Abstract
Dicer is a central enzymatic player in RNA interference (RNAi) pathways that acts to regulate gene expression in nearly all eukaryotes. Although the cytoplasmic function of Dicer is well-documented in mammals, its nuclear function remains obscure. Here we show that Dicer is present in both the nucleus and cytoplasm, but that its nuclear levels are tightly regulated. In its nuclear manifestation, Dicer interacts with RNA polymerase II (Pol II) at actively-transcribed gene loci. Loss of Dicer causes the appearance of endogenous dsRNA, leading to induction of the interferon response pathway and consequent cell death. Our results suggest that Pol II-associated Dicer restricts endogenous dsRNA formation from overlapping non-coding RNA transcription units. Failure to do so has catastrophic effects on cell function.
Introduction
The central role of RNA interference (RNAi) in regulating gene expression, is a general feature of eukaryotes1-3, and begins with formation of double strand RNA (dsRNA). Endogenous dsRNA derives from overlapping convergent transcription of either distinct protein coding genes4-6 or natural antisense non-coding RNA7. dsRNA may also enter the cell in the form of an RNA virus where it triggers a cytoplasmic antiviral mechanism called the interferon response pathway8. This leads to cellular apoptosis and consequent destruction of virally-infected cells9. Another major source of dsRNA is inverted repeat transcription, resulting in formation of RNA hairpin structures. Two RNAi-associated endoribonucleases process hairpin-derived dsRNA in higher eukaryotes. Nuclear hairpin dsRNA is recognised by the RNAse III type endonuclease, Drosha, together with an RNA binding protein DGCR810. Co-transcriptional RNA cleavage at the base of the stem loop releases hairpin RNA, which is then transported from the nucleus to cytoplasm11. Here, the second RNAse, Dicer, cleaves the dsRNA into short dsRNAs12, forming microRNAs or siRNAs that, together with argonaute proteins and associated factors, abrogate target mRNA by either degradation or translational inhibition2,12. Such RNAi is referred to as post-transcriptional gene silencing (PTGS) as it acts on cytoplasmic mRNA. In many eukaryotes, and exemplified by S.pombe, a second RNAi mechanism also exists where nuclear Dicer generates siRNA associated with a nuclear argonaute complex called RITS13. This recruits histone and DNA methyl transferase14,15 to target homologous chromatin and generates repressed chromatin structures, a process called transcriptional gene silencing (TGS).
The prevailing view has been that mammalian RNAi relies exclusively on PTGS through the action of microRNAs. Furthermore, mammalian Dicer lacks obvious nuclear localisation signals (NLS)17-19, and over-expression of GFP-tagged Dicer shows a restricted cytoplasmic localisation20. This argues that Dicer has a solely cytoplasmic function. However, it has recently been demonstrated that the C-terminal dsRNA binding domain of human Dicer possesses a noncanonical NLS implying a nuclear function21. Indeed, a number of studies over the past decade indicate that nuclear RNAi occurs in mammalian cells as in other eukaryotes22-27,5.
In light of uncertainties relating to nuclear Dicer, we set out to determine if Dicer localises to the nucleus, and, if so, what its function is in this cellular compartment. We show that Dicer is detectible in the nucleus of mammalian cells, but that its levels appear regulated. Furthermore, Dicer interacts with Pol II and chromatin genome-wide, and Dicer binding loci are associated with both dsRNA and small RNAs, presumably related to Dicer activity. We then looked for the nuclear function of Dicer and show that its absence correlates with accumulation of dsRNA, which triggers the interferon response pathway, leading to cellular apoptosis. We have, therefore, identified a major nuclear role for Dicer in restricting the formation of dsRNA from convergent transcription. Failure to perform this task leads to interferon activation and cellular apoptosis.
Results
Dicer nuclear location
We employed a Dicer antibody (Abcam: 13D6) to investigate the cellular distribution of Dicer in HEK293 cells harbouring a chromosomally-integrated inducible Dicer shRNA expression cassette28. Western analysis revealed 200kDa Dicer protein in lysates from HEK293 cells, but not following induction of Dicer shRNA (Supplementary Fig. 1a). Complete loss of Dicer cannot be achieved with this system, as Dicer itself is needed to process the shRNA that leads to Dicer knockdown.
We next carried out immunofluorescence analysis on these HEK293 cells using confocal microscopy. As shown in representative cell images (Fig. 1a), strong Dicer signals were detected especially around the cytoplasmic side of the nuclear periphery, where pre-microRNAs are processed into mature microRNAs2. However, a clear punctuate pattern was also visible throughout the nucleoplasm; these signals were absent following anti-Dicer shRNA induction.
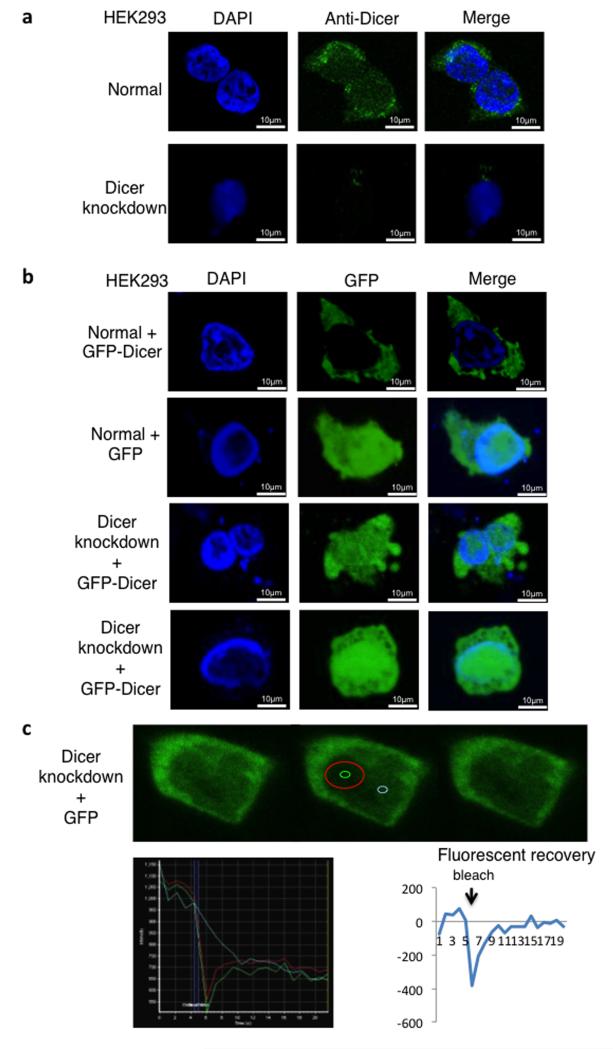
Figure 1. Dicer localizes in both nucleus and cytoplasm of HEK293 cells.
a) Immunofluorescence analyses of HEK293 cells with an integrated inducible Dicer shRNA expression cassette, using DAPI staining (blue) and anti-Dicer antibody (green). Top panels: Uninduced cells (normal),showing cytoplasmic and nuclear localization of Dicer Bottom panels: Induction of the shRNA cassette (Dicer knockdown), Dicer signals are absent (lower panels).
b) Expression of GFP-Dicer or GFP alone (green) in HEK293 cells; nucleus stained by DAPI (blue). Uninduced cells (normal) show no colocalization of GFP-Dicer signals with DAPI staining. In contrast GFP colocalizes with DAPI (top two rows). Following Dicer knockdown (bottom panels), GFP-Dicer, like GFP, localizes in both cytoplasm and nucleus. Although GFP-Dicer is ultimately down-regulated by Dicer shRNA expression, it is expressed at detectable levels during experimental time frame.
c) FRAP analysis measuring GFP-Dicer nuclear dynamics in Dicer knockdown cells transfected with Dicer-GFP. Top, fluorescence microscopy shows GFP-Dicer in green. Left, the cell before bleaching. Middle, the red circle corresponds to bleached area; fluorescent recovery was measured in the green circle; fluorescence background values were measured in the blue circle. Right, the cell after recovery. Bottom left, absolute levels of fluorescence; bottom right, relative levels of fluorescence (subtracting background fluorescence). All experiments described in Fig. 1a-c were independently replicated three times.
Note that the images shown are single sections to eliminate cytoplasmic signals overlapping the nucleus. Western blot analysis of cellular fractions indicated that Dicer is present in both nucleus and cytoplasm, while mitochondrial Grp75 was only in the cytoplasm, confirming that the nuclear fraction is cytoplasm-free (Supplementary Fig. 1b).
