Abstract
The mechanisms of hantavirus-induced modulation of host cellular immunity remain poorly understood. Recently, microRNAs (miRNAs) have emerged as a class of essential regulators of host immune response genes. To ascertain if differential host miRNA expression toward representative hantavirus species correlated with immune response genes, miRNA expression profiles were analyzed in human endothelial cells, macrophages and epithelial cells infected with pathogenic and nonpathogenic rodent- and shrew-borne hantaviruses. Distinct miRNA expression profiles were observed in a cell type- and viral species-specific pattern. A subset of miRNAs, including miR-151-5p and miR-1973, were differentially expressed between Hantaan virus and Prospect Hill virus. Pathway analyses confirmed that the targets of selected miRNAs were associated with inflammatory responses and innate immune receptor-mediated signaling pathways. Our data suggest that differential immune responses following hantavirus infection may be regulated in part by cellular miRNA through dysregulation of genes critical to the inflammatory process.
Keywords: Hantavirus, miRNA expression, Innate immune response
Introduction
Hantaviruses (genus Hantavirus), unlike other members of the family Bunyaviridae, have no known arthropod vector, and instead are hosted by small mammals, which are persistently infected. Hantavirus diseases in humans, known as hemorrhagic fever with renal syndrome (HFRS) and hantavirus cardiopulmonary syndrome (HCPS), are characterized by increased vascular permeability. Hantaan virus (HTNV), Dobrava virus (DOBV), Seoul virus (SEOV) and Puumala virus (PUUV) are the principal HFRS-causing hantaviruses, whereas Sin Nombre virus (SNV) and Andes virus (ANDV) are prototypic hantaviruses that cause HCPS. Prospect Hill virus (PHV) and Tula virus (TULV) represent two of the many putatively nonpathogenic rodent-borne hantaviruses (Easterbrook and Klein, 2008; Klein and Calisher, 2007).
Thottapalayam virus (TPMV), initially an unclassified virus isolated from the Asian house shrew (Suncus murinus) (Carey et al., 1971), is now known to be among the most genetically and phylogenetically divergent of all hantaviruses (Song et al., 2007a). Phylogenetic analysis of Imjin virus (MJNV), recently isolated from the Ussuri white-toothed shrew (Crocidura lasiura) (Song et al., 2009), and other newly identified hantaviruses in shrews (Arai et al., 2007, 2008a, 2012; Kang et al., 2011a, 2011b; Song et al., 2007a, 2007b; Song et al., 2009), moles (Arai et al., 2008b; Kang et al., 2009a, 2009b, 2011a) and insectivorous bats (Sumibcay et al., 2012; Weiss et al., 2012) indicates an ancient origin and early evolutionary divergence from rodent-borne hantaviruses.
Most hantaviruses are nonpathogenic, with only a limited number of rodent-borne hantavirus species causing disease in humans. Currently, the pathogenicity of the newly identified hantaviruses harbored by shrews, moles and bats is unknown. Previously, Gavrilovskaya et al. (1998) demonstrated that pathogenic and nonpathogenic hantaviruses use β3 and β1 integrins, respectively, to enter endothelial cells. Moreover, usage and regulation of β3 integrins by HFRS- and HCPS-causing pathogenic hantaviruses, compared to nonpathogenic hantaviruses, appear to contribute to pathogenesis (Gavrilovskaya et al., 2002)
Recently, cellular miRNAs have been implicated in various viral infections and are known to play a key role in the regulation of cellular gene expression (Gottwein and Cullen, 2008; Skalsky and Cullen, 2010). Studies indicate that ANDV infection of human endothelial cells results in altered level of specific miRNAs, and these miRNAs regulate angiogenesis and vascular integrity (Pepini et al., 2010). Although endothelial cells are considered to be the principal targets for hantaviruses, various other cell types, such as macrophages and epithelial cells, have also been used to study the pathogenicity of hantaviruses (Shim et al., 2011; Shin et al., 2012).
In the absence of known diseases or clinical syndromes associated with non-rodent-borne hantaviruses, we attempted to gain insights into their pathogenic potential by assessing the cell type- and virus species-specific changes in cellular miRNA signatures in human endothelial, epithelial and macrophage cells experimentally infected with two rodent-borne hantaviruses (pathogenic HTNV and nonpathogenic PHV) and two shrew-borne hantaviruses (putatively nonpathogenic TPMV and MJNV of unknown pathogenicity). We identified a group of miRNAs whose expression patterns differentiated the host response to pathogenic vs. nonpathogenic hantaviruses. Our work provides a global view of cell type- and viral species-specific miRNA profiles and a target map of miRNAs, which is expected to contribute to future investigations of miRNA regulatory mechanisms in the pathogenesis of hantavirus infection and disease.
Results
Unique cellular miRNA expression profiling
To assess the host cellular miRNA expression in response to hantavirus infection, three cell types (known to be susceptible to hantaviruses) were infected with four hantavirus species. Among 1205 miRNAs present on the arrays, 319 miRNAs were reliably expressed (excluding all the raw data values less than 10). The global expression patterns of differentially expressed miRNAs between HTNV, MJNV, TPMV and PHV-infected cells relative to a mock control are shown by hierarchical clustering analysis in Fig. 1. Global expression changes in miRNAs occurred depending on the cell type. Notably, the number of differentially expressed miRNAs, showing greater than 2-fold changes, was higher in human umbilical vein endothelial cells (HUVEC), regardless of viral species, compared to A549 or THP-1 cells. Thus, our data suggest that distinct miRNA expression patterns are observed in a cell type-specific manner in response to hantavirus infection.
Fig. 1.

