Abstract
MicroRNAs (miRNAs) are a class of gene regulators originating from non-coding endogenous RNAs. Altered expression, both up - and down-regulation, of miRNAs plays important roles in many human diseases. Correcting miRNA dysregulation by either inhibiting or restoring miRNA function may provide therapeutic benefit. However, efficient, nontoxic miRNA delivery systems are in need. Cell penetrating peptides (CPPs) have been widely exploited for protein, DNA, and RNA delivery. Few have examined CPP transfection efficiency with single stranded anti-miRNA. The R8 peptide condensed both siRNA and anti-miRNA. Greater than 50% of cells had anti-miRNA/R8 complexes associated and in these cells 68% of anti-miRNA escapes the endosome/lysosome. Single -stranded antisense miR-21 inhibitor (anti-miR-21) administered using the R8 peptide elicited efficient downstream gene upregulation. Glioblastoma cell migration was inhibited by 25% compared to the negative control group. To our knowledge, this is the first demonstration of miRNA modulation with anti-miR-21/R8, complexes which has laid the groundwork for further exploring octaarginine as intracellular anti-miRNAs carrier.
Keywords: miRNAs, miRNA-21, glioblastoma, anti-miRNAs, cell penetrating peptide
1. Introduction
Glioblastoma multiforme (GBM) is a fatal brain tumor with an annual incidence of approximately 5 in 100,000 people, equivalent to 17,000 new diagnoses per year [32]. The deadly threat posed by GBM resides in the explosive growth characteristics, extremely invasive behavior, difficulty in treatment due to the blood-brain barrier, and intrinsic resistance to current therapies [52]. The current standard treatment for glioblastoma is surgery and radiotherapy combined with temozolomide. This approach doubles the 2-year survival rate to 27%, but overall prognosis remains poor [32]. New therapies that provide highly specific treatment based on disease pathology characteristics are urgently needed.
One possible new therapeutic option for GBM is microRNA (miRNA) therapy. MiRNAs are short noncoding RNAs that post-transcriptionally regulate gene expression of multiple target genes through seed pairing with 3′ untranslated region (UTR) of mRNA. By binding to the 3′ UTR as part of the miRISC complex, miRNAs block mRNA translation and lead to mRNA degradation [4, 5, 18]. MiRNAs play a causative role in the development of cancer [47]. Depending upon the specific miRNA, a gain or a loss of miRNA can occur in the tumor compared to normal tissue [9, 16]. MicroRNA works by forming miRNA induced silencing complex (miRISC) and simultaneously modulating several downstream gene expression [49]. The dysregulation may result in both the up- and down-regulation of a network of proteins due to the mechanism of miRNA activity [3, 37, 49].
Recently, the delivery of miRNA mimics to supplement lost miRNAs and anti-miRNAs to block elevated miRNAs has attracted enormous attention as new cancer therapies [3, 37, 49]. Anti-miRNAs are single stranded nucleic acid sequence that hybridize with mature miRNAs in the cytoplasm and prevent the miRNAs from recognizing their mRNA targets while not causing degradation of the RNA [27]. Some view miRNAs as the new “beacon of hope” for cancer patients [6]. A recent analysis on clinical patient samples showed that miR-21 is consistently overexpressed in glioblastoma tumor cells, but not in adjacent normal brain parenchyma, and the miR-21 levels correlated significantly with the grade of glioma [23].
MiR-21 targets key matrix metalloproteinases (MMPs) regulators, promoting glioma invasiveness [19]. MiRNA-21 has also been shown to function as an oncogenic miRNA in glioblastoma through modulating a network of key tumor-suppressive pathways in glioblastoma cells [10, 29, 33]. Therefore, a new therapy targeting miR-21 could potentially slow glioma progression and kill the tumor cells. Due to the poor cellular uptake characteristics of polyanionic oligonucleotides, a formulation that can mediate sufficient cellular uptake is needed to achieve efficient miR-21 knockdown. In addition, minimizing toxicity associated with most cationic carriers for gene delivery, such as PEI and dendrimers, is paramount to achieve a clinical delivery system for miRNA [12].