We next investigated the pattern of GFP-tagged Dicer signal following transfection into HEK293 cells. Since GFP-Dicer is sensitive to the shRNA, we performed experiments within 24 hours of transfection. Whereas GFP expression showed equal nuclear and cytoplasmic signal, all GFP–Dicer signal was restricted to the cytoplasm in un-induced HEK293 cells (Fig. 1b). Thus, exogenous Dicer is actively restricted to the cytoplasm in normal HEK293 cells, as confirmed by Western blot analysis of nuclear and cytoplasmic fractions from cells transfected with GFP-tagged Dicer (Supplementary Fig. 1b), supporting previous analysis (ref 18). RFP-tagged Dicer was also restricted to the cytoplasm under normal conditions (Supplementary Fig. 1b).
When GFP-Dicer was expressed in HEK293 cells knocked down for endogenous Dicer, GFP signals were now clearly present in the nucleus. The same phenomenon was observed when GFP-Dicer was transfected in normal and Dicer-null murine ESC29 (Supplementary Fig. 1c). This implies that either the nuclear transport machinery has a higher affinity for the unmodified protein, or that the levels of nuclear Dicer are carefully titrated so that, in normal cells, additional transfected Dicer is blocked from nuclear import. In the latter scenario, only under Dicer knockdown conditions (anti-Dicer shRNA induction) will exogenous Dicer enter the nucleus.
To examine the dynamics of nuclear Dicer, we performed fluorescence recovery after photo-bleaching (FRAP) experiments on nuclear GFP-Dicer expressed in Dicer knockdown HEK293 cells. Remarkably, a photo-bleached GFP-Dicer signal at a specific nuclear location recovered within 10 seconds (Fig. 1c and Supplementary Fig. 1d), indicating high Dicer mobility within the nucleus.
Overall, our data demonstrate the nuclear presence of Dicer. Both the previous lack of a reliable Dicer antibody and the apparent saturation of endogenous nuclear Dicer may account for the previous view that Dicer is a cytoplasmic specific protein.
Dicer associates with Pol II and specific chromatin regions
To examine the role of nuclear Dicer, HeLa cell nuclear extracts were immuno-depleted for either Pol II or Dicer. Strikingly, immuno-depletion of Dicer caused a reduction in Pol II levels, and, similarly, depletion of Pol II reduced Dicer levels (Fig. 2a). Furthermore, immuno-precipitation (IP) of Pol II from nuclear extracts co-IPed Dicer (25% of input signal), while IP of Dicer co-IPed Pol II (17% of input signal). In the latter case, transcriptionally-active Pol IIO form was co-IPed by Dicer, indicating that Dicer preferentially associates with elongating Pol II (Fig. 2b). Both a core Pol II antibody (N20) and the large Pol II subunit C terminal domain (CTD)-specific antibody (8WG16)30 were used.
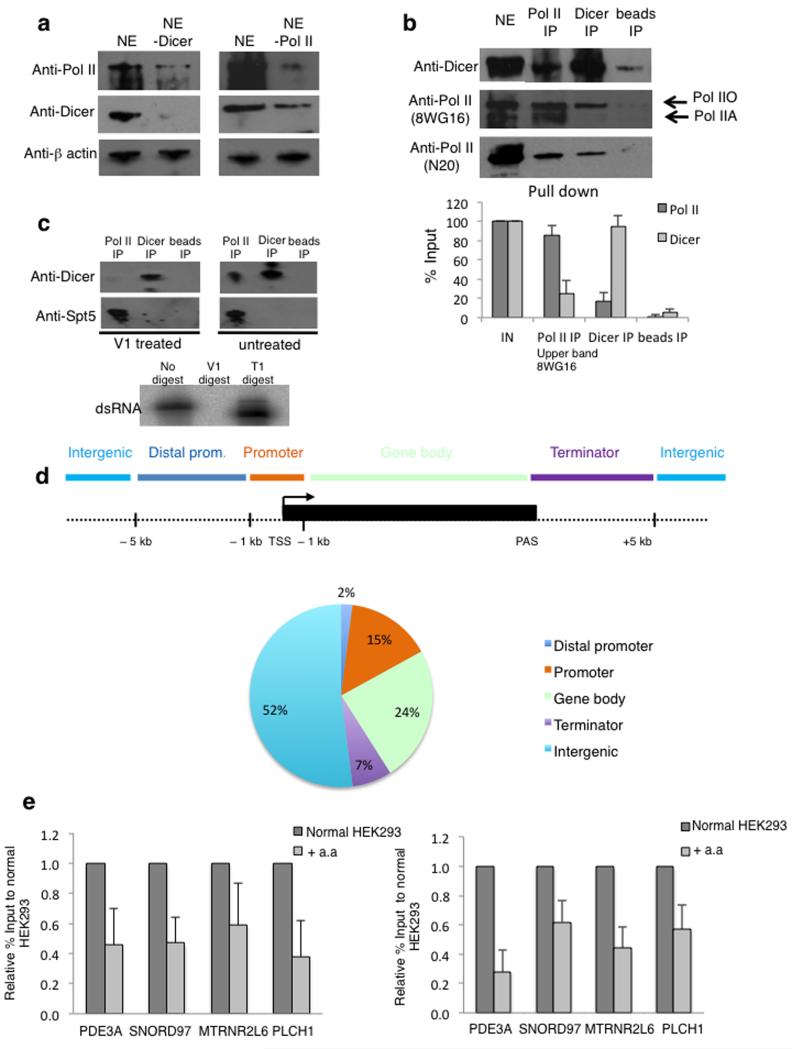
Figure 2. Dicer interaction with Pol II is dsRNA dependent and Dicer chromatin association is transcription dependent.
a) Immunodepletion analysis of nuclear extracts using anti-Pol II and anti-Dicer specific antibodies were followed by Western blot. β-actin was used as a control.
b) Co-immunoprecipitation of Pol II and Dicer in nuclear extracts, using specific antibodies followed by Western blot of Pol II and Dicer. Hyper-phosphorylated form of Pol II is depicted by Pol IIO, and un-phosphorylated as Pol IIA. Bottom; Pol II quantitation is based on the upper band observed with the 8WG16 antibody and the average values ± s.d. are from three independent biological experiments. See Supplementary Fig. 9 for uncropped blot images.
c) Nuclear extracts treated with dsRNA specific nuclease V1, followed by co-IP as in b). Western blot was performed using anti-Dicer and anti-Spt5 antibodies. Bottom; in vitro synthesized dsRNA digested with either V1 or T1 ribonucleases. All experiments described in Fig. 2a-c, were independently replicated three times. See Supplementary Fig. 9 for uncropped blot images.
d) Distribution of Dicer binding sites (ChIP-seq peaks) relative to UCSC RefGene coordinates. See Online Methods for details.
e) Pol II ChIP (left) and Dicer ChIP (right) over 4 Dicer binding loci in normal and α-amanitin (a.a) treated HEK293 cells. Pol II and Dicer levels are expressed as relative % of input normalized to untreated cells. Error bars, s.e.m. (n=3 cell cultures).
We next investigated whether the Dicer:Pol II association is dependent on the presence of dsRNA. Pre-treatment of nuclear extract with V1 double-strand specific nuclease caused a substantial reduction of Dicer association with Pol II. As a control, Pol II interaction with the elongation factor Spt5 was unaffected by V1 treatment (Fig. 2c). The efficiency and specificity of V1 treatment, was confirmed with in vitro synthesised 32P-dsRNA (see Online Methods), which was digested to completion by V1 nuclease, but not by single-strand specific T1 ribonuclease (Fig. 2c lower panel). Overall, these biochemical experiments argue that Dicer interacts with Pol IIO through a dsRNA moiety that is also Pol II associated.
Genomic analysis of Dicer association with chromatin (ChIP-seq) isolated from HEK293 cells reveals that Dicer associates with multiple human loci (Fig. 2d). The top 12 highest scoring Dicer:Pol II overlapping peaks are presented (Supplementary Fig. 2a), and 4 loci with different mRNA expression levels were selected for detailed study (Supplementary Fig. 2b). A metagene analysis of the top Dicer peaks showed its enrichment over transcription start-sites (TSS) and poly-A signals (PAS) (Supplementary Fig. 2c). We also investigated repetitive genomic elements in the Dicer ChIP-seq data. Long or short interspersed sequence (LINES and SINES) and long terminal repeat (LTR) elements were underrepresented, whilst Satellite, rRNA and tRNA DNA sequences were overrepresented (Supplementary Fig. 2d).