Hierarchical clustering analysis showing viral species-specific regulation of miRNAs during hantavirus infection. A549, HUVEC and THP-1 cells were infected with HTNV, MJNV, TPMV or PHV (MOI of 0.5) or mock infected. RNA was collected at day 3 post-infection. Heat maps depicting the miRNAs that are differentially expressed in A549, HUVEC and THP-1 cells after infection are shown. Colors indicate log2 ratios of infected vs. mock-infected control, according to the specified scale. Red denotes up-regulation, while green indicates down-regulation, highlighting significantly deregulated miRNAs at the corresponding viral infection. Clustering program: MeV 4.7.1 was used; Clustering method: Euclidean distance, average linkage, gene tree.
Viral species- and cell type-specific miRNA expression profiling changes
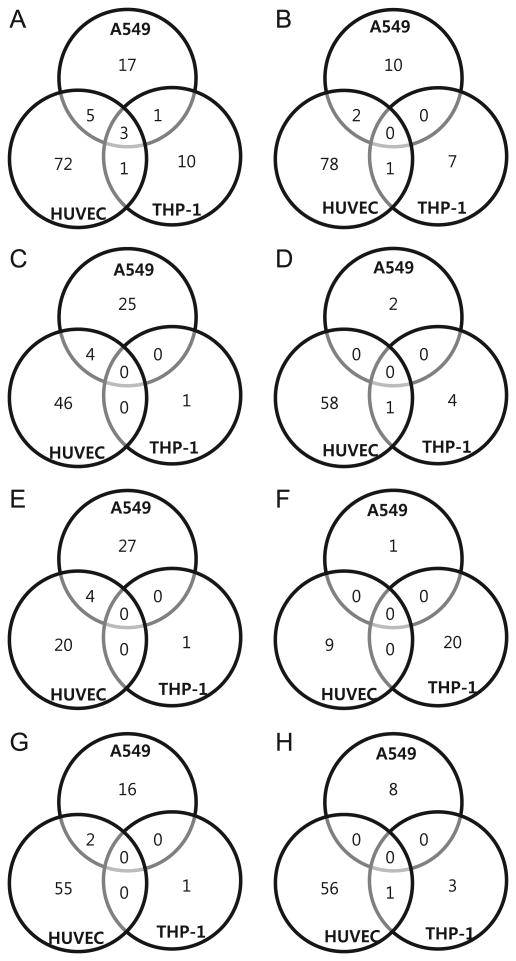
The number of differentially expressed miRNAs was compared in cells infected with HTNV, MJNV, TPMV and PHV (shown as Venn diagrams in Fig. 2). Only miRNAs that were altered at least 2-fold were considered significant candidates. Using these strict criteria, 26 up-regulated and 16 down-regulated miRNAs were identified in HTNV-infected HUVEC, compared to 1 up-regulated and no down-regulated miRNA in PHV-infected HUVEC (Fig. 2C and D). Similarly, increased numbers of up- or down-regulated miRNAs were identified in A549 and THP-1 cells infected with HTNV, compared to PHV. For example, in A549 cells, there were 5 up-regulated and 6 down-regulated miRNAs with HTNV, and 0 up-regulated and 1 down-regulated miRNA with PHV, whereas in THP-1 cells, there were 16 up-regulated and 4 down-regulated miRNAs with HTNV, and no up- or down-regulated miRNAs with PHV. In general, a limited number of miRNAs were detected in PHV-infected cells. The numbers of miRNAs that showed down-regulation in all virus samples was similar to the numbers of up-regulated miRNAs.
Fig. 2.

Venn diagrams of overlapping miRNA profiles for different cell types. The miRNA differential expression in HTNV, MJNV, TPMV and PHV is depicted in four overlapping circles for 2-fold up-regulation and 2-fold down-regulation for (A and B) A549, (C and D) HUVEC, and (E and F) THP-1. The numbers indicate the miRNA counts in the indicated area.
One up-regulated and 3 down-regulated miRNAs from HUVEC showed overlapping expression for HTNV, MJNV, TPMV and PHV. The Venn diagrams showed no miRNAs that were common in THP-1 cells infected with all four hantavirus species. In A549 cells, 16 up-regulated vs. 0 down-regulated miRNAs showing overlapping expression were detected (Fig. 3). No overlapping expression of miRNAs was detected with different cell types, suggesting cell type-specific host responses after hantavirus infection. Thus, the data indicate that infections with pathogenic and nonpathogenic hantaviruses induce distinct cellular miRNA expression patterns in different cells.
Fig. 3.
Venn diagrams of overlapping miRNA profiles for different hantaviruses. The miRNAs differentially expressed in A549, HUVECs and THP-1 cells are depicted in three overlapping circles for 2-fold up-regulation and 2-fold down-regulation, respectively, for (A and B) HTNV, (C and D) MJNV, (E and F) TPMV and (G and H) PHV. The numbers indicate the miRNA counts in the indicated area.