Cell penetrating peptides are small peptides (6 to 30 amino acid residues) that have membrane translocation activity. They constitute an important category of transfection reagent due to their low toxicity and low immunogenicity [46]. Arginine-rich peptides are one of the most investigated categories of CPPs, and include TAT, protamine and oligoarginine [28, 38, 45]. In particular, the guanidinium groups on arginine are efficient at mediating cellular entry while maintaining low cytotoxicity [22]. Although oligoarginine-complexes of several types have been investigated as carriers for gene therapy and siRNA delivery [14], we are interested in understanding the entry oligoarginine-RNA complexes to rationally design protease-activated sytems, similar to our design of small-molecule delivery systems [14]. By understanding the cellular entry, interactions, and activity for the peptide-RNA complexes we, and others, can better design systems in the future.
Aiming to lower the miR-21 level in glioblastoma, we set out to use octaarginine (R8) to form complexes with single stranded anti-miR-21. Single stranded anti-miRNAs hybridize with mature miRNAs in the cytoplasm and prevent the miRNAs from recognizing their mRNA targets [27]. Double stranded siRNA has been delivered using TAT and R9 [43]; however, little is known concerning the differences between double stranded siRNA and single stranded anti-miRNA influence interaction with CPPs, the understanding of which will facilitate the use of CPPs as a RNA delivery vehicle. To this end, we sought to investigate the R8-mediated delivery efficiency of single stranded anti-miRNA and double stranded siRNA. Further, we sought to examine anti-miR-21/R8 complex efficiency of gene silencing and inhibiting glioblastoma cell migration. Given the high fatality rate of glioblastoma multiforme, an anti-miR-21/R8 formulation with low toxicity could be easily administered by intracranial infusion to brain tumor patients or in the tumor void as a post-operative treatment after resection surgery suggesting that miRNA/R8 complexes hold potential alone, as a cell-entry molecule for nanoparticles, [17] or as part of a locally activated drug delivery system [8, 34, 39, 40, 44, 50].
2. Materials and methods
2.1. Materials
Acetyl-RRRRRRRR-amide (R8) was synthesized by UIC research supply center. MirVana™ miRNA-21 inhibitor (miRBase Accession# MIMAT0000076, mature miRNA sequence UAGCUUAUCAGACUGAUGUUGA), miRNA inhibitor negative control #1, and Silencer® siRNA negative control #1 were purchased from Ambion (Austin, TX). Cy3-labeled anti-miRNA, Cy3-labeled siRNA and LysoTracker yellow HCK-123 were obtained from Invitrogen. Human glioblastoma U251 cell line was received from Dr. Lena Al-Harthi (Rush University). The cell line was maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS), 1% nonessential amino acids, 1 mM sodium pyruvate, 100 U/mL penicillin, and 100 U/mL streptomycin. Cells were incubated at 37 °C in humidified air and 5% CO2.
2.2. Preparation and characterization of complexes
The anti-miRNA/R8 or siRNA/R8 was prepared by combining equal volumes of RNA (100 pmol) and peptide at predetermined positive to negative (+/−) charge ratios in RNase free phosphate buffered saline (PBS) to make complexes, vortexed for 15 sec, and then incubated at room temperature for 20 min before use. The resulting solutions were analyzed by electrophoresis using a 20% non-denaturing acrylamide gel for 1 h at 100 V in 1× TBE buffer (89 mM Tris-borate, 2 mM EDTA). Following ethidium bromide (0.5 μg/mL) staining, the gel was visualized using a gel documentation system (GelDoc 2000, Bio-Rad, Hercules, CA). The size distribution and zeta potential of the complexes were measured using a Nicomp 380 Zeta Potential/Particle Sizer in RNAse free water (Particle Sizing Systems, Santa Barbara, CA).