To test the dependency of Dicer binding to these Dicer positive loci on transcription, chromatin was isolated from HEK293 cells grown with or without the transcription inhibitor α-amanitin. Both Pol II and Dicer levels were reduced on chromatin at the 4 selected gene loci with α-amanitin (Fig. 2e), implying that they co-associate during transcription and that Dicer chromatin binding is transcription-dependent. As a control, we tested the effect of α-amanitin treatment on H3 levels over these loci (Supplementary Fig. 2e). Although we saw a small reduction in the levels of H3, the reduction in Pol II and Dicer levels was substantially higher. The Dicer-binding MTRNR2L6 gene was examined in greater detail. Two probes, one before and one after the Dicer-binding region showed the predicted pattern in Pol II and H3 levels while Dicer binding is at background levels at these positions (Supplementary Fig. 2f).
We next investigated the effect of Dicer knockdown on these 4 Dicer loci. Each gene generated at least ten-fold more nascent sense transcript (detected using intron specific primers) than antisense transcript based on strand specific RT-qPCR using specific primers (Fig. 3a). Interestingly, both sense and antisense transcripts were enhanced following Dicer knockdown (Fig. 3b and 3c). This effect was sensitive to dsRNA specific V1 ribonuclease (Fig. 3d and 3e), suggesting dsRNA formation that is restricted by Dicer. As negative controls GAPDH and 28S rDNA were similarly tested. No change in either sense or antisense transcript levels was detected (Supplementary Fig. 3a and 3b), ruling out the possibility of a general effect of Dicer knockdown on transcription.
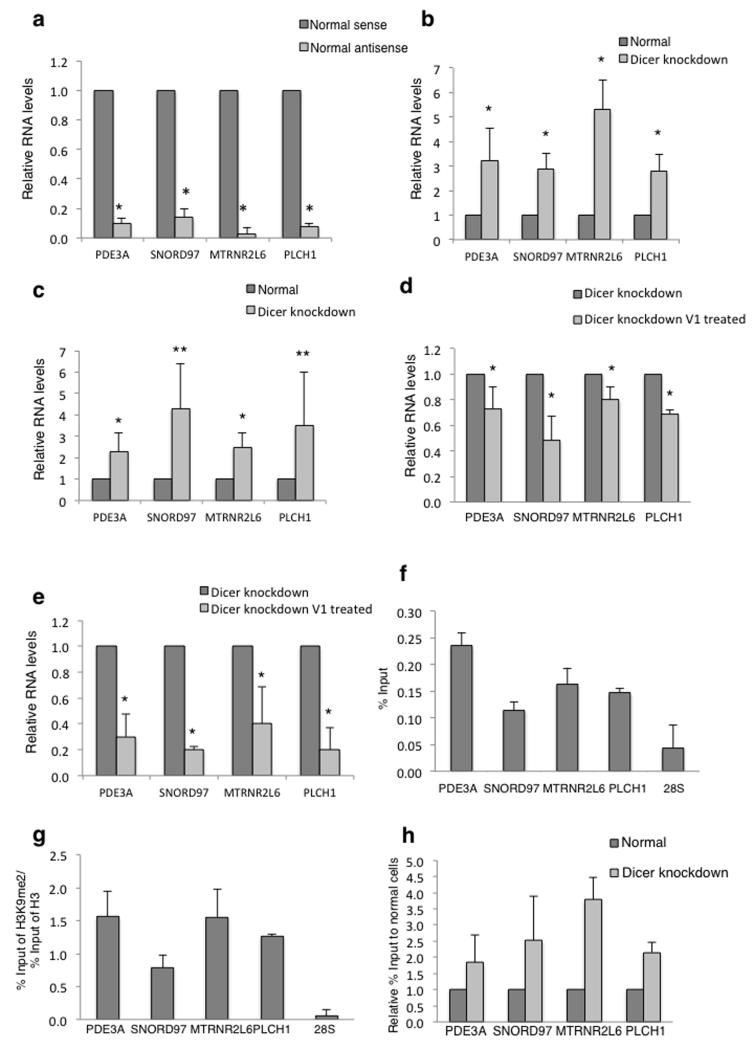
Figure 3. Dicer affects levels of nascent transcripts and TGS.
a) qRT-PCR analysis of sense and antisense transcripts from Dicer binding loci. Signals are normalized to the levels of sense transcript, set at 1. Error bars, s.d. (n=3 cell cultures). *P < 0.05 by two-tailed Student’s t test.
b) qRT-PCR analysis of sense transcripts of Dicer binding loci. Signals are normalized to the levels of sense transcript in normal cells, set at 1. Error bars, s.d. (n=3 cell cultures). *P < 0.05 by two-tailed Student’s t test.
c) qRT-PCR as in b) detecting antisense transcripts. Error bars, s.d. (n=3 cell cultures). *P < 0.05; **P < 0.12 by two-tailed Student’s t test.
d) qRT-PCR analysis of RNA isolated from Dicer knockdown HEK293 cells and treated with or without V1 nuclease. Signals are normalized to the levels of transcripts in untreated samples, set as 1. Error bars, s.d. (n=3 cell cultures). *P < 0.05; by two-tailed Student’s t test.
e) qRT-PCR as in d) detecting antisense transcripts. Error bars, s.d. (n=3 cell cultures). *P < 0.05; by two-tailed Student’s t test.
f) ChIP analysis of Ago1 levels Levels over 28S were used as a negative control. Error bars, s.e.m. (n=3 cell cultures).
g) Ratio of H3K9me2 ChIP signal versus H3 as in f).
h) ChIP analysis of Pol II levels on 4 tested genes in normal and Dicer knockdown HEK293 cells. Pol II levels are expressed as relative % of input normalised to normal cells. Error bars, s.e.m. (n=3 cell cultures).
Since these 4 Dicer positive loci generate dsRNA, we tested whether they were also subjected to TGS effects. Argonaute family proteins are required for TGS31 and Ago1 has been shown to direct siRNA-mediated TGS in human cells32. ChIP analysis revealed Ago1 signals enriched over each loci in comparison to 28S, suggesting Ago1-directed TGS (Fig. 3f). Since TGS leads to repressive chromatin formation, we also showed enrichment of H3K9me2 over each Dicer positive locus, as compared to 28S (Fig. 3g). H3K9me2 levels were normalised to total H3 signals to rule out variability in nucleosome density. Finally, TGS should lead to lower transcription levels. Indeed, all loci showed higher Pol II levels following Dicer knockdown (Fig. 3h), implying Dicer-dependent transcriptional silencing. Overall, our results indicate that Dicer associated with these loci, together with Ago1, promote the formation of H3K9me2 repressive marks through the processing of dsRNA.
Dicer restricts dsRNA accumulation in nuclei
We used a dsRNA specific antibody (J2) to directly visualise dsRNA accumulation by cellular immunofluorescence. To confirm J2 specificity, we tested its in vitro binding to dsRNA versus a hairpin pre-micro RNA, and, single strand (ss) RNA (see Online Methods). As shown by immunoprecipitation experiments, dsRNA was efficiently bound to J2 antibody (43.7% of input signal) as compared to greatly reduced binding with either a premicroRNA-like hairpin RNA (4.7% of input signal) or ssRNA (8.9% of input signal), each of similar lengths (Fig. 4a). Although some pre-microRNA-like hairpin RNA and ssRNA were precipitated, this was detected in oversaturated experimental samples. These results confirmed that J2 is dsRNA-selective. Furthermore, J2-immunoprecipitated dsRNA was highly sensitive to dsRNA-specific V1 ribonuclease versus ss nucleic acid specific S1 nuclease (Fig. 4b).