Distinct miRNA expression profiling between HTNV and PHV
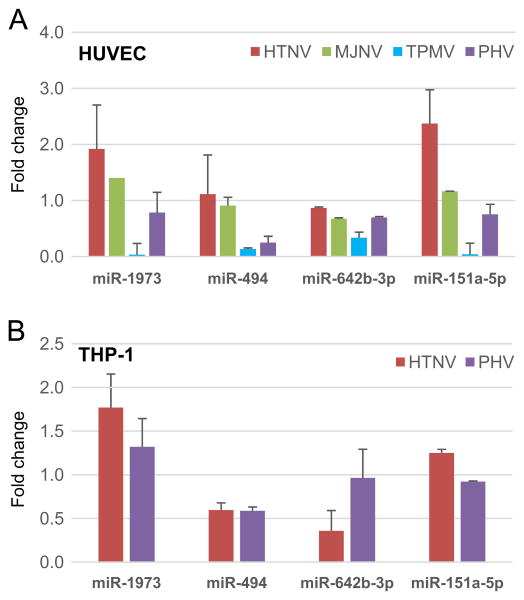
Quantitative RT-PCR was performed to validate miRNA microarray analysis. Among several cellular miRNAs showing distinct expression patterns between HTNV and PHV, we focused on four miRNAs (miR-494, miR-642b-3p, miR-151a-5p, and miR-1973). These miRNAs had different expression patterns between HTNV and PHV, including directly opposite regulation (up in one and down in the other) or regulation in the same direction but to differing degrees (Fig. 4). For example, miR-1973 and miR-151a-5p were up-regulated ∼2-fold and > 2-fold, respectively, HTNV-infected HUVEC, while no increased expression in these miRNA was detected during PHV infection. In contrast, in THP-1 cells, these miRNAs tended to be more highly expressed during HTNV than PHV infection, while the expression of miR-494 and miR-642b-3p did not differ between HTNV and PHV. Taken together, the expression levels of several miRNAs were found to be opposite in HTNV- and PHV-infected HUVEC and THP-1 cells.
Fig. 4.
qRT-PCR analysis of miRNA expression during hantavirus infection. The results of qRT-PCR analyses for (A) HUVEC and (B) THP-1 cells are shown. A total of four miRNA expressions were determined, including miR-151-5p, miR-642b-3p, miR-494 and miR-1973, after HTNV, MJNV, TPMV and PHV infection, respectively. Each graph represents the mean absolute fold change of triplicate experiments for each miRNA compared to mock-infected controls collected at day 3 post-infection. All qRT-PCR data are represented as means ± the SEM.
Predicted targets of miRNAs affected by hantavirus infection
To fully inspect the function of the differentially expressed miRNAs, we selected several miRNAs differentially expressed between samples and analyzed predicted mRNA targets, using GO term and KEGG pathway annotation. We decided to focus on previously characterized or potentially important receptor and signaling pathways regulated by pathogenic hantaviruses in cells (Gavrilovskaya et al., 1999, 2008; Handke et al., 2009; Lee et al., 2011). The following receptor and signaling pathways were included for analysis: vascular endothelial growth factor (VEGF), chemokine signaling pathway, innate immune receptors (integrin, retinoic acid inducible gene-I (RIG-I)-like receptor (RLR), nucleotide-binding oligomerization domain-containing protein (NOD)-like receptor (NLR), Toll-like receptor (TLR) signaling pathway)), inflammatory signaling pathways, including Janus kinase (JAK)-signal transducer and activator of transcription (STAT), phosphoinositide 3-kinase (PI3K)-Akt, mitogen activated protein kinase (MAPK). Many of the selected miRNAs whose expression was opposite between HTNV and PHV infection were shown to have mRNA targets involved in innate immune signaling receptors and inflammatory signaling pathways (Table 1). These pathways were also found to be regulated by reliably expressed miRNAs by ingenuity pathway analysis (IPA) (Fig. S1).
Table 1.
Biological pathways associated with targets of miRNAs differentially expressed in hantavirus infection. Differential expression of miRNAs during hantavirus infection was subjected to target prediction and pathway enrichment analyses. All pathways were obtained from the KEGG and target analysis was performed using Tarbase, TargetScan, miRanda, microRNA.org and microcosm.
| miRNA | Integrin signaling pathway |
Chemokine signaling pathway |
RIG-I-like receptor signaling pathway |
NOD-like receptor signaling pathway |
Toll-like receptor signaling pathway |
Jak-STAT signaling pathway |
PI3K-Akt signaling pathway | VEGF signaling pathway |
MAPK signaling pathway |
|---|---|---|---|---|---|---|---|---|---|
| miR-494 | FZD3, DNAJC, MYNN, APLN | STAT1, GNG10, RAP1B, PAK1, IKBKB, MAP2K1 | IKBKB, IL12B | IKBKB | IKBKB, IL12B, STAT1, MAP2K1, TLR6 | IL12B, STAT1, IL6ST, IL5RA, STAM, IL26, SOCS5, SOCS4, MYC | IKBKB, GNG10, MAP2K1, FGFR2, CDK6, CDK4, MYC, IGF1R | MAP2K1, PTGS2 | IKBKB, RAP1B, PAK1, MAP2K1, FGFR2, IL1R1, PPM1A, MAP4K4, IL1A, JUND, MYC, STK3 |
| miR-1225-5p | NR1H4 | IFNG | |||||||
| miR-642-3p | ASB2, C12orf4 | CYLD, CASP10 | MAP2K6 | CBL, IL11 | FLT1, LPAR4, SYK, ANGPT1, ITGB8 | PPM1A, PAK2, MAP2K6 | |||
| miR-151a-5p | NTRK2 | ||||||||
