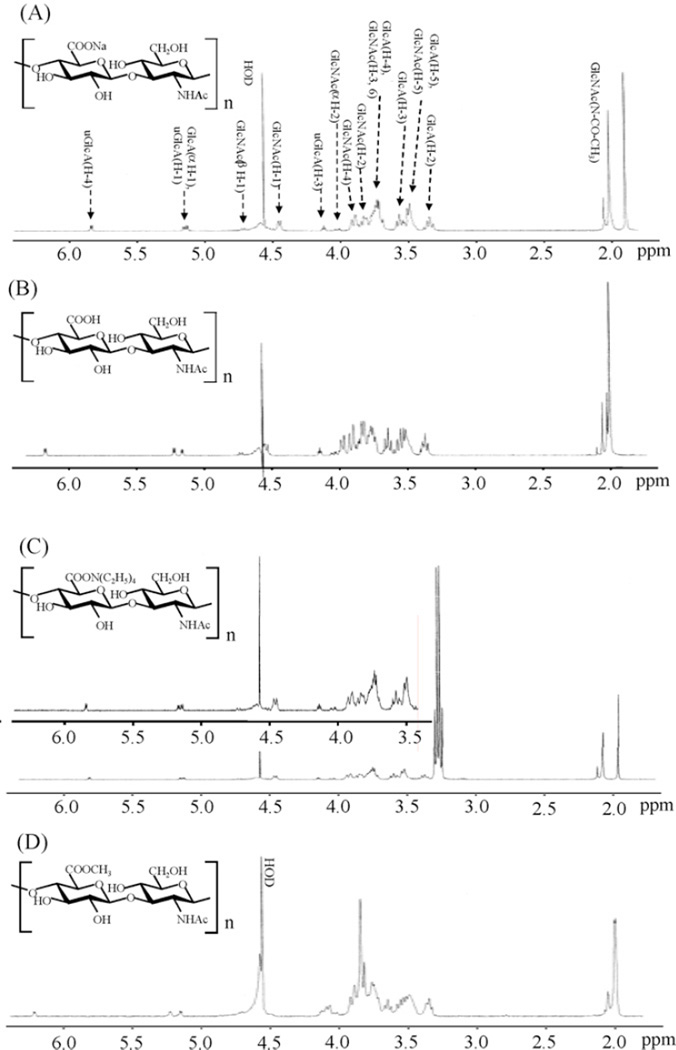
The sample digested for 18 h, corresponding to 60% reaction completion, was selected for the preparation of the organic ammonium salt, the acidic of and the methyl esterified forms of HA oligosaccharides for NMR experiments in large scale sample preparation. The 1H NMR spectra of each form of the 60% digested HA oligosaccharide mixtures (2–3 mg each) were obtained next (Fig. 2). Differences in the spectra of each form of HA oligosaccharides are clearly evident. As expected, sodium salt (—COONa), acidic form (—COOH) or methyl esterified carboxyl groups of glucuronate (GlcA) residues of each HA oligosaccharide sample show different effects on the chemical shift of the surrounding protons. For example, the chemical shift of GlcA H-5 signal GlcA was significantly affected by situation of carboxyl group. The O-methyl of GlcA carboxyl ester shows a singlet signal at 3.85 ppm with intensity comparable to that of N-acetylmethyl signal resonating at 2.0 ppm. No unassignable signals were observed in the spectrum as shown in Fig. 2. Some differences in the chemical shift values were observed in each spectrum. The expected differences in the anisotropic effect of carboxyl group of each HA oligosaccharide form are observed, however, the negative charge of the carboxyl groups produce a repulsive force between each other that also impact the chemical shifts of GlcA and GlcNAc ring protons.
Fig. 2.
1H NMR spectra of various preparation of hyaluronan oligosaccharides. (A) Hyaluronan oligosaccharide sodium salts, (B) acidic hyaluronan oligosaccharides, (C) hyaluronan oligosaccharides TEA salts and (D) methyl esterified hyaluronan oligosaccharides.