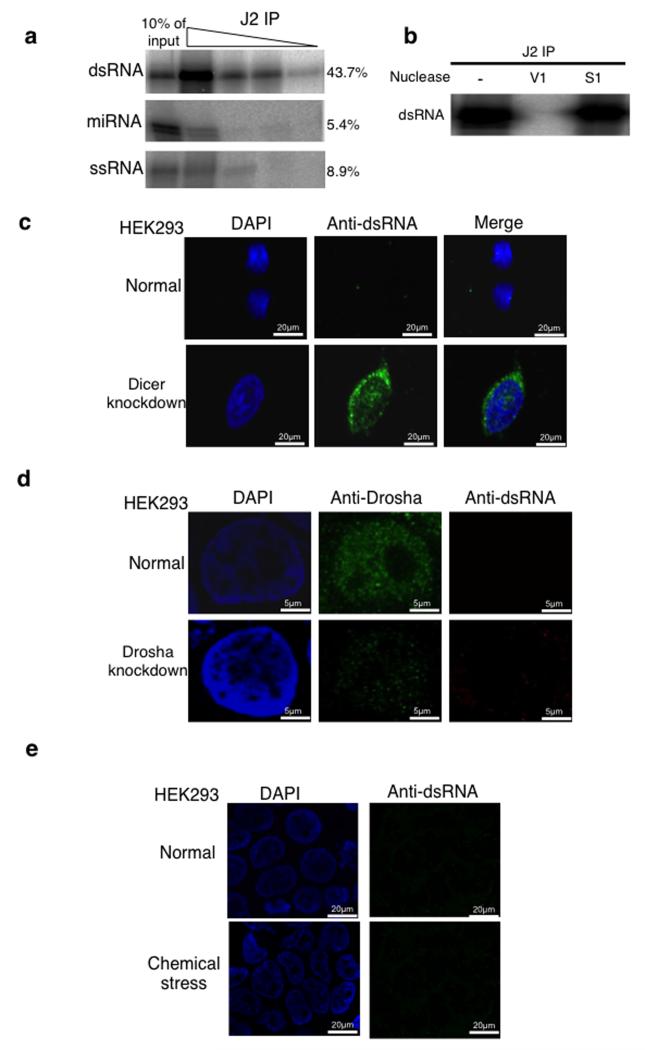
Figure 4. Loss of Dicer leads to accumulation of long dsRNA.
a) Immunoprecipitation analysis of uniformly 32P-radiolabelled dsRNA, miRNA and, ssRNA (See Online Methods) at 10-fold decreasing dilutions using J2 antibody. Signals were visualized and quantitated by PhosphoImager. The level of signal for the first concentration is shown on the right-hand side of the blot and it is expressed as a % of input. This experiment was replicated twice. See Supplementary Fig. 9 for uncropped blot images.
b) IP of dsRNA as in (a) and treated with V1, S1 or not treated. See Supplementary Fig. 9 for uncropped blot images.
c) Immunofluorescence (IF) analysis using J2 antibody showing dsRNA (green) in normal and Dicer knockdown HEK293 cells. Nuclei were stained with DAPI (blue).
d) IF analysis using anti-Drosha and J2 antibodies (green) in normal and Drosha knockdown HEK293 cells. Nuclei were stained with DAPI (blue).
e) IF analysis using J2 antibody on normal cells and cells treated with 2mg/ml acivicin for 48 hours. All experiments described in Fig. 4c-e, were independently replicated three times.
Immunofluorescence experiments on normal HEK293 cells gave little dsRNA signal in either nucleus or cytoplasm. In contrast, Dicer knockdown cells showed a clear accumulation of dsRNA signal, especially around the nuclear envelope, but also within the nuclear compartment (Fig. 4c). We tested the effect of Drosha knockdown on dsRNA accumulation, as loss of Dicer might have indirect effects on dsRNA-accumulation through loss of microRNA production. HEK293 cells were knocked down for Drosha expression, either by transfection with a plasmid expressing shRNA against Drosha mRNA (Fig. 4d) or by Drosha-specific siRNA treatment (Supplementary Fig. 4a). In both cases, Drosha expression was reduced to about 10% of normal cells, as shown by immunofluorescence (Fig. 4d). To ensure that at this level of Drosha knockdown there is an increase in primiRNAs, we analysed the levels of pri-miRNA15a and showed that its levels doubled upon Drosha shRNA-directed knockdown (Supplementary Fig. 4b). Even though Drosha levels were substantially reduced in these knockdown cells, no detectible accumulation of J2-specific dsRNA was apparent. This argues that accumulation of dsRNA through Dicer knockdown is not due to the accumulation of pre-microRNA through loss of Dicer activity. We also tested whether loss of Dicer induces dsRNA accumulation through general cellular stress. Treatment of HEK293 cells with acivicin, an inhibitor of γ-glutamyl transpeptidase, failed to induce dsRNA accumulation (Fig. 4e). Finally, we tested the affect of Dicer loss on an unrelated cell type to HEK293; ES cells derived from Dicer knock-out mice29. We again detected nuclear Dicer signals in ES cells which were lost in knock out cells (Supplementary Fig. 5). Notably, Dicer accumulates in nucleoli of ES cells possibly reflecting the different expression patterns of this cell type. Nonetheless, we demonstrate the presence of nuclear Dicer in a different mammal and cell type. We also confirmed that these Dicer knockout cells display a clear J2-specific dsRNA signal in both the nucleus and cytoplasm.
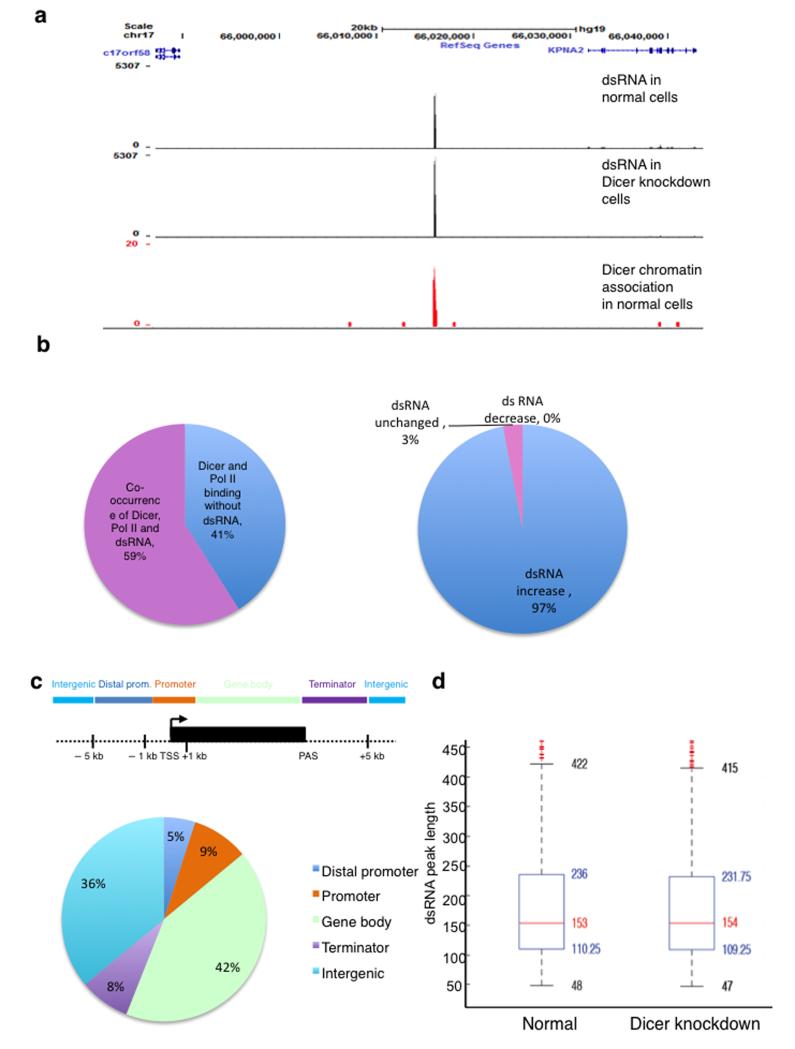
We sequenced dsRNA isolated from normal and Dicer knockdown cells, treated with T1 nuclease to remove ssRNA by genomic RNA sequencing (RNA-seq). As shown for specific loci (Fig. 5a; 5′ flanking region of KPNA2) and genome-wide, there is a remarkable correspondence between Dicer associated loci and dsRNA. Moreover, 67% of loci showing Dicer and Pol II co-occurrence (see Online Methods) are also associated with dsRNA (Fig. 5b). Levels of dsRNA detected in Dicer-depleted cells, as compared to normal cells, indicates that in most cases dsRNA accumulates upon Dicer depletion at sites where Pol II, Dicer and dsRNA co-localise (Fig. 5b, right). The median length of dsRNA peaks, in both normal and Dicer depleted cells, is approximately 150 nt (Fig. 5d), and a detailed numerical analysis of dsRNA levels at Dicer associated loci shows that dsRNA levels often increase substantially following Dicer knockdown (Supplementary Fig. 6a-c). Repetitive genomic elements including LINEs, SINEs, LTRs and Satellites are significantly underrepresented in the dsRNA data-set. In contrast, rRNAs and tRNAs are significantly overrepresented among dsRNA peaks (Supplementary Fig. 6d). Metagene analysis of top dsRNA peaks (see Online Methods) showed an accumulation before TSS and broader distribution around PAS (Supplementary Fig. 6e).