| miR-29c-3p | PCDHA10, YPEL2 | CDC42 | DDX3X | IGF1, SGK1, CDK2, LAMC1 | CDC42 | CDC42 | |||
| miR-1973 | IL8 | IL8 | IL8 | IL8 | CACNB1 | ||||
| miR-210 | CACNA1C | ||||||||
| miR-3656 | PXN | PXN | |||||||
| miR-1915-3p | KCNC1, | ADCY1, FOXO3, | MAVS, IKBKE | IKBKE, IRAK1, | SOCS7, PTPN6, | FOXO3, GSK3B, PDGFRB, BCL2, FLT4, SYK, | SRF, PDGFRB, CACNG2, CACNG1, FGFR3 | ||
| VAMP1 | GSK3B | MYD88 | IL20RB, IL2RB | IL2RB, FGFR3 | |||||
| miR-1268a | TOLLIP | ||||||||
| miR-199a-5p | ATP9A, ZNF563 | GSK3B | DDX3X | IL13 | GSK3B, CDKN1B | TGFB2, MAP3K11, STK4 | |||
| miR-193a-3p | ST6GALNAC3, PLAU | OSMR | OSMR, MCL1, LAMC2 | STMN1, CACNA1E, NF1 | |||||
| miR-1207-5p | 41531, SBNO1 | CX3CL1, SRC | CASP10 | RIPK2 | MAP2K6, TLR6 | STAT6, CBL, IL6ST | CHAD, CSF1, LAMC3, FLT1, CHRM1, PDPK1, ITGB3 | SRC, MAPKAPK2 | MAPKAPK2, CACNA2D1, MAP2K6, JUND, TGFBR2, TGFBR1, CACNG7, PPM1B, CACNB3, RAPGEF2, MAPK8IP3 |
| miR-222-3p | CBWD2, MEGF9 | JAK3 | CYLD, AZI2 | FOS | JAK3, PRLR | JAK3, KIT, CDKN1B, KDR, MDM2, PRLR | KDR | NTF3, CACNB4, NLK, STMN1, FOS | |
| miR-181b-5p | SPP1, ITGA2, ITGB8, GP5 | KRAS, RAP1B, NRAS, GNB4, MAP2K1 | DDX3X, ATG5 | CARD8 | MAP2K1, SPP1, TLR4, MAP2K4, MAP3K8, IRF5 | IL2, LIFR, CSF2RB, PRLR, SOCS4 | KRAS, NRAS, GNB4, MAP2K1, CREB1, SPP1, ITGA2, IL2, CREB5, PRLR, FLT1, PDGFRA, KITLG, TLR4, ITGB8, SGK3, DDIT4 | KRAS, N RAS, MAP2K1, PTGS2 | KRAS, RAP1B, NRAS, MAP2K1, TGFBR1, PDGFRA, IL1A, ZAK, MAP2K4, GNA12, MAP3K8, CACNA2D4, MAP3K3 |
To further characterize the function of differentially expressed miRNAs, IPA was used to generate gene lists known to have direct or indirect functional relationships with these miRNAs. There was a significant enrichment of genes involved in immune cell trafficking, cell-mediated immune response, and inflammatory response (Fig.S2). The interaction of miRNAs which were differentially expressed (either up-regulated or down-regulated only in HTNV or PHV) between HTNV and PHV was analyzed for four mRNA genes: myxovirus resistance gene A (MxA), interferon-β (IFN-β interferon gamma-induced protein-10 (IP-10), and Regulated upon Activation, Normal T-cell Expressed, and Secreted (RANTES). These genes were selected based on our previous results, which suggested differential expression of these genes in response to HTNV, compared to PHV (Shin et al., 2012). Furthermore, in addition to mRNA levels, IP-10 protein expression levels in hantavirus-infected cell supernatants were confirmed in Fig. S3. In HUVEC, two miRNAs (miR-3132 and miR-146a-5p) were found to target these genes (Fig. 5). miR-3132 was only up-regulated in PHV and unchanged in HTNV, while miR-146a-5p was only up-regulated in HTNV and unchanged in PHV. miR-3132 targets CXCL-10 (IP-10), whereas miR-146a-5p targets CCL5 (RANTES) and IFN-β. Our data suggest an inverse relationship between these miRNAs and their targets in that up-regulation of miR-3132 in PHV led to less production of CXCL-10 in comparison to HTNV. Similarly, increased expression of miR-146a-5p led to less expression of IFN-β in HTNV, compared to PHV. Furthermore, the interaction of miRNAs which were differentially expressed between HTNV-infected HUVEC, A549 cells or THP-1 cells also revealed an inverse relationship between these miRNAs and their target mRNA genes. miR-3652, miR-3656 and miR-4271 were down-regulated in HUVEC and their target CCL5 was highly up-regulated in HUVEC. Also, miR-15b-5p was down-regulated in A549 and its target CXCL-10 was up-regulated in these cells (Fig. 6A). Similarly, miR-503 was down-regulated in THP-1 cells and its targets CCL5 and CXCL-10 were highly up-regulated in THP-1 cells, as compared to HUVEC (Fig. 6B). Therefore, our network analysis supports the miRNA and mRNA expression data of anti-viral and inflammatory molecules differentially induced by these hantaviruses.
Fig. 5.
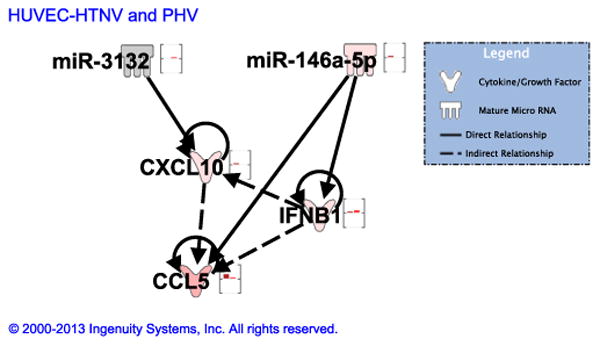
miRNA–mRNA association network for HTNV and PHV in HUVEC. miRNA–mRNA interaction network between reliably differentially regulated miRNAs and anti-viral and pro-inflammatory gene mRNAs [MxA, IFN-β, CXCL-10 (IP-10), CCL5 (RANTES)] in HUVEC between HTNV and PHV. Bars represent differential regulation: green bar, down-regulated; pink, up-regulated; no bar, not changed. First bar, HTNV; second bar, PHV.
