Abstract
BACKGROUND:
The virulent factors of Escherichia coli (E.coli) play an important role in the process of pathopoiesis. The study aimed to compare drug-resistant genes and virulence genes between extended spectrum β-lactamases (ESBLs)-producing E.coli and non-ESBLs-producing E.coli to provide a reference for physicians in management of hospital infection.
METHODS:
From October 2010 to August 2011, 96 drug-resistant strains of E.coli isolated were collected from the specimens in Qingdao Municipal Hospital, Qingdao, China. These bacteria strains were divided into a ESBLs-producing group and a non-ESBLs-producing group. Drug sensitivity tests were performed using the Kirby-Bauer (K-B) method. Disinfectant gene, qacEΔ1-sull and 8 virulence genes (CNF2, hlyA, eaeA, VT1, est, bfpA, elt, and CNF1) were tested by polymerase chain reaction (PCR).
RESULTS:
Among the 96 E.coli isolates, the ESBLs-producing E.coli comprised 46 (47.9%) strains and the non-ESBLs-producing E.coli consisted of 50 (52.1%) strains. The detection rates of multiple drug-resistant strain, qacEΔ1-sull, CNF2, hlyA, eaeA,VT1, est, bfpA, elt, and CNF1 in 46 ESBLs-producing E.coli isolates were 89.1%, 76.1%, 6.5%, 69.6%, 69.6%, 89.1%, 10.9%, 26.1%, 8.7%, and 19.6%, respectively. In the non-ESBLs-producing E.coli strains, the positive rates of multiple drug-resistant strain, qacEΔ1-sull, CNF2, hlyA, eaeA, VT1, est, bfpA, elt, and CNF1 were 62.0%, 80.0%, 16.0%, 28.0%, 64.0%, 38.0%, 6.0%, 34.0%, 10.0%, and 24.0%, respectively. The difference in the detection rates of multiple drug-resistant strain, hlyA and VT1 between the ESBLs-producing E.coli strains and the non-ESBLs-producing E.coli strains was statistically significant (P<0.05).
CONCLUSION:
The positive rate of multiple drug-resistant strains is higher in the ESBLs-producing strains than in the non-ESBLs-producing strains. The expression of some virulence genes hlyA and VT1 varies between the ESBLs-producing strains and the non-ESBLs-producing strains. Increased awareness of clinicians and enhanced testing by laboratories are required to reduce treatment failures and prevent the spread of multiple drug-resistant strains.
KEY WORDS: ESBLs-producing Escherichia coli, Non-ESBLs-producing E.coli, Drug-resistant genes, Virulence genes, Multiple drug-resistant
INTRODUCTION
Escherichia coli (E.coli), one of the most common gram-negative blood culture isolates in hospitalized patients, has become a very important pathogen associated with hospital infection.[1] The deadly E.coli outbreak killed persons and spread across Europe in May 2011. Multidrug resistant E. coli (MDR-ECO) strains have increased with the application of broad-spectrum antibiotics. A large number of drug-resistant genes have been reported.[2] Since ESBLs-producing Enterobacteriaceae were first reported in 1983 from Germany, a steady increase of these strains has been reported worldwide.[3] ESBLs producers are clinically resistant to many β-lactams,[4] while non-ESBLs producers still keep a good sensitivity to most β-lactams. E.coli possess many kinds of virulence factors such as intimin (encoded by the eaeA gene),[5] cytotoxic necrotizing factor 1 (CNF1), cytotoxic necrotizing factor 2 (CNF2), hemolysin (encoded by the hlyA gene),[6] bundle-forming pilli (encoded by the bfpA gene),[7] heat-labile toxin (encoded by the elt gene), heat-stable toxin (encoded by the est gene),[8] and verotoxin1 (encoded by the VT1 gene).[9]
The virulent factors of E.coli play an important role in the process of pathopoiesis. This study was undertaken to compare the difference in drug-resistant genes and virulence genes between the ESBLs-producing strains and the non ESBLs-producing strains from clinical samples by using polymerase chain reaction (PCR).