Figure 5. Loss of Dicer leads to the accumulation of dsRNA that co-localizes with chromatin-associated Dicer loci.
a) Snapshot of the UCSC genome browser displaying dsRNA and Dicer ChIP-seq data; the example shows intergenic region neighbouring KPNA2 on chromosome 17. Top: dsRNA in normal cells (3677 reads), middle: dsRNA in Dicer knockdown cells (5307 reads) and, bottom: Dicer-chromatin association at the same locus.
b) Left: Distribution of Dicer binding sites that co-occur with Pol II binding sites and sites of dsRNA accumulation upon Dicer depletion. Right: Division of loci where Dicer, Pol II and dsRNA co-localise, according to dsRNA levels increasing (blue) or remaining the same (red) upon Dicer depletion.
c) Distribution of dsRNA (dsRNA top peaks) relative to UCSC RefGene coordinates as in Figure 2d.
d) Box Plot of dsRNA length in normal and Dicer depleted cells. The red mark indicates the median, at a value of 153 and 154 respectively. The lower quartile q1 (25th percentile) is at 110.25 and 109.25 respectively, and the upper quartile q3 (75th percentile) at 236 and 231.75 respectively (blue box).
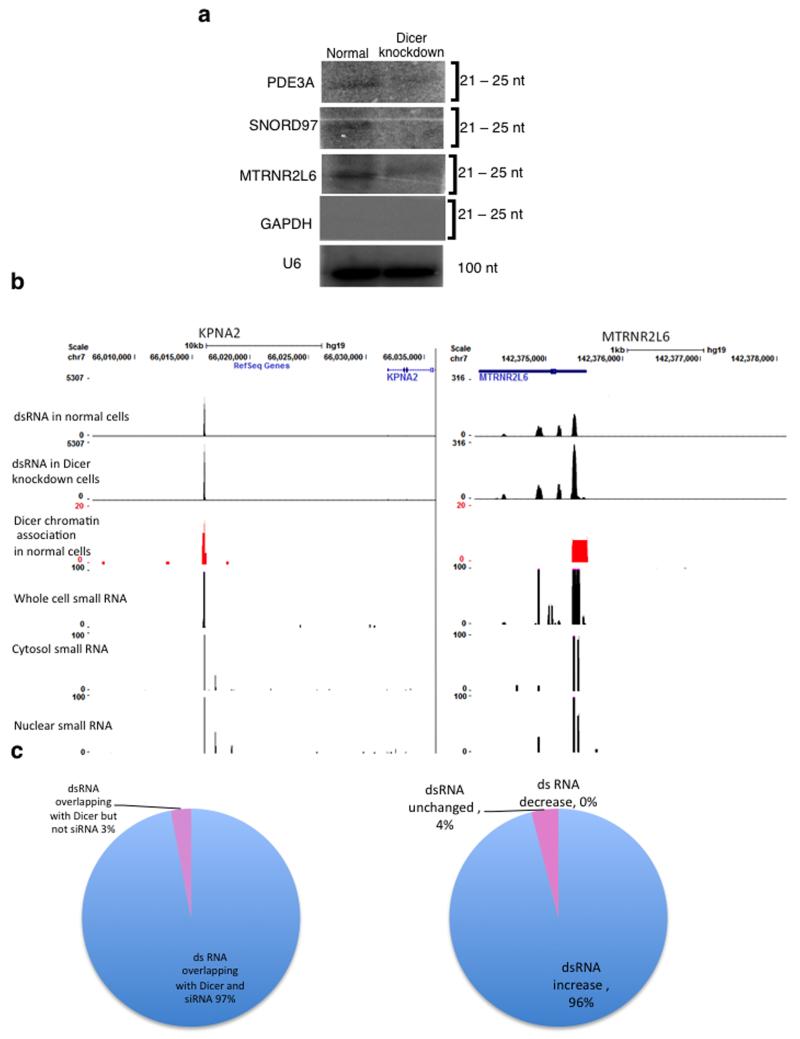
The co-occurrence of dsRNA and Dicer genome-wide, and the fact that dsRNA accumulates upon Dicer depletion, suggests that dsRNA is processed into siRNA. For three of the four Dicer-positive loci, tested in these studies (Fig. 2 and 3), siRNAs were detected above background levels using a chemical cross-linking Northern blot technique. Furthermore, these weak siRNA signals were reduced following Dicer knockdown (Fig. 6a). No RdRP activity has been observed in mammals, unlike in S. pombe and plants, where this RNA polymerase acts to amplify the levels of siRNA allowing their ready detection3,14. In spite of this, small 5′ monophosphate RNAs have been detected by genomic sequencing in various cell lines33. Examination of these small RNAs from IMR90 cells revealed the presence of small RNAs associated with the co-localising Dicer binding sites and dsRNA loci (Fig. 6b). We note that these small RNA clusters appear consistent between the various cell lines tested33. Bioinformatic analysis revealed that there is substantial overlap (97%) between Dicer binding sites, dsRNA loci and sites of small RNA detection (Fig. 6c) suggesting that these small RNAs are siRNAs. Furthermore, Dicer depletion leads to accumulation of dsRNA in 94% of cases, implying an RNAi associated mechanism (Fig. 6c).
Figure 6. Loss of Dicer leads to a reduction of siRNA that co-localise with chromatin-associated Dicer sites and dsRNA sites.
a) Northern Blot analysis of small RNA isolated from normal and Dicer knockdown HEK293 cells and hybridized to specific 32P-radiolabelled probes. Probes against GAPDH and U6 snRNA were used as negative and loading controls respectively. Signals were visualised by PhosphoImager. See Supplementary Fig. 9 for uncropped blot images.
b) Snapshot of the three data sets in the UCSC genome browser; the examples show the intergenic region of KPNA2 and the terminator region of MTRNR2L6 (as in Fig. 5a). The last three panels show small RNAs isolated from whole cell, cytosolic and nuclear fractions of IMR90 cells respectively.
c) Left: Distribution of Dicer binding sites that overlap with sites of dsRNA and siRNA based on small RNA RNA-seq data. Right: Division of loci where Dicer, dsRNA and siRNA co-localise, according to dsRNA levels increasing (blue) or remaining the same (red) upon Dicer depletion.
Overall, we have demonstrated that loss of Dicer causes dsRNA accumulation corresponding to the majority of Dicer chromatin binding sites. This effect can be correlated with nuclear dsRNA synthesis from convergent transcription units. Normally, this dsRNA is rapidly processed by Pol II-associated Dicer, but when Dicer is ablated, it accumulates in the nucleus and then the cytoplasm.
Loss of Dicer triggers the interferon response and apoptosis
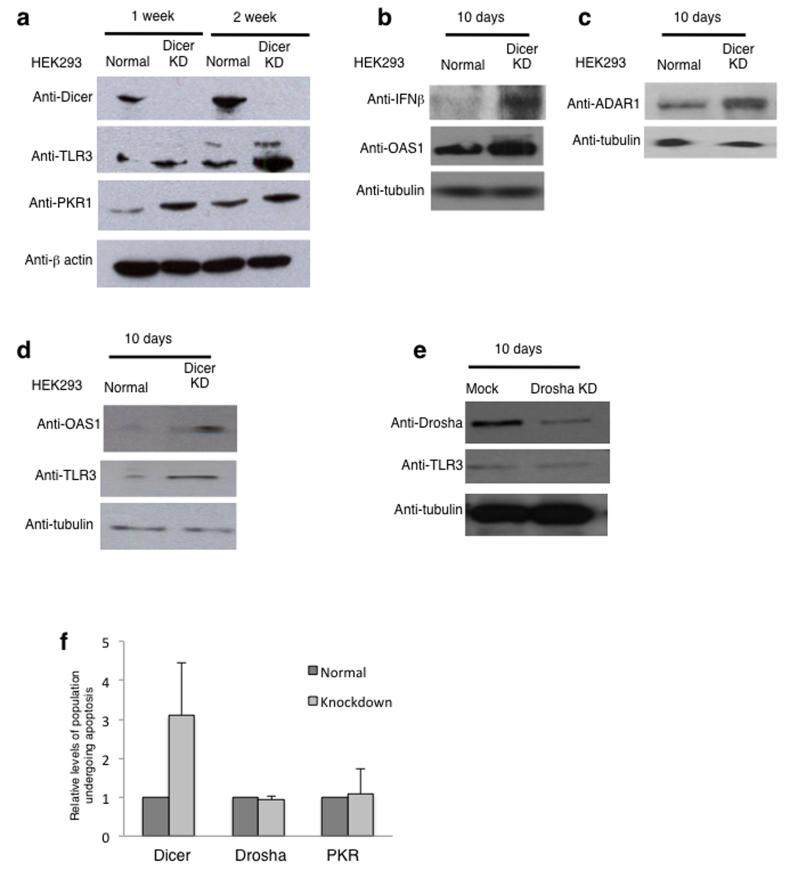
Mammalian somatic cells (though not stem cells34) possess a potent antiviral mechanism, the interferon response pathway35, triggered by cytoplasmic dsRNA. Interferon leads to cellular apoptosis, thus eliminating virally-infected cells. Since loss of Dicer leads to accumulation of dsRNA, we tested if this also results in an interferon induction response. HEK293 cells were grown for 1 or 2 weeks under Dicer shRNA induction conditions and two key proteins in the interferon response pathway, TLR336 and PKR19, were clearly elevated by Dicer knockdown based on quantitative Western blotting (Fig. 7a and Supplementary Fig. 7a). PKR1 activation by Dicer knockdown was also confirmed by immunofluorescence using the PKR1 specific antibody (Supplementary Fig. 7b). Additional interferon-induced proteins, INF-β and OAS1, were also strongly up-regulated in Dicer knockdown cells (Fig. 7b), as was ADAR1, an enzyme involved in dsRNA editing. Note the long form of this protein is interferon inducible (Fig. 7c). To control for the possibility of off-target effects, the levels of OAS1 and TLR3 were examined in Dicer depleted cells using an siRNA targeted to a different sequence on Dicer mRNA. Again, induction of the interferon response was evident, suggesting that this is a Dicer specific effect (Fig. 7d). As a negative control, we analysed the levels of TLR3 following shRNA–directed Drosha knockdown, and no change was detected (Fig. 7e).