Fig. 6.
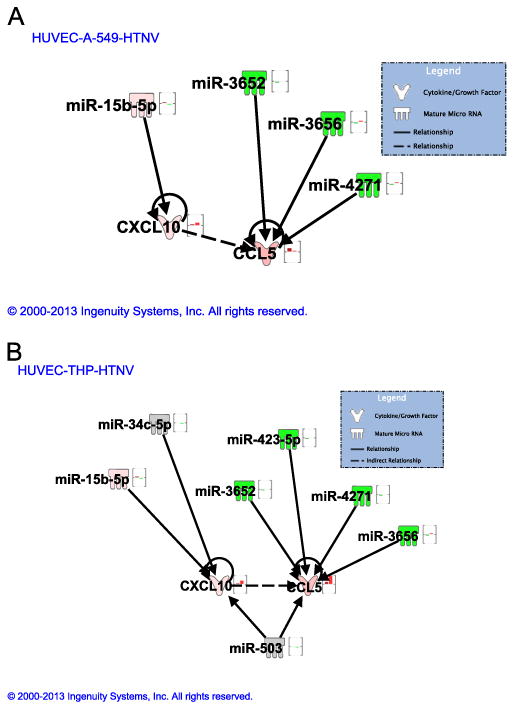
miRNA-mRNA associated network for HTNV in HUVEC, A549 and THP-1 cells. miRNA-mRNA interaction network between reliably differentially regulated miRNAs and anti-viral and pro-inflammatory gene mRNAs in HTNV-infected HUVEC and its comparison with (A) A549 cells and (B) THP-1 cells. Bars represent differential regulation: green bar, down-regulated; pink, up-regulated; no bar, not changed. First bar, HUVEC; second bar, A549 (in A) and THP-1 (in B).
Discussion
Host miRNAs appear to play critical roles in virus–host interactions. Host miRNA profiling has been well described for several viruses, including hepatitis C virus (HCV) (Murakami et al., 2006), human immunodeficiency virus type 1 (HIV-1) (Gupta et al., 2011, 2012), influenza viruses (Loveday et al., 2012), human T-cell lymphotropic virus type I (HTLV-I) (Ruggero et al., 2010; Sampey et al., 2012) and human cytomegalovirus (HCMV) (Li et al., 2011). In contrast, the involvement of miRNAs during hantavirus infection is largely unexplored. Here, we provide experimental evidence demonstrating the complex cell type- and virus species-specific regulation of host miRNAs by pathogenic and nonpathogenic hantaviruses in different human cells. We found that several miRNAs were differentially expressed between pathogenic HTNV and putatively nonpathogenic PHV and that these miRNAs were found to have targets associated with innate immune receptor signaling and inflammatory pathways.
Because the differential pathogenicity between rodent-borne and shrew-borne hantaviruses may stem from differences in host miRNA expression, we decided to analyze hantaviral species-specific patterns of host miRNA expression. Increased number of differentially expressed miRNAs in cells infected with HTNV was detected by hierarchical clustering heat map than in PHV-infected cells. The down-regulation of miRNAs was more pronounced with the highly pathogenic HTNV than with the nonpathogenic PHV. Increased numbers of miRNAs unique to HUVEC were found to be significantly up- or down-regulated, compared with miRNAs unique to A549 or THP-1 cells, indicating the cell type-specific changes in miRNA expression profiling.
Our previous study showed that all the studied hantaviruses replicated best in HUVEC and less productively in THP-1 and A549 cells (Shin et al., 2012). More distinct changes in miRNA expression in HUVEC may result from higher kinetics and efficiency of viral replication. It is possible that differences in hantavirus replication and expression of viral proteins could contribute to differences in miRNA expression that may be more related to how robust the infection is rather than the particular virus–cell type combination. Our previous data also indicate that HTNV and MJNV replicated more efficiently in HUVEC and THP-1 cells, compared to PHV and TPMV, and also caused the highest induction of anti-viral and pro-inflammatory responses.
Our miRNA–mRNA interaction network supports our previous findings in that differential expression pattern of mRNAs (such as anti-viral responsive genes and pro-inflammatory genes) is regulated by miRNAs oppositely expressed in HTNV vs. PHV. Up-regulation of miR-3132 in PHV caused less expression of CXCL-10 in comparison to HTNV. Similarly, increased expression of miR-146a-5p decreased expression of IFN-β in HTNV in comparison to PHV. Differential induction of anti-viral and pro-inflammatory responses between HTNV and PHV has been previously reported and our data suggest that this regulation may be mediated by specific miRNAs (Handke et al., 2009; Shim et al., 2011; Shin et al., 2012). miR-146a is probably one of the most characterized miRNAs so far, and it is known to negatively regulate signal transduction pathways leading to NF-κB activation and thus, its dysregulation is associated with diseases, such as cancer, viral infections and autoimmune diseases (Labbaye and Testa, 2012; Ma et al., 2011; Taganov et al., 2006).
MJNV and TPMV are representative shrew-borne hantaviruses of unknown pathogenicity in humans. We previously reported that shrew-borne hantaviruses differentially induce anti-viral and pro-inflammatory innate immune responses, compared with nonpathogenic PHV (Shin et al., 2012). We showed that TPMV and PHV replicated less efficiently than HTNV and MJNV in both HUVEC and THP-1 cells and this differential viral replication pattern may explain the differential innate immune responses triggered by differentially expressed miRNAs by each of these viruses. Thus, this may affect expression of miRNAs involved in the regulation of anti-viral and pro-inflammatory pathways. Our current data with miRNA expression profiling indicate that restricted numbers of miRNAs were changed in TPMV-infected cells, compared with HTNV-infected cells. This supports the notion that the pathogenicity of TPMV differs from that of HTNV in humans.