METHODS
Bacterial strains
Drug-resistant E.coli isolates were collected from clinical specimens at Qingdao Municipal Hospital within 11 months between Octomber 2010 and August 2011. They were identified by different physiological and biochemical standard methods.[8] Drug sensitivity tests were performed by the Kirby-Bauer (K-B) method which was recommended by the WHO, and the judgement was made by the explaination standard of Cinical and Laboratory Standards Institue (CLSI). Multiple drug- resistant strain was resistant to three (e.g. aminoglycosides, sulphonamides, macrolides) or more than three antibiotics. Selected bacteria stains were stored at –70 ºC. E.coli O157:H7 were used as a reference strain in the study.
DNA extraction
Boiling with double distilled water was adopted for DNA extraction. Two to 3 colonies of isolated bacterial strains were placed into a tube containing 100 μL double distilled water. DNA was extracted at 100 ºC for 10 minutes, then the cells were pelleted by centrifugation. The supernatant containing DNA was taken out and stored at –20 ºC.
Polymerase chain reaction (PCR)
DNA supernatant was used for PCR analysis. PCR was performed in a final reaction volume of 25 μL, which comprised 15.3 μL sterile milliQ water, 2.5 μL 10×reaction buffer, 2 μL deoxynucleoside triphosphates (dNTPs), 1 μL (each) reverse and forward primers, 0.2 μL of Taq DNA polymerase, and 3 μL of bacterial lysate (supernatant with template DNA). Amplifications were performed with the GeneAmp PCR System 9600 (Perkin-Elmer Applied Biosystems, Inc., Foster City, Calif.). PCR amplification was started with initial denaturation at 94 ºC for 5 minutes, then 35 cycles of denaturation at 94 ºC for 30 seconds, annealing at 54 ºC for 30 seconds, extension at 72 ºC for 1 minute. A final elongation at 72 ºC for 10 minutes was also conducted. Finally, PCR amplification products were stored at 4 ºC. Primers used and annealing temperature in PCR are listed in Table 1.
Table 1.
Oligonucleotide primer pairs used in this study
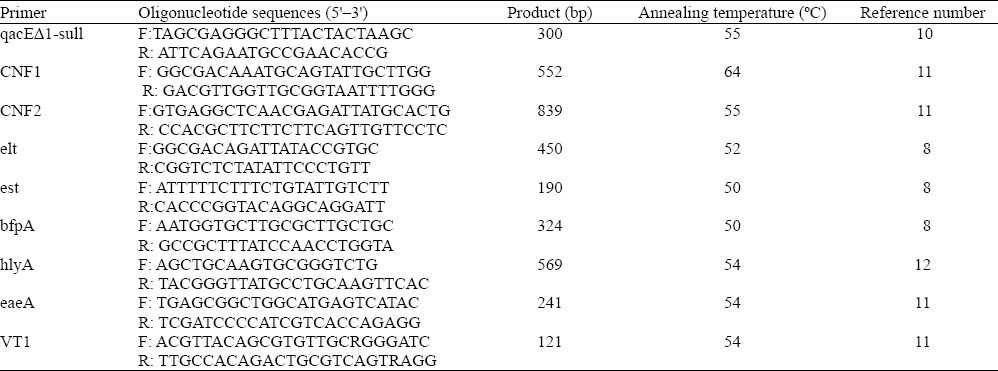
The PCR amplification product was separated by submarine gel electrophoresis on 1.2% agarose, stained with ethidium bromide,and gel image was captured digitally with UV transillumination.
Statistical analysis
The data of the study were analyzed by using the SPSS 17.0. The Chi-square test was used to compare the difference of virulence between the ESBLs-producing E.coli and the Non-ESBLs-producing E.coli. P<0.05 was considered statistically significant.
RESULTS
Multiple resistance analysis
A total of 96 E.coli isolates were obtained from clinical samples, including urine (n=49), sputamentum (n=30), blood (n=6), gastric fluid (n=1), bile (n=3), drain (n=1), liquor puris (n=1), synovia (n=1), pucture fluid (n=1), pharynx swab (n=1), and ascites (n=2). Among the 96 patients, 40 were male and 56 were female, with a mean age of 65.2 years.