3.2. Comparison of HA oligosaccharides salts and methylester for MALDI-TOFMS
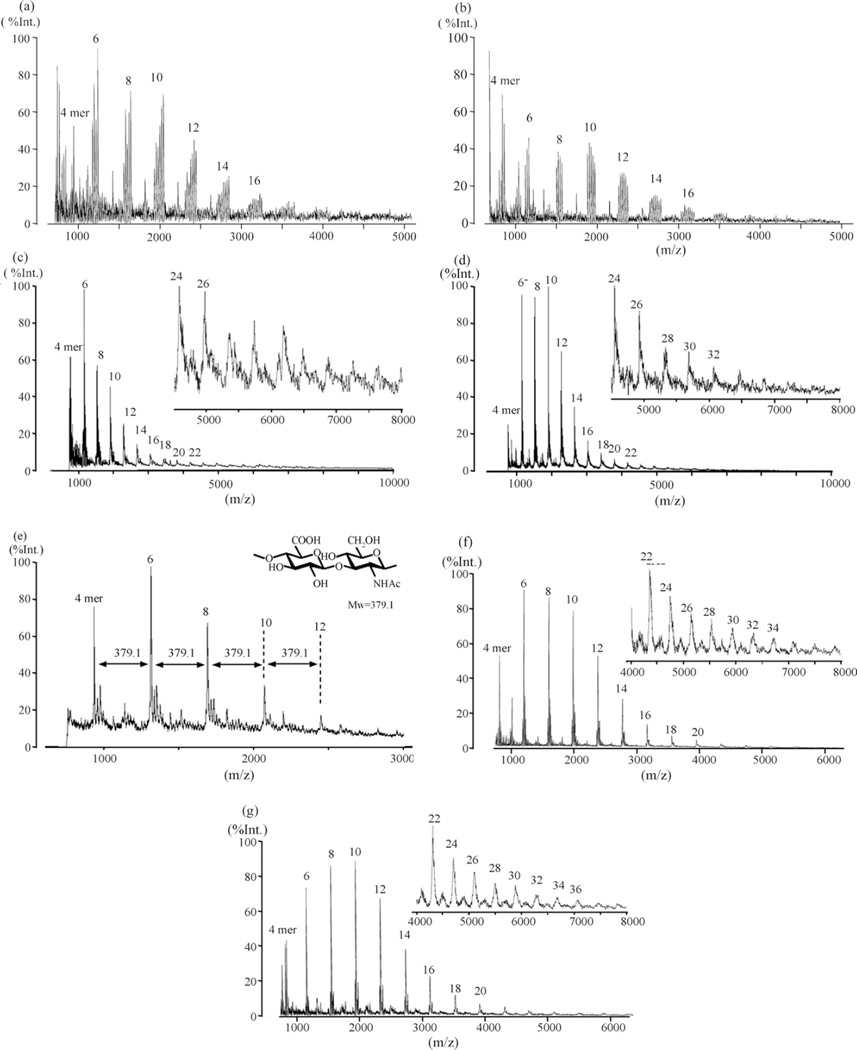
We next evaluated several matrices for the analysis of the sodium salt form of HA oligosaccharides by MALDI-TOFMS. These included 2,5-dihydroxybenzoic acid (DHBA), which has been used recently for the analysis of HA oligomers [30,31], but this matrix did not generate the strong signals previously reported by Mahoney et al. [32]. In the positive-ion mass spectra, the major peaks were generally a mixture of metal ion adducts, such as [M+Na]+ and [M+K]+, whereas [M−H]− was the predominant species in the negative-ion mode (data not shown) as described previously [33]. Consequently, a new strategy was designed to control the formation of counter-ion species of uronate residues in the MALDI-TOF mass spectra of HA oligosaccharides.