Figure 7. Loss of Dicer triggers the interferon response.
a) Western blot analysis of cell extracts isolated from normal and Dicer knockdown HEK293 cells, cultured for 1 or 2 weeks, using antibodies specific to Dicer, TLR3, PKR1 and β-actin. Anti-TLR3 antibody detects two bands and according to size markers, the upper band is likely to correspond to modified protein. Quantitation is shown in Supplementary Figure 7. See Supplementary Fig. 9 for uncropped blot images.
b) Western blot analysis of cell extracts isolated from normal and Dicer knockdown HEK293 cells, cultured for 10 days using antibodies specific to IFN-β, OAS1 and tubulin. See Supplementary Fig. 9 for uncropped blot images.
c) Western blot analysis as in (b) using antibodies specific to ADAR1 and tubulin. See Supplementary Fig. 9 for uncropped blot images.
d) Western blot analysis of cell extracts isolated from normal and siRNA-directed Dicer knockdown HEK293 cells using antibodies specific to OAS1, TLR3 and tubulin. See Supplementary Fig. 9 for uncropped blot images.
e) Western blot analysis of cell extracts isolated from normal and Drosha knockdown HEK293 cells using antibodies specific to Drosha, TLR3 and tubulin. See Supplementary Fig. 9 for uncropped blot images.
f) Flow cytometry based quantitation of the % of cell population undergoing apoptosis in normal and Dicer, Drosha or PKR knockdown cells. % of cell population values are based on average values ± s.e.m. from three independent biological experiments. All experiments in the figure were independently repeated 3 times.
To further confirm that Dicer knockdown cells undergo apoptosis, we used flow cytometric analysis on cells stained with Annexin V, an apoptotic marker, and 7-aminoactinomycin D (7-AAD), which binds to DNA. Annexin V stains cells that are actively undergoing apoptosis while 7-AAD binds dead cells (see Online Methods). The cell population which is Annexin V positive and 7-AAD negative is indicative of apoptotic cells. A three-fold increase in cells undergoing apoptosis in the Dicer knockdown cells was observed (Fig. 7f). To control for the possibility that apoptosis occurs through deregulation of the microRNA pathway or simply by nonspecific shRNA effects, the proportion of cells undergoing apoptosis after shRNA–directed Drosha or PKR knockdown was measured. Neither Drosha nor PKR knockdown cells showed increased levels of apoptosis (Fig. 7f).
We finally compared the cellular morphology of HEK293 cells with or without Dicer expression by electron microscopy (Supplementary Fig. 8). As shown by representative images, Dicer knockdown cells displayed disrupted cellular morphology with abnormal membrane blebbing (Supplementary Fig. 8a), and aberrant nuclear chromatin, indicating extensive DNA fragmentation (Supplementary Fig. 8b). Cytoplasmic mitochondria also appeared abnormally elongated, indicating organelle fusion (Supplementary Fig. 8c). All of these morphological abnormalities are hallmarks of cells undergoing apoptosis. Together, these results establish that loss of Dicer in HEK293 cells leads first to accumulation of dsRNA, which, in turn, causes interferon response activation and consequent cell death by apoptosis.
Discussion
Dicer has a well established role in siRNA biogenesis37 and in mammalian cells, has been considered a purely cytoplasmic protein. In contrast we detected Dicer in the nucleus, but, when Dicer was over-expressed from an episomal plasmid, it was only transported into the nucleus, if the endogenous protein had also been depleted. Possibly a chaperone (either protein or potentially RNA) is required for the nuclear localisation of Dicer. This proposed chaperone is under active investigation. The presence of nuclear Dicer in mammalian cells opens up an interesting question about its biological role in this cellular compartment. A recent study also shows nuclear localisation of RNAi proteins, further pointing to a biological function for TGS in human cells39.
A striking feature of mammalian transcriptome analysis is that the high density of transcription units leads to multiple situations where overlapping transcription may occur. There is an extensive network of generally unstable long non-coding (lnc) RNAs, many antisense to protein coding gene transcripts or to other lncRNAs40. The capacity for such transcripts to anneal and generate dsRNA is commonplace across the mammalian genome. Repetitive sequence elements such as LINES and SINES that often derive from retroposons are also transcriptionally active and may generate dsRNA. Hence dsRNA synthesis is a possible outcome for large tracts of the human genome. The connection between Dicer and SINE-derived dsRNA was made by studies on the molecular basis of macular degeneration in the human retina41. SINE RNA was detectible as dsRNA and signals were evident in diseased eyes. Importantly, loss of Dicer was directly associated with this defect. These results are highly resonant with our studies on Dicer knockdown and dsRNA accumulation. Reduced Dicer levels have also been observed in both cancer cell lines, as well as cells derived from aging animals, possibly implying a breakdown in the normal turnover of dsRNA in such cells42.
A key aspect of nuclear Dicer function in mammalian cells is likely to relate to its TGS function as described in other eukaryotes1,3. Much of the mammalian somatic cell genome is encased in repressive heterochromatin, and can be classified as either constitutive or facultative. Constitutive or pericentric heterochromatin is associated with repetitive elements such as satellite sequence and is generally defined by CG methylation and H3K9me3 histone marks that, in turn, recruit heterochromatic proteins such as HP1 isoforms. Among the methyl transferases responsible for this mark are Suv39h1 and Suv39h2, as their gene deletion in transgenic mice results in loss of pericentric heterochromatin44,45. Recent studies have associated the establishment of this class of heterochromatin with features of actively-transcribed DNA such as transcription factor occupancy46. Several reports have indicated that this category of repressed chromatin displays low-level transcriptional activity, resulting in dsRNA synthesis47. Facultative heterochromatin may be a more dynamic repressed state, and, in this case, is associated with different H3K9me2 histone marks, which are likely added by the heterodimeric methyl transferase G9a-GLP48. Several studies have indicated that this type of repressed chromatin can be induced at localised positions by exogenous siRNAs23,26, or may exist endogenously, where it acts to regulate gene transcription and transcript RNA processing49. Based on the identification of H3K9me2 marks in Dicer-associated loci, we suggest that there is a Dicer-mediated conversion of locally synthesised dsRNA into an RNAi response through Ago1 that, in turn, sets up and maintains heterochromatin structure.
Finally, we showed that Dicer knockdown perturbs the normal cycle of low-level dsRNA synthesis and Dicer-dependent TGS resulting in accumulation of dsRNA throughout the cell. We predict that dsRNA initially formed in the nucleus escapes into the cytoplasm and triggers the interferon response leading to cellular apoptosis (Fig. 8 model). Numerous studies show that Dicer loss has severe consequences to the cell. Thus, mice lacking Dicer fail to develop beyond the embryonic stage. However, while ES cells derived from these knockout mice are viable (albeit growth impaired), ES cells lack the interferon response pathway, so explaining their viability. In contrast, this study shows that for cultured mammalian somatic cells, loss of Dicer leads to dsRNA accumulation and consequent apoptosis. We therefore predict that a major role of nuclear Dicer is to set up the right balance between heterochromatin and euchromatin. Failure of this process through Dicer knockout results in dsRNA accumulation. This results in a “last ditch” process of cellular apoptosis to eliminate misregulated cells that would otherwise lead to cell pathologies such as cancer.