These isolated bacterial strains were divided into a ESBLs-producing E.coli group and a non-ESBLs-producing E.coli group according to the extended spectrum β-lactamases. The ESBLs-producing E.coli group consisted of 46 strains, and the non-ESBLs-producing E.coli group 50 strains. Multiple drug resistance was detected by antibiotic sensitivity test (Table 2). Of the 96 isolates,73 (76.0%) were multiple drug-resistant, and the rest were non multiple drug-resistant. In E.coli with ESBLs, 91.3% strains were multiple drug-resistant, while in E.coli without ESBLs 62.0% were multiple drug-resistant (P<0.05). Consequently, the rate of multiple drug resistance in the ESBLs-producing E.coli group was higher than that in the non-ESBLs-producing E.coli group.
Table 2.
Distributions of multiple drug-resistant strain in E.coli with ESBLs and without ESBLs

Comparison of gene detection rate between the ESBLs-producing E.coli group and the non-ESBLs-producing E.coli group
The results PCR in this study are shown in Figure 1. Among the examined 96 drug resistant strains, qacEΔ1-sull was the most frequently detected gene, accounting for 78.1%. Thirty-five (76.1%) of the 46 ESBLs-producing E.coli strains were qacEΔ1-sull positive; the 40 (80.0%) of the 50 non ESBLs-producing E.coli strains were qacEΔ1-sull positive. The detection rate of the qacEΔ1-sull gene was not statistically different (χ2=0.215, P>0.500). The positive number of CNF2, hlyA, eaeA, VT1, est, bfpA, elt, and CNF1 in the ESBLs-producing E.coli group was 3, 32, 32, 41, 5, 12, 4, and 9, respectively. The positive number of CNF2, hlyA, eaeA, VT1, est, bfpA, elt, and CNF1 in the non- ESBLs-producing E.coli group was 8, 14, 32, 19, 3, 17, 5, and 12, respectively (Table 3). The positive rates of hlyA and VT1 in the ESBLs-producing E.coli group were significantly higher than those in the non-ESBLs- producing E.coli group (P<0.05). The detection of other six virulence genes between the two groups of E.coli was not statistically significant.
Figure 1.
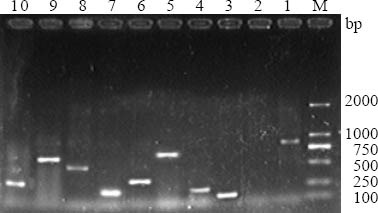
Polymerase chain reaction (PCR)-amplified products. M: DL2000 DNA marker; 1: CNF2 (839bp); 2: negative control; 3: VT1 (121bp); 4: eaeA (241bp); 5: hlyA (569bp); 6: bfpA (324bp); 7: est (190bp); 8: elt (450bp); 9: CNF1 (552bp); 10: qacEΔ1-sull (300bp).
Table 3.
The detection of virulence genes in E.coli with ESBLs and without ESBLs
DISCUSSION
Disinfection is an important measure to prevent and control hospital infection.[13] In the condition of the disinfectant genes expressed by E.coli, disinfection would not live up to the expected effect.[14,15] E.coli are a common pathogen of nosocomial infection.[16] With the wide use of antibiotics, drug resistance becomes more and more serious. But it is more difficult to prevent nosocomial infection if it is caused by mutiplle drug resistance of E.coli. Since multiple drug-resistant ESBLs-producing E.coli infection is increased gradually, it is very difficult to cure.[17] More effort thus should be taken to prevent nosocomial infection outbreak. Studies indicated that the drug resistance rate of ESBLs-producing E.coli was higher than that of non-ESBLs-producing E.coli.[18,19] It was also reported that the rate of multiple drug-resistant strain was higher in ESBLs-producing E.coli than in non ESBLs-producing E.coli, and that multiple drug-resistant strains occurred frequently in drug-resistant strains.[20] Hence more attention should be paid clinically to ESBLs-producing E.coli, and antibiotics should be used rationally according to the results of antimicrobial susceptibility tests. Multiple drug-resistance may be related to the extensive use of spectrum β-lactamases. However, its mechanism is still uncertain. Nosocomial infection is due to E.coli virulence genes.[21] The strains that are subjected to continuous antimicrobial challenges become resistant to some drugs but are likely to become more virulent than their predecessors. Thus it is very important to underststand the virulence genes of E.coli. In the present study we compared several virulence genes between the ESBLs-producing E.coli group and the non-ESBLs-producing E.coli group. After verification, the positive rates of hlyA and VT1 in the ESBLs- producing E.coli group were higher than those in the non ESBLs-producing E.coli group. This study revealed drug-resistance and virulence of the ESBLs-producing E.coli group were more serious than those of the non ESBLs-producing E.coli group. Chacón et al[22] found that virulence factors may play an important role in pathogensis, and that the detection of virulence genes is a crucial step in determining the potential pathogenicity. Since clinical laboratories only do drug sensitive test without analysis of bacteria virulence, clinicians could not estimate the toxicity of E.coli. This study revealed the virulence of ESBLs-producing E.coli provides evidence for clincians.