Using CHCA as a matrix reagent, the various forms of the oligosaccharides (60% digestion) were examined in MALDI-TOFMS experiments (Fig. 3). The spectra of HA oligosaccharides containing non-esterified acidic form of GlcA residues show strong intensity signals in a simple pattern (Fig. 3a–d) compared with those of our preliminary results on the sodium salt form of HA oligosaccharides using DHBA as matrix (not shown). Characteristic multiple ions were observed from each HA oligosaccharide sodium salts in the both positive and negative detection modes (Fig. 3a and b). For example, five signals and four signals from 8-mer were detected in positive and negative ion spectra, respectively. In the positive detection mode these signals were identified as [M+Na]+, [M−H+2Na]+, [M−2H + 3Na]+, [M−3H + 4Na]+ and [M−4H + 5Na]+. Unfortunately, because of a number of reasons, including the presence of multiple adduct peaks these data could not be used quantitatively. The relative peak intensities in these mass spectra did not correlate to their concentrations determined in CE. While the acidic form of HA oligosaccharides showed single peaks for each oligosaccharide (Fig. 3c and d) when the acidic samples were stored at −80 °C for periods of 60 days, their spectra were altered becoming similar to those obtained on the sodium form of HA oligosaccharide mixture (data not shown) showing a complicated mixture of metal ion adducts. The spectra of the tetraethyl ammonium (TEA) form of HA oligosaccharides showed a pattern of peaks spaced at m/z = 379.1 (Fig. 3e). The production of multiple metal/ammonium adducts of HA oligosaccharides could not be controlled under these conditions. The methyl ester of HA oligosaccharide mixture afforded excellent results (Fig. 3f and g) and clearly shows improved sensitivity. In contrast to the salt forms, no degradation/fragmentation of the HA oligosaccharides by laser irradiation for MALDI was observed (data not shown). The reproducibility of the analyses was acceptable (both intra-and inter day variation were below 10%). Table 1 shows mass assignments for the methyl esterified HA oligosaccharide mixture in both positive-ion and negative-ion MALDI-TOFMS. The major peaks observed in the positive ion spectra were the sodium adducts [M′+Na]+. Unexpectedly, the major peaks observed in the negative ion mode were the [M′−CH3]− ions. In all cases, the positive-ion detection mode was superior to the negative-ion detection mode for HA oligosaccharide mixtures.
Fig. 3.
Effects of salts and methylesterification on hyaluronan oligosaccharides for MALDI-TOF mass spectrometry. (a) Sodium salts of carboxy groups (—COONa) of HA oligosaccharides, positive ion mode; (b) sodium salts of carboxy groups of HA oligosaccharides, negative ion mode; (c) acidic form of carboxy groups (—COOH) of HA oligosaccharides, positive ion mode; (d) acidic form of carboxy groups of HA oligosaccharides, negative ion mode; (e) tetraethyl amination on carboxy groups of HA oligosaccharides, negative ion mode; (f) methylesterification on carboxy groups of HA oligosaccharides, positive ion mode; and (g) methylesterification on carboxy groups of HA oligosaccharides, negative ion mode. All spectra were collected by the linear mode.
Table 1.