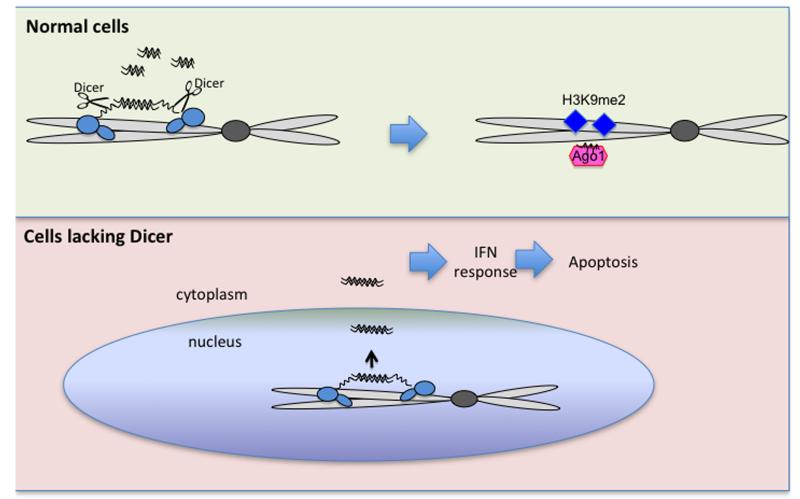
Figure 8. Model of nuclear Dicer function.
Under normal conditions, Dicer is recruited to loci of endogenous overlapping transcription through association with Pol II and dsRNA. Dicer then cleaves co-transcriptionally dsRNA into siRNA leading to Ago1 recruitment and establishment of the H3K9me2 mark. In cells lacking Dicer, endogenous dsRNA accumulates resulting in interferon induction and consequent cell death.
Online Methods
Primers
| Name | Sequence |
|---|---|
| PDE3A fw | 5′gacatggccttccacatcta |
| PDE3A rev | 5′ccatcttcgccttcttcttg |
| SNORD97 fw | 5′gaaaatgccctgttttatcctt |
| SNORD97 rev | 5′ctttgaatgtccagcgtcct |
| MTRNR2L6 fw | 5′gggataacagcgcaatccta |
| MTRNR2L6 rev | 5′tagatgggaggtgtggagga |
| 5′ MTRNR2L6 fw | 5′tccaacacaggcatgctcta |
| 5′ MTRNR2L6 rev | 5′aagagacagctgaaccctcg |
| 5′ MTRNR2L6 fw | 5′tgcctggagtcctagttttagt |
| 3′ MTRNR2L6 rev | 5′tctgcaaatgaggaccccat |
| PLCH1 fw | 5′ttggctaaggcaatgaaag |
| PLCH1 rev | 5′cctcctgggttcagtgtgtc |
| 28S fw | 5′gcaggaggtgtcagaaaagttaccacag |
| 28S rev | 5′ctaacctgtctcacgacggtctaaaccc |
| GAPDH fw | 5′gatgacatcaagaaggtggt |
| GAPDH rev | 5′ttgacaaagtggtcgttgag |
| pri-miRNA 15a fw | 5′accttggagtaaagtagcagcac |
| pri-miRNA 15a rev | 5′tattttcttcagaagatcag |
Tissue culture
Human HEK293, Dicer-kd/2b228 and mouse Dicer knockout ESC29 cell lines were maintained under standard conditions. All cell lines were tested for mycoplasma contamination. Induction of shRNA was carried out by addition of 10μg/ml of doxycycline for 7, 10 or 14 days. Unless otherwise stated, cells were treated with doxycycline for 10 days. Transfections of plasmids expressing shRNA directed against Drosha or PKR 50 were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions every 72 hours for a total of 10 days. Transcription inhibition was carried out by addition of 2.5μg/ml α-amanitin for 48 hours. Stress induction was carried out by addition of 2μg/ml acivicin for 48 hours. Transient transfections carried out using Lipofectamin 2000 (Invitrogen).
Immunofluorescence and Microscopy
Dicer nuclear localization was analysed by immunofluorescence and FRAP experiments, using anti-Dicer 13D6 (Abcam-ab14601), according to standard protocol using a confocal microscope (Olympus). Anti-PKR1 (Abcam-ab28943), anti-Drosha (Abcam-ab12286) and J2 antibody (Scicon,10010200) (Weber et al., 2006) were also used in immunofluorescence experiments. Note that all presented images show signals at one specific Z-axis. All antibodies were used at 1:1500 dilutions and validation for use in immunofluorescence is provided on the manufacturer’s website with the exception of J2.
Chromatin analysis
ChIP experiments were performed using formaldehyde-crosslinked chromatin. Antibodies employed were anti-Dicer 13D6 (Abcam-ab14601), anti-Pol II N20X (Santa Cruz Biotechnology-sc-899X), anti-Ago1 (Millipore-04-083) (Kim et al., 2006), anti-H3K9me2 (Abcam-ab1220) and anti-H3 (Abcam-ab1791). 5μg of antibody was used per reaction and validation for all abcam antibodies is provided on the manufacturer’s website. Immuno-precipitated, non-precipitated and input DNA was analyzed by qRT-PCR. For ChIP-sequencing (seq), the eluted ChIP DNA was used for library preparation and cluster generation using Illumina kits, following the manufacturer’s instructions. After passing Solexa CHASTITY quality filter, the reads were mapped to the human genome (hg19) using BOWTIE, allowing maximum 2 mismatches. 990,363 uniquely mapped reads were obtained. 1957 enriched regions were identified by peak calling using MACS V1.4.0 with the default p-value threshold of 10−5. For downstream analysis, only 119 high-confidence, top-scoring regions (score>100) were considered, referenced in the manuscript as “Dicer ChIP-seq peaks”. For annotation, genomic coordinates of hg19 RefGene transcription start sites (TSS) and gene bodies were downloaded from UCSC table browser (http://genome.ucsc.edu/). Promoters were defined as 2kb regions flanking TSS 1kb upstream and downstream. Gene bodies were defined as genic regions excluding previously defined promoters. Terminators were defined as regions 5kb downstream of annotated polyadenylation sites (PAS) that do not overlap with any promoter or gene body. Distal promoters were defined as regions between 1kb and 5kb upstream of a promoter, not overlapping with any of the previous categories. The rest of the genome was considered intergenic. The peak summit coordinate determined the annotation. The coordinates of Pol II binding regions (raw signal as well as called peaks) in HEK293 cell line were obtained from ENCODE Transcription Factor Binding Sites by ChIP-seq from Stanford/Yale/USC/Harvard (GEO accession GSE31477) 51. Dicer and Pol II overlap was calculated in two different ways. In the first case, Dicer ChIP-seq peaks were eliminated from comparison if signal was lower than an Input control from HEK293 cells, and the remaining 49 peaks were assessed for overlap based on Pol II ChIP-seq signal which also displayed a higher level of signal than the input control. In the second case, Dicer and Pol II peaks overlap was calculated using PeakAnalyzer 1.4, using 106 random datasets as a control (used for Supplementary Fig. 2).
RNA analysis
RNA isolation was carried out using TRIzol reagent (Invitrogen) and reverse transcribed with SuperScript III Reverse Trancriptase (Invitrogen) using gene-specific primers. RNA to prepare dsRNA, ssRNA and miRNA was in vitro transcribed from PCR-generated templates of either exon 2 of the human β-globin gene or the mir22 sequence, using T7 or T3. RNA was then gel purified and for dsRNA and miRNA was hybridized by denaturation at 95oC and then cooling slowly to allow RNAs to anneal, before being used for IP with 1μg J2 antibody (Scicon,10010200). The binding % numbers (Fig. 4a) are based on quantitation of the input and the IP signal from 100% of input (lane 2) and calculated as follows: (IP/ (Input * 10))*100. Digestions with V1, T1 and S1 were carried out according to manufacturer specification (Ambion). For RNA-seq, total RNA was treated with T1 nuclease according to manufacturer instruction and remaining dsRNA was purified and used for library preparation using NEBNext Small RNA Library Prep Set for Illumina, following manufacturer instruction. RNA reads were mapped to the human genome as described for ChIP-seq. Peaks derived from normal and Dicer depleted cells were cross-compared: multiple peaks in one dataset which overlapped only one peak in the other, were combined into one peak. This was done iteratively until no change was observed within the datasets. For downstream analysis, we imposed a cut off of 1500 in both dsRNA samples, and only the remaining peaks were considered (dsRNA top peaks). Metagene analysis was carried out as described for ChIP-seq using the defined dsRNA top peaks. The length box plots for dsRNA top peaks display points as outliers (red crosses), if they are larger than q3 + 1.5(q3 – q1) or smaller than q1 – 1.5(q3 – q1), where q1 and q3 are the 25th and 75th percentiles (the blue box), respectively. The default whisker length of 1.5 corresponds to approximately ±2.7σ and 99.3% coverage if the data are normally distributed. The plotted whisker extends to the adjacent value, which is the most extreme data value that is not an outlier. Loci where Dicer and Pol II colocalise were then overlapped with dsRNA peaks to look for co-occurrence using Perl 5.14.2 (Figure 5b). Small RNA mapped binary data was downloaded from UCSC genome browser for IMR90 cells33. Any signal above 0 was considered as presence of small RNA. Peaks were overlapped with dsRNA peaks dataset and with Dicer ChIP-seq top (118) peaks dataset to look for co-occurrence using Perl 5.14.2 (Fig. 6c). siRNA isolation was carried out using PEG precipitation and separated on a 20% PAGE, transferred using a semi-dry blot apparatus and chemically crosslinked (using EDC52) before being probed with 32P-labelled PCR products of tested gene loci. Probes were labelled using the DECAprime kit (Ambion).