In this study, we explored the integration of microarray technology, qRT-PCR and target prediction and pathway enrichment analyses. These studies allowed us to perform a robust comparative bioinformatics study to reveal the host miRNA molecular signatures associated with diverse hantavirus species. We identified a unique pattern of cell type- and viral species-specific host molecular responses and our data provide key insights into miRNA expression patterns dynamically regulated by HTNV vs. PHV infection. Although this study only observed miRNA expression changes at day 3 post-infection, it will be interesting to do temporal specific analysis of miRNA microarrays, since viral replication and resulting pathogenicity differ between early (day 1 post-infection) vs. later (day 3–7 post-infection) time points during infection (Handke et al., 2009; Shim et al., 2011, Shin et al., 2012).
In conclusion, the expression profiles provide unique insights into the miRNAs that are differentially expressed during infection with hantaviruses of known and unknown pathogenicities. The specific role of each of these miRNAs during hantavirus infection is yet to be determined and is beyond the scope of the present study, but our data suggest that the virus species-specific changes may be another important determinant of hantavirus-associated pathology. Our findings also raise the possibility of identifying, in the very near future, circulating miRNA biomarkers to predict clinical progression and/or disease severity in hantavirus-infected patients.
Materials and methods
Cells and reagents
A549 cells (human epithelial lung cell, ATCC CCL-185) were maintained in RPMI-1640 (Lonza) supplemented with 5% fetal bovine serum (FBS), 2 mM l-glutamine and antibiotics (penicillin–streptomycin) at 37 °C at 5% CO2. Induced pluripotent stem cell HUVEC was purchased from Inopharmascreen (Asan, Korea) and maintained in M199 (Gibco), containing penicillin–streptomycin, 25 mM HEPES, 10 unit/mL heparin, 2.2 g/L sodium bicarbonate, 20% FBS, basic fibroblast growth factor (20 ng/mL) and passaged up to seven times. The human monocytic cell line, THP-1, obtained from the Korean Cell Line Bank (Seoul, Korea), was grown in RPMI-1640 (Lonza) supplemented with 5% FBS, 2 mM l-glutamine and antibiotics (penicillin–streptomycin) at 37 °C at 5% CO2. To induce monocyte-to-macrophage differentiation, THP-1 cells were cultured for 24 h in standard culture medium supplemented with 100 nM phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich).
Virus infection
TPMV (strain VRC 66412) (Carey et al., 1971), HTNV (strain 76–118) (Lee et al., 2004), PHV (strain PHV-1) (Lee et al., 1985) and MJNV strain 04–55 (Song et al., 2009) were propagated in Vero E6 cells (Shim et al., 2011). A549, HUVEC and THP-1 cells were infected at MOI of 0.5 for 90 min at 37 °C. After adsorption, the cells were washed with phosphate buffered saline (PBS) and maintained in complete medium.
MicroRNA microarray analysis
Total RNA was isolated followed by quality checks of both total RNA and small RNA by using a 2100 Bioanalyzer and software that detects 28 S and 18 S ribosomal RNA ratio, total RNA integrity number, and small RNA and miRNA concentrations. miRNA expression profiling was performed using the human microRNA microarrays 8 × 60 K (V16) (Agilent Technologies, Inc). The microarray was designed based on Sanger miRBase (release 16.0) and contained probes for 1205 human and 144 human viral miRNAs. Hundred nanogram of total RNA was labeled using the Agilent miRNA Complete Labeling and Hybridization Kit, according to the manufacturer's instructions.
Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was used to validate miRNA expression changes. Reverse transcription was performed with GenoExplorer miRNA First-strand cDNA Core kit (Genosensor), according to the manufacturer's instructions. Reverse transcription was carried out in a volume of 20 μL with 1 μg of total RNA. Real-time PCR was performed in a StepOne Plus machine (Applied Biosystems) using Power SYBR Green (Applied Biosystems), according to the manufacturer's instructions, in a total volume 15 μL. Cycling conditions for 4 target miRNA (hsa-miR-494, hsa-miR-642b-3p, hsa-miR-151a-5p, hsa-miR-1973) and U6 small RNA (internal control) were 10 min at 95 °C, 40 cycles of 15 s at 95 °C, and 30 s at optimal Tm (59 °C). Quantification was carried out with StepOne software v2.2.2 (Applied Biosystems). Relative levels of target miRNAs were expressed as the ratio of comparative threshold Cycle (Ct) to internal control small RNA.
Bioinformatics prediction of mRNA targets and gene ontology
A list of common gene targets from at least 5 prediction databases (Tarbase, TargetScan, miRanda, microRNA.org and microcosm) was obtained for each differentially expressed miRNA and these target prediction softwares were used to predict the target genes. To predict gene ontology (GO) and biological pathways associated with targets of selected miRNAs, the GO package in R http://www.r-project.org/, canonical Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways maps and Panther protein classification tools, was used to annotate the functions of the miRNA targets.
Ingenuity pathways analysis (IPA)
IPA (Ingenuity System Inc, USA) was used to determine miRNA-mediated networks, biological functions and canonical signaling pathways, functional significance for the pathways targeted by miRNAs. Biofunctions were grouped in: immune cell trafficking, cell-mediated immune response, and inflammatory response. In a similar way canonical pathways were grouped in ERK/MAPK, integrin, NF-kB, VEGF, JAK/STAT, role of pattern recognition receptors in recognition of bacteria and viruses, role of RIG-I-like receptors in anti-viral innate immunity, and interferon signaling pathways.