Theoretical and experimental masses for methylated hyaluronan oligosaccharide mixture
| Positive mode [M′ + Na]+ | Negative mode [M′ −CH3]− | |||
|---|---|---|---|---|
| Theoretical m/z |
Experimental m/z |
Theoretical m/z |
Experimental m/z |
|
| 4 mer | 809.7 | 809.7 | 771.6 | 771.4 |
| 6 mer | 1203.0 | 1203.2 | 1165.0 | 1165.2 |
| 8 mer | 1596.3 | 1596.5 | 1558.3 | 1558.1 |
| 10 mer | 1989.7 | 1989.9 | 1951.6 | 1951.5 |
| 12 mer | 2383.0 | 2383.2 | 2345.0 | 2344.7 |
| 14 mer | 2776.3 | 2776.2 | 2738.3 | 2738.1 |
| 16 mer | 3169.6 | 3169.4 | 3131.6 | 3131.0 |
| 18 mer | 3563.0 | 3563.1 | 3525.0 | 3524.2 |
| 20 mer | 3956.3 | 3956.3 | 3918.3 | 3918.2 |
| 22 mer | 4349.6 | 4349.9 | 4311.6 | 4310.5 |
| 24 mer | 4743.0 | 4742.6 | 4705.0 | 4705.0 |
| 26 mer | 5136.3 | 5135.7 | 5098.3 | 5098.4 |
| 28 mer | 5529.6 | 5530.8 | 5491.6 | 5491.0 |
| 30 mer | 5923.0 | 5923.4 | 5885.0 | 5884.6 |
| 32 mer | 6316.3 | 6315.2 | 6278.3 | 6279.4 |
| 34 mer | 6709.6 | 6707.9 | 6671.6 | 6670.6 |
| 36 mer | 7103.0 | n.da | 7065.0 | 7064.1 |
| 38 mer | 7496.3 | n.d. | 7458.3 | n.d. |
Not identified (Signal to noise ratios were below 2).
4. Conclusions
MALDI-TOFMS has become a popular method for biopolymer analysis but its utility has not yet been established in the structural determination of acidic carbohydrates. While there have been previous attempts to analyze HA oligosaccharides by MALDI-TOFMS [30,31], these have generally been applied to smaller oligosaccharides. A relatively simple derivatization method involving methyl esterification of carboxyl groups of uronate residues affords simplified mass spectra, improved sensitivity that give spectral intensity that more closely parallels the quantity of oligosaccharides measured by CE polyanionic saccharides by MALDI-TOFMS. This new approach may also be useful for the analysis and sequencing of more complex glycosaminoglycans such as chondro/dermatan sulfates, and heparin/heparan sulfate. The application of this technique on such molecules is currently ongoing.
References
- 1.Laurent TC, Fraser JR. Hyaluronan. FASEB J. 1992;6:2397–2404. [PubMed] [Google Scholar]
- 2.Fraser JR, Laurent TC, Laurent J. Hyaluronan: its nature, distribution, functions and turnover. J. Intern. Med. 1997;242:27–33. doi: 10.1046/j.1365-2796.1997.00170.x. [DOI] [PubMed] [Google Scholar]
- 3.Sherman L, Sleeman J, Herrlich P, Ponta H. Hyaluronate receptors: key players in growth, differentiation, migration and tumor progression. Curr. Opin. Cell. Biol. 1994;6:726–733. doi: 10.1016/0955-0674(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 4.Estes JM, Adzick NS, Harrison MR, Longaker MT, Stern R. Hyaluronate metabolism undergoes an ontogenic transition during fetal development: implications for scar-free wound healing. J. Pediatr. Surg. 1993;28:1227–1231. doi: 10.1016/s0022-3468(05)80303-3. [DOI] [PubMed] [Google Scholar]
- 5.Brown JJ, Papaioannou VE. Ontogeny of hyaluronan secretion during early mouse development. Development. 1993;117:483–492. doi: 10.1242/dev.117.2.483. [DOI] [PubMed] [Google Scholar]
- 6.Meyer MF, Kreil G. Cells expressing the DG42 gene from early Xenopus embryos synthesize hyaluronan. Proc. Natl. Acad. Sci. U.S.A. 1996;93:4543–4547. doi: 10.1073/pnas.93.10.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gakunga P, Frost G, Shuster S, Cunha G, Formby B, Stern R. Hyaluronan is a prerequisite for ductal branching morphogenesis. Development. 1997;124:3987–3997. doi: 10.1242/dev.124.20.3987. [DOI] [PubMed] [Google Scholar]