2.3. Fluorescence quenching
The fluorescence quenching assay was conducted as previously reported [51]. Briefly, 60 pmol of Cy3-labeled anti-miRNA or Cy3-labeled siRNA was complexed with R8 at different charge ratios. Cy3-labeled anti-miRNA or Cy3-labeled siRNA without R8 was used to normalize (100% uncondensed) the loss of fluorescence intensity due to condensation. Pure R8 solution of the highest concentration in experimental groups was used as negative control. The samples were incubated for 20 to 30 mins after mixing before being transferred to a Corning black 96 well plate and measured on BioTek Synergy 2 Multi-Mode Microplate Reader (Winooski, VT).
2.4. Cell association and uptake
To measure cell association and uptake, 2×105 cells were seeded in 12-well plate and incubated overnight. Complexes prepared at different charge ratios with the final anti-miRNA or siRNA concentration maintained constant at 100 nM were added to OPTI MEM medium to make the total media volume 1 mL prior to an addition incubation of 4 hours. Cells were then washed with cold PBS twice followed by trypsinization and centrifugation at 1500 rpm for 5 mins. The pellets were then washed twice with cold PBS and centrifugation before resuspending in 200 μL of 1% formaldehyde and analyzing on MoFlo Legacy cell sorter (Beckman Coulter, Brea, CA) at excitation 543 nm and emission 570 nm.
To further confirm cell internalization, U251 cells were seeded in Lab-Tek™ 8-chambered coverglass (Nunc; Thermo Fisher Scientific, Skokie, IL) at 3×104 cells per chamber in 200 μL growth media 24 hours before an experiment. The medium was exchanged with OPTI MEM and complexes were applied to the chamber. Four hours later, cells were rinsed twice with PBS. The nuclei and endosomes were stained with Hoechst 33258 for 5 min and 50 nM LysoTracker yellow for 15 min before CLSM imaging. CLSM images were acquired using a Zeiss LSM 510 META (Carl Zeiss, Germany) with a water immersion 63× objective (C-Apochromat, Carl Zeiss). Excitation wavelengths were 405 nm (Diode 405), 488 nm (argon laser) and 543 nm (HeNe laser) for Hoechst 33258, LysoTracker, and Cy3, respectively. In order to obtain the 3D information of the endosome escape efficiency, 20 slices for 20 cells were obtained with CLSM Z-stack and 20 cells were counted as previously reported [1, 15]. The colocalization ratio between Cy3-labeled anti-miRNA or Cy3-labeled siRNA and endosome was measured with Mender’s coefficient, M, using ImageJ, which calculates the percentage of red pixels (Cy3) colocalizing with green pixels (lysoTracker) with an automatic threshold [7]. The endosome escape efficiency, εee, was calculated, Equation 1.
| Equation 1 |
2.5. Transfection
U251 cells were seeded in a 24-well plate at a density of 1.4×105 cells/cm2 and cultivated with 1 mL of DMEM growth medium for 24 h until 60–70% confluent. The cells in each well were replaced with 250 μL OPTI MEM medium containing 27.5 μL of anti-miR-21/R8 complexes solution (charge ratio = 50/1) with a final anti-miR-21 concentration is 55 nM. Four hours after transfection, media was replaced with fresh DMEM growth media.