In conclusion, the results of this study suggest that drug resistance and expression of virulence genes in ESBLs-producing E.coli are more serious than in non -ESBLs-producing E.coli. Therefore, attention should be paid to the ESBLs-producing E.coli strains in order to prevent the outbreak of nosocomial E.coli infection in related departments.
ACKNOWLEDGMENTS
The authors are grateful to the technicians of Clinical Laboratory of Qingdao Municipal Hospital for their cooperation in collecting samples, and also to Pro. Bing Luo from Department of Medical Microbiology, Qingdao University School of Medicine for his support in data presentation and manuscript preparation.
Footnotes
Funding: None.
Ethical approval: The study was approved by the Ethical Committee of Qingdao University Medical College, Qingdao, China.
Conflicts of interest: The authors have no financial or other conflicts of interest regarding this article.
Contributors: Li S proposed and wrote the study. All authors read and approved the final manuscript
REFERENCES
- 1.Hilali F, Ruimy R, Saulnier P, Barnabé C, LeBouguénec C, Tibayrenc M, et al. Prevalence of virulence genes and clonality in Escherichia coli strains that cause bacteremia in cancer patients. Infect Immun. 2000;68:3983–3989. doi: 10.1128/iai.68.7.3983-3989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsui K, Wong SS, Lin LC, Tsai CR, Chen LC, Huang CH. Laboratory identification, risk factors, and clinical outcomes of patients with bacteremia due to Escherichia coli and Klebsiella pneumoniae Producing Extended-Spectrum and AmpC type β-Lactamases. J Microbiol Immunol Infect. 2012;45:193–199. doi: 10.1016/j.jmii.2011.11.003. Epub 2012 May 12. [DOI] [PubMed] [Google Scholar]
- 3.Paterson DL, Bonomo RA. Extended-spectrum betalactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacoby GA. Extended-spectrum beta-lactamases and other enzymes providing resistance to oxyimino-beta-lactams. Infect Dis Clin North Am. 1997;11:875–887. doi: 10.1016/s0891-5520(05)70395-0. [DOI] [PubMed] [Google Scholar]
- 5.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Afset JE, Bruant G, Brousseau R, Harel J, Anderssen E, Bevanger L, et al. Identification of virulence genes linked with diarrhea due to atypical enteropathogenic Escherichia coli by DNA microarray analysis and PCR. J Clin Microbiol. 2006;44:3703–3711. doi: 10.1128/JCM.00429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke SC, Haigh RD, Freestone PP, Williams PH. Virulence of Enteropathogenic Escherichia coli, a Global Pathogen. Clin Microbiol Rev. 2003;16:365–378. doi: 10.1128/CMR.16.3.365-378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraborty S, Deokule JS, Garg P, Bhattacharya SK, Nandy RK, Nair GB, et al. Concomitant infection of enterotoxigenic escherichia coli in an outbreak of cholera caused by Vibrio cholerae O1 and O139 in Ahmedabad, India. J Clin Microbiol. 2001;39:3241–3246. doi: 10.1128/JCM.39.9.3241-3246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pabst WL, Altweqq M, Kind C, Mirjanic S, Hardegger D, Nadal D. Prevalence of Enteroaggregative Escherichia coli among children with and without diarrhea in Switzerland. J Clin Microbiol. 2003;41:2289–2293. doi: 10.1128/JCM.41.6.2289-2293.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin ZP, Cheng YB, Wang QS, Wang Q, Ge YJ. Drug resistance of Acinetobacter baumanii isolated from children in the pediatric intensive care unit. Zhongguo Dang Dai Er Ke Za Zhi. 2012;14:229–230. [PubMed] [Google Scholar]