- 8.Wang KK, Nemeth IR, Seckel BR, Chakalis-Haley DP, Swann DA, Kuo JW, Bryan DJ, Cetrulo CL., Jr Hyaluronic acid enhances peripheral nerve regeneration in vivo. Microsurgery. 1998;18:270–275. doi: 10.1002/(sici)1098-2752(1998)18:4<270::aid-micr11>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 9.Chen WY, Abatangelo G. Function of hyaluronan in wound repair. Wound Repair Regen. 1999;7:79–89. doi: 10.1046/j.1524-475x.1999.00079.x. [DOI] [PubMed] [Google Scholar]
- 10.Edelstam GA, Laurent UB, Lundkvist OE, Fraser JR, Laurent TC. Concentration and turnover of intraperitoneal hyaluronan during inflammation. Inflammation. 1992;16:459–469. doi: 10.1007/BF00918972. [DOI] [PubMed] [Google Scholar]
- 11.Knudson W. Tumor-associated hyaluronan. Providing an extracellular matrix that facilitates invasion. Am. J. Physiol. 1996;148:1721–1726. [PMC free article] [PubMed] [Google Scholar]
- 12.Lokeshwar VB, Obek C, Soloway MS, Block NL. Tumor-associated hyaluronic acid: a new sensitive and specific urine marker for bladder cancer. Cancer Res. 1997;15:773–777. [PubMed] [Google Scholar]
- 13.Hayen W, Goebeler M, Kumar S, Riessen R, Nehls V. Hyaluronan stimulates tumor cell migration by modulating the fibrin fiber architecture. J. Cell Sci. 1999;112:2241–2251. doi: 10.1242/jcs.112.13.2241. [DOI] [PubMed] [Google Scholar]
- 14.Itano N, Sawai T, Miyaishi O, Kimata K. Relationship between hyaluronan production and metastatic potential of mouse mammary carcinoma cells. Cancer Res. 1999;59:2499–2504. [PubMed] [Google Scholar]
- 15.Brecht M, Mayer U, Schlosser E, Prehm P. Increased hyaluronate synthesis is required for fibroblast detachment and mitosis. Biochem. J. 1986;239:445–450. doi: 10.1042/bj2390445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoneda M, Yamagata M, Suzuki S, Kimata K. Hyaluronic acid modulates proliferation of mouse dermal fibroblasts in culture. J. Cell Sci. 1988;90:265–273. doi: 10.1242/jcs.90.2.265. [DOI] [PubMed] [Google Scholar]
- 17.West DC, Kumer S. Hyaluronan and angiogenesis. Ciba Found. Symp. 1989;143:187–201. doi: 10.1002/9780470513774.ch12. [DOI] [PubMed] [Google Scholar]
- 18.Deed R, Rooney P, Kumar P, Norton JD, Smith J, Freemont AJ, Kumar S. Early-response gene signalling is induced by angiogenic oligosaccharides of hyaluronan in endothelial cells. Inhibition by non-angiogenic, high-molecular-weight hyaluronan. Int. J. Cancer. 1997;71:251–256. doi: 10.1002/(sici)1097-0215(19970410)71:2<251::aid-ijc21>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 19.Furuya T, Yamagata S, Shimoyama Y, Fujihara M, Morishima N, Ohtsuki K. Biochemical characterization of glycyrrhizin as an effective inhibitor for hyaluronidases from bovine testis. Biol. Pharm. Bull. 1997;20:973–977. doi: 10.1248/bpb.20.973. [DOI] [PubMed] [Google Scholar]
- 20.Li MW, Yudin AI, VandeVoort CA, Sabeur K, Primakoff P, Overstreet JW. Inhibition of monkey sperm hyaluronidase activity and heterologous cumulus penetration by flavonoids. Biol. Reprod. 1997;56:1383–1389. doi: 10.1095/biolreprod56.6.1383. [DOI] [PubMed] [Google Scholar]