2.6. Quantitative real-time PCR analysis
Total RNA was extracted using the TRIzol® reagent (Invitrogen) 24-hour post anti-miR-21/R8 transfection. To maintain a constant RNA purity, PureLink™ RNA Mini Kit (Invitrogen) was used in combination with DNAse (Invitrogen) according to manufacturer’s instructions. High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) was used to reverse transcribe the purified RNA to cDNA. The PCR reactions were performed on an Applied Biosystems StepOnePlus™ PCR machine. The PCR mixture was composed of 5 μL SYBR® Green PCR Master Mix (Applied Biosystems), 2 μL sequence specific primers (0.5 mM, final concentration) and 3 μL cDNA. The conditions for PCR reaction were previously reported [25]. Briefly, 95°C for 10 min followed by 40 cycles of 15 s of denaturation at 95°C and 60 s of annealing and elongation at 60°C. Primer sequences used in the PCR were as follows: Programmed cell death 4 (PDCD4): 5′-CAGTTGGTGGGCCAGTTTATTG-3′ (sense), 5′-AGAAGCACGGTAGCCTTATCCA-3′ (antisense); Serpin peptidase inhibitor, clade B (ovalbumin), member 5 (SERPINB5): 5′-ACAGTGGACTAATCCCAGCACCAT-3′ (sense), 5′-ATTTGATAGGGCCACTCCCTTGGT-3′ (anti-sense); Glyceraldehyde 3-phosphate dehydrogenase (GAPDH): 5′-TTC GAC AGT CAG CCG CAT CTT CTT-3′ (sense), 5′-GCC CAA TAC GAC CAA ATC CGT TGA-3′ (anti-sense). The relative gene expression level of the gene of interest was determined using delta-delta-Ct method normalized to the endogenous reference gene, GAPDH.
2.7. In vitro cell migration assay
The influence of anti-miR-21/R8 transfection on cell migration potential was measured using a wound healing assay [26]. Cells were seeded and transfected as previously described. Twenty-four hour post transfection, a wounding area was made along the cell monolayer using a 200 μL pipette tip. Cells were carefully washed with pre-warmed PBS without dislodging the cell monolayer followed by further incubation in DMEM growth medium for another 48 hours until images were taken. Cell migration was measured, Equation 2, as the recovery ratio, P, of wound area at 48-hour post scratching, a48, relative to the initial wound area, a0, for the treatment, a0,t and a48,t, relative to the untreated group, a0,c and a48,c.
| Equation 2 |
2.8. Statistical analysis
Data were presented as the mean plus or minus (±) standard error of the mean (S.E.M.). Three independent replicates of each experiment were performed. Statistics was determined using Student’s t-test or ANOVA using GraphPad Prism v.4.0 (GraphPad Software, San Diego, CA). Post hoc analysis was performed using Turkey’s test when p-value was less than 0.05. Statistical significance was expressed as P<0.05 (single symbol), P<0.01 (double symbol), and P<0.001 (triple symbol).
3. Results
3.1. Physicochemical characterization of complexes
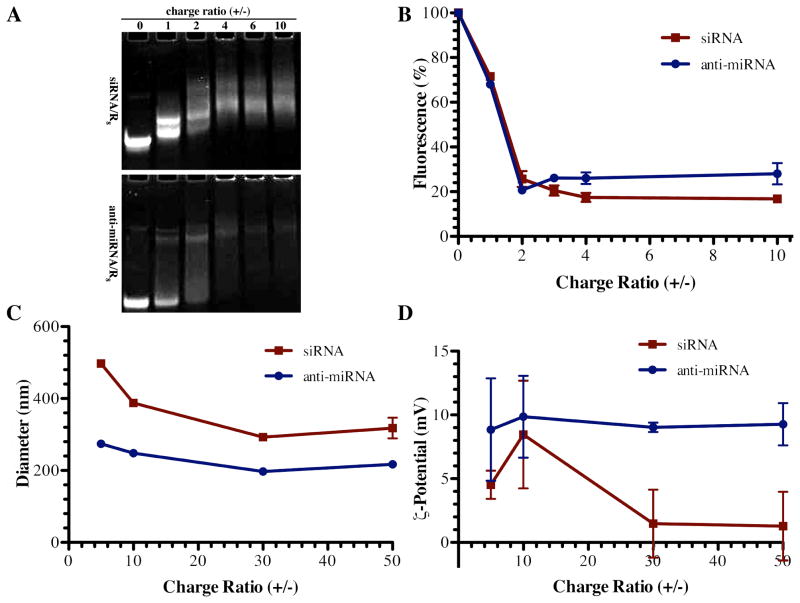
To compare the condensing capacity of R8 for single stranded anti-miRNA and double stranded siRNA, a gel shift assay was conducted by mixing 100 pmol of anti-miRNA or siRNA with R8 at different positive to negative charge ratios (+/−). The R8 peptide successfully formed complexes with both anti-miRNA and siRNA. The mobility of siRNA was retarded at lower charge ratio (Figure 1A) suggesting that double stranded siRNA was more readily interacting with the peptide.