Protein analysis
Immunoprecipitation and immunodepletion experiments were performed on nuclear extracts using specific antibodies (Pol II-8WG16(Abcam-ab24758), Dicer-13D6 (Abcam-ab14601)). 10ml of antibody were used per reaction and validation of their use in immunoprecipitation is provided on the manufacturer’s website. Western blot experiments were performed according to standard protocols using the following antibodies: anti-Pol II 8WG16 (Abcam-ab24758), anti-Pol II N20 (Santa Cruz Biotechnology-sc-899), anti-Dicer 13D6 (Abcam- ab14601), anti-β actin (Sigma-A3853), anti-Spt5 (Millipore-ABE443), anti-TLR3 (Millipore-06-008), anti-PKR1 (Millipore-07-151), anti-IFN-β (Abcam-ab6979), anti-OAS1 (Abcam-ab82666), anti-ADAR1 (Abcam-ab88574), anti-Drosha (Abcamab12286), anti-tubulin (Sigma-T5168), anti-Grp75 (Abcam-ab2799). Antibodies were used at 1:500 dilution except anti-tubulin which was used at 1:10000. Validation for all antibodies is provided on the manufacturer’s website.
Scanning Electron Microscopy (SEM)
Cells grown on glass cover slips were fixed in 2.5% glutaraldehyde/0.1M sodium cacodylate at pH7.2, followed by secondary fixation in 1% osmium tetroxide. Samples were dehydrated sequentially in 30%, 50%, 70%, 80%, 90%, 95% and 100% ethanol and dried using Critical Point Dryer. Finally, cells were gold plated and viewed under the SEM.
Transmission Electron Microscopy (TEM)
Cells grown on cover slips were fixed as in SEM. Dehydration was followed by epoxy resin infiltration and embedding. Resin blocks with cells on top were polymerized overnight at 60 °C. To remove cover slips, sample blocks were submerged in liquid nitrogen. Blocks with samples were cut for ultrathin sections (70nm) and transferred to a 200 copper mesh grid. Grids were post-stained with 2% uranyl acetate and lead citrate then viewed under TEM.
Flow cytometry
Normal and Dicer, Drosha or PKR depleted cells were Annexin V APC (Ebioscience) and 7-AAD (Ebioscience) labeled and then measured by flow cytometry according to the manufacturer’s instructions.
Relative levels of pull down were measured by image-Quant software, are expressed as a % of input and shown in the lower-panel graph.
Supplementary Material
Acknowledgements
We are grateful to E. Johnson for the help with TEM and SEM experiments. This work was supported by grants from the Wellcome Trust (091805/Z/10/Z N.J.P.) and E.P.Abraham Trust grant to NJP, and by an MRC Career Development Award (MR/K006606/1 M.G.) and L’Oreal-UNESCO woman in science UK and Ireland award to M.G.
Footnotes
Databases: Genomic ChIP-seq and RNA-seq data are located at http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=MGlab&hgS_otherUserSessionName=hg19
References
- 1.Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev. 2010;20:134–141. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 4.Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132:983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- 5.Gullerova M, Proudfoot NJ. Convergent transcription induces transcriptional gene silencing in fission yeast and mammalian cells. Nat Struct Mol Biol. 2012 doi: 10.1038/nsmb.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minchiotti G, Di Nocera PP. Convergent transcription initiates from oppositely oriented promoters within the 5′ end regions of Drosophila melanogaster F elements. Mol Cell Biol. 1991;11:5171–5180. doi: 10.1128/mcb.11.10.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Han J, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 11.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Jaskiewicz L, Filipowicz W. Role of Dicer in posttranscriptional RNA silencing. Curr Top Microbiol Immunol. 2008;320:77–97. doi: 10.1007/978-3-540-75157-1_4. [DOI] [PubMed] [Google Scholar]
- 13.Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 14.Buhler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- 15.Moazed D, et al. Studies on the mechanism of RNAi-dependent heterochromatin assembly. Cold Spring Harb Symp Quant Biol. 2006;71:461–471. doi: 10.1101/sqb.2006.71.044. [DOI] [PubMed] [Google Scholar]
- 16.Maida Y, Masutomi K. RNA-dependent RNA polymerases in RNA silencing. Biol Chem. 2011;392:299–304. doi: 10.1515/BC.2011.035. [DOI] [PubMed] [Google Scholar]
- 17.Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci U S A. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Provost P, et al. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotaja N, et al. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci U S A. 2006;103:2647–2652. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakymiw A, et al. Overexpression of dicer as a result of reduced let-7 MicroRNA levels contributes to increased cell proliferation of oral cancer cells. Genes Chromosomes Cancer. 2010;49:549–559. doi: 10.1002/gcc.20765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doyle M, et al. The double-stranded RNA binding domain of human Dicer functions as a nuclear localization signal. RNA. 2013;19:1238–1252. doi: 10.1261/rna.039255.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haussecker D, Proudfoot NJ. Dicer-dependent turnover of intergenic transcripts from the human beta-globin gene cluster. Mol Cell Biol. 2005;25:9724–9733. doi: 10.1128/MCB.25.21.9724-9733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 24.Janowski BA, et al. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat Chem Biol. 2005;1:216–222. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 25.Saint-Andre V, Batsche E, Rachez C, Muchardt C. Histone H3 lysine 9 trimethylation and HP1gamma favor inclusion of alternative exons. Nat Struct Mol Biol. 2011;18:337–344. doi: 10.1038/nsmb.1995. [DOI] [PubMed] [Google Scholar]
- 26.Allo M, et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- 27.Ameyar-Zazoua M, et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol. 2012;19:998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- 28.Schmitter D, et al. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006;34:4801–4815. doi: 10.1093/nar/gkl646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nesterova TB, et al. Dicer regulates Xist promoter methylation in ES cells indirectly through transcriptional control of Dnmt3a. Epigenetics Chromatin. 2008;1:2. doi: 10.1186/1756-8935-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris DP, Michelotti GA, Schwinn DA. Evidence that phosphorylation of the RNA polymerase II carboxyl-terminal repeats is similar in yeast and humans. J Biol Chem. 2005;280:31368–31377. doi: 10.1074/jbc.M501546200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 32.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 33.Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R, et al. Mouse Embryonic Stem Cells Are Deficient in Type I Interferon Expression in Response to Viral Infections and Double-stranded RNA. J Biol Chem. 2013;288:15926–15936. doi: 10.1074/jbc.M112.421438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 36.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barraud P, et al. An extended dsRBD with a novel zinc-binding motif mediates nuclear retention of fission yeast Dicer. EMBO J. 2011;30:4223–4235. doi: 10.1038/emboj.2011.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. RNAi Factors Are Present and Active in Human Cell Nuclei. Cell Rep. 2014;6:211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 41.Kaneko H, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Passon N, et al. Expression of Dicer and Drosha in triple-negative breast cancer. J Clin Pathol. 2012 doi: 10.1136/jclinpath-2011-200496. [DOI] [PubMed] [Google Scholar]
- 43.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peters AH, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 45.Fodor BD, et al. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bulut-Karslioglu A, et al. A transcription factor-based mechanism for mouse heterochromatin formation. Nat Struct Mol Biol. 2012;19:1023–1030. doi: 10.1038/nsmb.2382. [DOI] [PubMed] [Google Scholar]
- 47.Martens JH, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fagegaltier D, et al. The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc Natl Acad Sci U S A. 2009;106:21258–21263. doi: 10.1073/pnas.0809208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aagaard L, et al. A facile lentiviral vector system for expression of doxycycline inducible shRNAs: knockdown of the pre-miRNA processing enzyme Drosha. Mol Ther. 2007;15:938–945. doi: 10.1038/sj.mt.6300118. [DOI] [PubMed] [Google Scholar]
- 51.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.