- 21.Toida T, Ogita Y, Suzuki A, Toyoda H, Imanari T. Inhibition of hyaluronidase by fully O-sulfonated glycosaminoglycans. Arch. Biochem. Biophys. 1999;370:176–182. doi: 10.1006/abbi.1999.1395. [DOI] [PubMed] [Google Scholar]
- 22.Kakehi K, Kinoshita K, Hayase S, Oda Y. Capillary electrophoresis of N-acetylneuraminic acid polymers and hyaluronic acid: correlation between migration order reversal and biological functions. Anal. Chem. 1999;71:1592–1596. doi: 10.1021/ac981248y. [DOI] [PubMed] [Google Scholar]
- 23.Nobel PW, McKee CM, Coweman M, Shin HS. Hyaluronan fragments activate an NF-kappa B/I-kappa B alpha autoregulatory loop in murine macrophages. J. Exp. Med. 1996;183:2373–2378. doi: 10.1084/jem.183.5.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cramer JA, Bailey LC. A reversed-phase ion-pair high-performance liquid chromatography method for bovine testicular hyaluronidase digestions using postcolumn derivatization with 2-cyanoacetamide and ultra violet detection. Anal. Biochem. 1991;196:183–191. doi: 10.1016/0003-2697(91)90137-i. [DOI] [PubMed] [Google Scholar]
- 25.Busse K, Averbeck M, Anderegg U, Arnold K, Simon JC, Schiller J. The signal-to-noise ratio as a measure of HA oligomer concentration: a MALDI-TOF MS study. Carbohydr. Res. 2006;341:1065–1070. doi: 10.1016/j.carres.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Hirano K, Sakai S, Ishikawa T, Avci FY, Linhardt RJ, Toida T. Preparation of the methyl ester of hyaluronan and its enzymatic degradation. Carbohydr. Res. 2005;340:2297–2304. doi: 10.1016/j.carres.2005.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephen LC, David JO. The separation of chondroitin sulfate disaccharides and hyaluronan oligosaccharides by capillary electrophoresis. Anal. Biochem. 1991;195:132–140. doi: 10.1016/0003-2697(91)90308-g. [DOI] [PubMed] [Google Scholar]
- 28.Toida T, Toyoda H, Imanari T. High-resolution proton nuclear magnetic resonance studies on chondroitin sulfates. Anal. Sci. 1993;9:53–58. [Google Scholar]
- 29.Toida T, Maruyama M, Ogita Y, Suzuki A, Toyoda H, Imanari T, Linhardt RJ. Preparation of anticoagulant activity of fully O-sulfonated glycosaminoglycans. Intern. J. Biol. Macromol. 1999;26:233–241. doi: 10.1016/s0141-8130(99)00088-4. [DOI] [PubMed] [Google Scholar]
- 30.Schiller J, Arnhold J, Benard S, Reichl S, Arnold K. Cartilage degradation by hyaluronate lyase and chondroitin ABC lyase: a MALDI-TOF mass spectrometric study. Carbohydr. Res. 1999;318:116–122. doi: 10.1016/s0008-6215(99)00063-4. [DOI] [PubMed] [Google Scholar]
- 31.Yeung B, Marecak D. Molecular weight determination of hyaluronic acid by gel filtration chromatography coupled to matrix-assisted laser desorption ionization mass spectrometry. J. Chromatogr. A. 1999;852:573–581. doi: 10.1016/s0021-9673(99)00647-0. [DOI] [PubMed] [Google Scholar]
- 32.Mahoney DJ, Aplin RT, Calabro A, Hascall VC, Day AJ. Novel methods for the preparation and characterization of hyaluronan oligosaccharides of defined length. Glycobiology. 2001;11:1025–1033. doi: 10.1093/glycob/11.12.1025. [DOI] [PubMed] [Google Scholar]
- 33.Holmbeck S, Lerner L. Separation of hyaluronan oligosaccharides by the use of anion-exchange HPLC. Carbohydr. Res. 1993;239:239–244. doi: 10.1016/0008-6215(93)84218-u. [DOI] [PubMed] [Google Scholar]