Figure 1. Physicochemical characterization of complexes.
(A) Mobility of siRNA (upper panel) and anti-miRNA (lower panel) was retarded in the presence of R8 peptide above a threshold charge (+/−) ratio. (B) Fluorescence was also quenched above a threshold charge (+/−) ratio, indicating double stranded siRNA (■) and single stranded anti-miRNA (◆) were condensed by the R8 peptide (mean ± S.E.M., n=3). Similarly, R8 complexes (C) diameters and (D) ζ potential were measured by dynamic light scattering (mean ± S.E.M., n=3) for siRNA/R8 complexes (■)and anti-miRNA/R8 complexes (◆) further indicating complex formation at the given charge (+/−) ratios (mean ± S.E.M., n=3).
To investigate whether there is difference between the binding behavior of double stranded siRNA and that of single stranded anti-miRNA with R8, we used dye quenching assay. The fluorescence intensity of fluorophore-labeled oligonucleotides is quenched by the close spatial proximity in complexes where many oligonucleotides are compacted. The trend of fluorescence quenching is the same between anti-miRNA/R8 and siRNA/R8. However, at higher charge ratios, from 4:1 to 10:1, the extent of fluorescence quenching is greater with double stranded siRNA than with single stranded anti-miRNA, suggesting there may be tighter binding between siRNA and the R8 peptide (Figure 1B). The particle size of siRNA/R8 was larger than anti-miRNA/R8 (Figure 1C) at all charge ratios tested.
Cell association of complexes
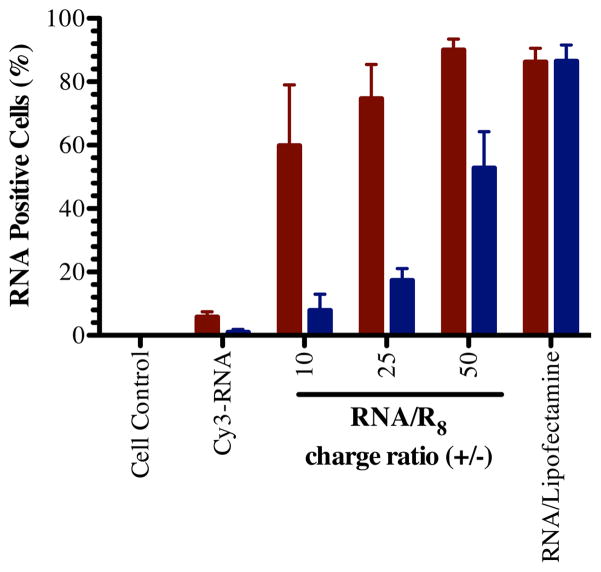
Cell association of siRNA/CPP complexes has been reported to depend on the complex conformation [43]. We compared the cell association efficiency of the two types of complexes using flow cytometry. SiRNA/R8 associated with cells to a greater extent than anti-miRNA/R8 at all of the charge ratios tested (Figure 2). It is worth to noting that there is no significant difference between the ζ potential of anti-miRNA/R8 and siRNA/R8, which were near neutral (Figure 1D). It should be noted that the dissociation methods used may detach or degrade surface-bound complexes from the surface of the cells before flow cytometry. The values presented, may underestimate the surface-bound RNA; however, as the internalized RNA will not be degraded or removed and the flow cytometry would overestimate the internalized RNA. The reproducibility of the data suggests that the method is consistent and able to detect a majority of cells with associated RNA. To confirm this observation and better understand the localization of the RNA, we next examined the internalization of the RNAs using confocal microscopy.