Supplementary Material
Acknowledgments
This study was supported by a grant of the TEPIK (Transgovernmental Enterprise for Pandemic Influenza in Korea), which part of Korea Healthcare technology R&D Project by Ministry of Health & Welfare, Republic of Korea (Grant No.: A103001). This work was supported in part by a Korea University Grant and US Public Health Service Grant R01AI075057 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, as well as by a grant from the 65th Medical Brigade/USAMEDDAC-Korea.
Footnotes
Appendix A. Supporting information: Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.virol.2013.07.036.
References
- Arai S, Bennett SN, Sumibcay L, Cook JA, Song JW, Hope A, Parmenter C, Nerurkar VR, Yates TL, Yanagihara R. Phylogenetically distinct hantaviruses in the masked shrew (Sorex cinereus) and dusky shrew (Sorex monticolus) in the United States. Am J Trop Med Hyg. 2008a;78:348–351. [PMC free article] [PubMed] [Google Scholar]
- Arai S, Gu SH, Baek LJ, Tabara K, Bennett SN, Oh HS, Takada N, Kang HJ, Tanaka-Taya K, Morikawa S, Okabe N, Yanagihara R, Song JW. Divergent ancestral lineages of newfound hantaviruses harbored by phylogenetically related crocidurine shrew species in Korea. Virology. 2012;424:99–105. doi: 10.1016/j.virol.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S, Ohdachi SD, Asakawa M, Kang HJ, Mocz G, Arikawa J, Okabe N, Yanagihara R. Molecular phylogeny of a newfound hantavirus in the Japanese shrew mole (Urotrichus talpoides) Proc Natl Acad Sci USA. 2008b;105:16296–16301. doi: 10.1073/pnas.0808942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S, Song JW, Sumibcay L, Bennett SN, Nerurkar VR, Parmenter C, Cook JA, Yates TL, Yanagihara R. Hantavirus in northern short-tailed shrew, United States. Emerg Infect Dis. 2007;13:1420–1423. doi: 10.3201/eid1309.070484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey DE, Reuben R, Panicker KN, Shope RE, Myers RM. Thottapalayam virus: a presumptive arbovirus isolated from a shrew in India. Indian J Med Res. 1971;59:1758–1760. [PubMed] [Google Scholar]
- Easterbrook JD, Klein SL. Immunological mechanisms mediating hantavirus persistence in rodent reservoirs. PLoS Pathog. 2008;4:e1000172. doi: 10.1371/journal.ppat.1000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilovskaya IN, Brown EJ, Ginsberg MH, Mackow ER. Cellular entry of hantaviruses which cause hemorrhagic fever with renal syndrome is mediated by beta3 integrins. J Virol. 1999;73:3951–3959. doi: 10.1128/jvi.73.5.3951-3959.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilovskaya IN, Gorbunova EE, Mackow NA, Mackow ER. Hantaviruses direct endothelial cell permeability by sensitizing cells to the vascular permeability factor VEGF, while angiopoietin 1 and sphingosine 1-phosphate inhibit hantavirus-directed permeability. J Virol. 2008;82:5797–5806. doi: 10.1128/JVI.02397-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilovskaya IN, Peresleni T, Geimonen E, Mackow ER. Pathogenic hantaviruses selectively inhibit beta3 integrin directed endothelial cell migration. Arch Virol. 2002;147:1913–1931. doi: 10.1007/s00705-002-0852-0. [DOI] [PubMed] [Google Scholar]
- Gavrilovskaya IN, Shepley M, Shaw R, Ginsberg MH, Mackow ER. beta3 Integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc Natl Acad Sci USA. 1998;95:7074–7079. doi: 10.1073/pnas.95.12.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Nagilla P, Le HS, Bunney C, Zych C, Thalamuthu A, Bar-Joseph Z, Mathavan S, Ayyavoo V. Comparative expression profile of miRNA and mRNA in primary peripheral blood mononuclear cells infected with human immunodeficiency virus (HIV-1) PLoS One. 2011;6:e22730. doi: 10.1371/journal.pone.0022730. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gupta A, Swaminathan G, Martin-Garcia J, Navas-Martin S. MicroRNAs, hepatitis C virus, and HCV/HIV-1 co-infection: new insights in pathogenesis and therapy. Viruses. 2012;4:2485–2513. doi: 10.3390/v4112485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handke W, Oelschlegel R, Franke R, Kruger DH, Rang A. Hantaan virus triggers TLR3-dependent innate immune responses. J Immunol. 2009;182:2849–2858. doi: 10.4049/jimmunol.0802893. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Bennett SN, Dizney L, Sumibcay L, Arai S, Ruedas LA, Song JW, Yanagihara R. Host switch during evolution of a genetically distinct hantavirus in the American shrew mole (Neurotrichus gibbsii) Virology. 2009a;388:8–14. doi: 10.1016/j.virol.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Bennett SN, Hope AG, Cook JA, Yanagihara R. Shared ancestry between a newfound mole-borne hantavirus and hantaviruses harbored by cricetid rodents. J, Virol. 2011a;85:7496–7503. doi: 10.1128/JVI.02450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Bennett SN, Sumibcay L, Arai S, Hope AG, Mocz G, Song JW, Cook JA, Yanagihara R. Evolutionary insights from a genetically divergent hantavirus harbored by the European common mole (Talpa europaea) PLoS One. 2009b;4:e6149. doi: 10.1371/journal.pone.0006149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kadjo B, Dubey S, Jacquet F, Yanagihara R. Molecular evolution of Azagny virus, a newfound hantavirus harbored by the West African pygmy shrew (Crocidura obscurior) in Cote d'Ivoire. Virol J. 2011b;8:373. doi: 10.1186/1743-422X-8-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein SL, Calisher CH. Emergence and persistence of hantaviruses. Curr Top Microbiol Immunol. 2007;315:217–252. doi: 10.1007/978-3-540-70962-6_10. [DOI] [PubMed] [Google Scholar]