- 11.Pass MA, Odedra R, Batt RM. Multiplex PCRs for identification of Escherichia coli virulence genes. J Clin Microbiol. 2000;38:2001–2004. doi: 10.1128/jcm.38.5.2001-2004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang G, Clark CG, Rodgers FG. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J Clin Microbiol. 2002;40:3613–3619. doi: 10.1128/JCM.40.10.3613-3619.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuenck RP, Dadalti P, Silva MG, Fonseca LS, Santos KR. Oxacillin-and mupirocin-resistant Staphylococcus aureus: in vitro activity of silver sulphadiazine and cerium nitrate in hospital strains. J Chemother. 2004;16:453–458. doi: 10.1179/joc.2004.16.5.453. [DOI] [PubMed] [Google Scholar]
- 14.Lee JY, Ko KS. OprD mutations and inactivation, expression of efflux pumps and AmpC, and metallo-β-lactamases in carbapenem-resistant Pseudomonas aeruginosa isolates from South Korea. Int J Antimicrob Agents. 2012 May;:25. doi: 10.1016/j.ijantimicag.2012.04.004. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 15.Pirnay JP, De Vos D, Cochez C, Bilocq F, Pirson J, Struelens M, et al. Molecular epidemiology of Pseudomonas aeruginosa colonization in a burn unit: persistence of a multidrug-resistant clone and a silver sulfadiazine-resistant clone. J Clin Microbiol. 2003;41:119–202. doi: 10.1128/JCM.41.3.1192-1202.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avazashvili N, Nozadze T, Chikviladze D, Gachechiladze Kh, Metreveli D, Mikeladze M. Microbial spectrum of nosocomial pneumonia in traumatological patients. Georgian Med News. 2011;194:76–79. [PubMed] [Google Scholar]
- 17.Helfand MS, Bonomo RA. Extended-spectrum beta-lactamases in multidrug-resistant Escherichia coli: changing the therapy for hospital-acquired and community-acquired infections. Clin Infect Dis. 2006;43:1415–1416. doi: 10.1086/508891. Epub 2006 Oct 25. [DOI] [PubMed] [Google Scholar]
- 18.Palasubramaniam S, Subramaniam G, Muniandy S, Parasakthi N. Extended-spectrum beta-lactam resistance due to AmpC hyperproduction and CMY-2 coupled with the loss of OMPK35 in Malaysian strains of Escherichia coli and Klebsiella pneumoniae. Microb Drug Resist. 2007;13:186–190. doi: 10.1089/mdr.2007.726. [DOI] [PubMed] [Google Scholar]
- 19.Palasubramaniam S, Subramaniam G, Muniandy S, Parasakthi N. Extended-spectrum beta-lactam resistance due to AmpC hyperproduction and CMY-2 coupled with the loss of OMPK35 in Malaysian strains of Escherichia coli and Klebsiella pneumoniae. Microb Drug Resist. 2007;13:186–190. doi: 10.1089/mdr.2007.726. [DOI] [PubMed] [Google Scholar]
- 20.Hussain M, Hasan F, Shah AA, Hameed A, Jung M, Rayamajhi N, et al. Prevalence of class A and AmpC β-lactamases in clinical Escherichia coli isolates from Pakistan Institute of Medical Science, Islamabad, Pakistan. Jpn J Infect Dis. 2011;64:249–252. [PubMed] [Google Scholar]
- 21.Pradel N, Bertin Y, Martin C, Livrelli V. Molecular analysis of shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome patients and dairy samples in France. Appl Environ Microbiol. 2008;74:2118–2128. doi: 10.1128/AEM.02688-07. Epub 2008 Feb 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chacón MR, Figueras MJ, Castro-Escarpulli G, Soler L, Guarro J. Distribution of virulence genes in clinical and environmental isolates of Aeromonas spp. Antonie van Leeuwenhoek. 2003;84:269–278. doi: 10.1023/a:1026042125243. [DOI] [PubMed] [Google Scholar]