- Labbaye C, Testa U. The emerging role of MIR-146 A in the control of hematopoiesis, immune function and cancer. J Hematol Oncol. 2012;5:13. doi: 10.1186/1756-8722-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Lee PW, Johnson KM. Isolation of the etiologic agent of Korean hemorrhagic fever. 1978 J Infect Dis. 2004;190:1711–1721. doi: 10.1093/infdis/190.9.1711. [DOI] [PubMed] [Google Scholar]
- Lee MH, Lalwani P, Raftery MJ, Matthaei M, Lutteke N, Kirsanovs S, Binder M, Ulrich RG, Giese T, Wolff T, Kruger DH, Schonrich G. RNA helicase retinoic acid-inducible gene I as a sensor of Hantaan virus replication. J Gen Virol. 2011;92:2191–2200. doi: 10.1099/vir.0.032367-0. [DOI] [PubMed] [Google Scholar]
- Lee PW, Amyx HL, Yanagihara R, Gajdusek DC, Goldgaber D, Gibbs CJ., Jr Partial characterization of Prospect Hill virus isolated from meadow voles in the United States. J Infect Dis. 1985;152:826–829. doi: 10.1093/infdis/152.4.826. [DOI] [PubMed] [Google Scholar]
- Li S, Zhu J, Zhang W, Chen Y, Zhang K, Popescu LM, Ma X, Lau WB, Rong R, Yu X, Wang B, Li Y, Xiao C, Zhang M, Wang S, Yu L, Chen AF, Yang X, Cai J. Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation. 2011;124:175–184. doi: 10.1161/CIRCULATIONAHA.110.012237. [DOI] [PubMed] [Google Scholar]
- Loveday EK, Svinti V, Diederich S, Pasick J, Jean F. Temporal- and strain-specific host microRNA molecular signatures associated with swine-origin H1N1 and avian-origin H7N7 influenza A virus infection. J Virol. 2012;86:6109–6122. doi: 10.1128/JVI.06892-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-kappaB signaling. J Mol Cell Biol. 2011;3:159–166. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- Pepini T, Gorbunova EE, Gavrilovskaya IN, Mackow JE, Mackow ER. Andes virus regulation of cellular microRNAs contributes to hantavirus-induced endothelial cell permeability. J Virol. 2010;84:11929–11936. doi: 10.1128/JVI.01658-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero K, Corradin A, Zanovello P, Amadori A, Bronte V, Ciminale V, D'Agostino DM. Role of microRNAs in HTLV-1 infection and transformation. Mol Aspects Med. 2010;31:367–382. doi: 10.1016/j.mam.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Sampey GC, Van Duyne R, Currer R, Das R, Narayanan A, Kashanchi F. Complex role of microRNAs in HTLV-1 infections. Front Genet. 2012;3:295. doi: 10.3389/fgene.2012.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim SH, Park MS, Moon S, Park KS, Song JW, Song KJ, Baek LJ. Comparison of innate immune responses to pathogenic and putative non-pathogenic hantaviruses in vitro. Virus Res. 2011;160:367–373. doi: 10.1016/j.virusres.2011.07.013. [DOI] [PubMed] [Google Scholar]
- Shin OS, Yanagihara R, Song JW. Distinct innate immune responses in human macrophages and endothelial cells infected with shrew-borne hantaviruses. Virology. 2012;434:43–49. doi: 10.1016/j.virol.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JW, Gu SH, Bennett SN, Arai S, Puorger M, Hilbe M, Yanagihara R. Seewis virus, a genetically distinct hantavirus in the Eurasian common shrew (Sorex araneus) Virol J. 2007a;4:114. doi: 10.1186/1743-422X-4-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JW, Kang HJ, Gu SH, Moon SS, Bennett SN, Song KJ, Baek LJ, Kim HC, O'Guinn ML, Chong ST, Klein TA, Yanagihara R. Characterization of Imjin virus, a newly isolated hantavirus from the Ussuri white-toothed shrew (Crocidura lasiura) J Virol. 2009;83:6184–6191. doi: 10.1128/JVI.00371-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JW, Kang HJ, Song KJ, Truong TT, Bennett SN, Arai S, Truong NU, Yanagihara R. Newfound hantavirus in Chinese mole shrew, Vietnam. Emerg Infect Dis. 2007b;13:1784–1787. doi: 10.3201/eid1311.070492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumibcay L, Kadjo B, Gu SH, Kang HJ, Lim BK, Cook JA, Song JW, Yanagihara R. Divergent lineage of a novel hantavirus in the banana pipistrelle (Neoromicia nanus) in Cote d'Ivoire. Virol J. 2012;9:34. doi: 10.1186/1743-422X-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S, Witkowski PT, Auste B, Nowak K, Weber N, Fahr J, Mombouli JV, Wolfe ND, Drexler JF, Drosten C, Klempa B, Leendertz FH, Kruger DH. Hantavirus in bat, Sierra Leone. Emerg Infect Dis. 2012;18:159–161. doi: 10.3201/eid1801.111026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.