Figure 2. Cell association of RNA/R8 complexes.
Relative association of anti-miRNA/R8 (blue bars) and siRNA/R8 (red bars) with U251 glioblastoma cells after 4-hour interaction in vitro measured by flow cytometry (mean ± S.E.M., n=3).
Cellular uptake and endosome escape
To illustrate the cellular uptake of the complexes and not just surface association, we observed the presence of Cy3-labeled anti-miRNA/R8 and Cy3-labeled siRNA/R8 with confocal microscopy (Figure 3). The complexes showed punctate signal in the cytoplasm of the majority of cells. The punctate nature suggested endocytosis-mediated cellular uptake. Endocytosis and macropinocytosis have been reported as the main mechanisms for the cellular uptake of arginine-rich CPPs [30, 31]. Given the difference in cell association between anti-miRNA/R8 and siRNA/R8 complexes, we speculated that there might also be differences in their interactions with the endosome, specifically escape and separation from the peptide.
Figure 3. Evaluation of endosomal escape efficiencies of anti-miRNA/R8 and siRNA/R8.
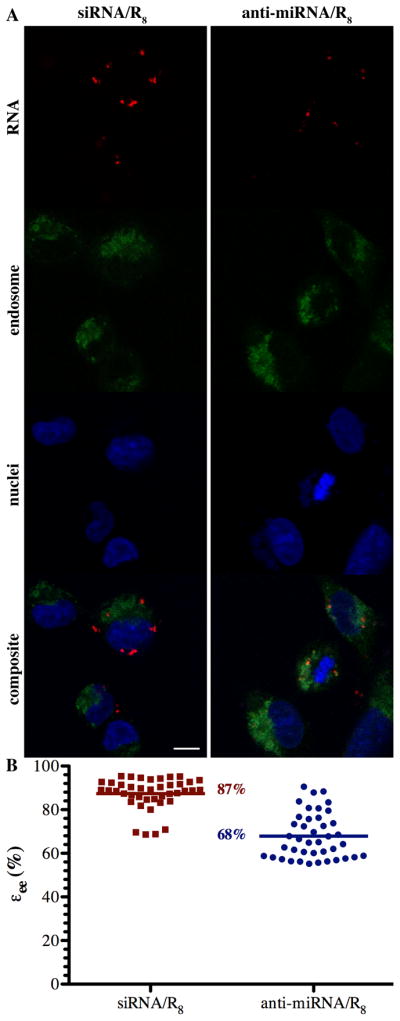
(A) Representative confocal micrographs (scale bar 10 μm) of siRNA/R8 (left) and anti-miRNA/R8(right) complex-endosome colocalization showing RNA (red), endosome (green), nuclei (blue), and the composite of the three pseudocolor images where the RNA concentration was 55 nM and mixed with R8 at a charge ratio of 50. (B) Quantitative comparison of endosome escape efficiency, εee, between anti-miRNA/R8 and siRNA/R8. Endosome escape efficiency was calculated for 20 random, individual cells (p < 0.001).
We compared their endosome escape efficiency by obtaining the colocalization efficiency of Cy3-labeled anti-miRNA or Cy3-labeled siRNA with LysoTracker yellow stained endosomes. By determining the colocalization coefficient in every Z-stack slice, the endosome escape in the cytoplasm measured, while avoiding the bias of utilizing single slice. SiRNA/R8 more efficiently escaped the endosomes compared to anti-miRNA/R8, consistent with the cell association. However, we did not control for constant cell entry in this experiment, i.e. the greater separation may be due to the greater amount of siRNA within the endosomes. This should be examined in more detail to better understand the influence of the properties of the complexes and their separation.
Anti-miRNA-21/R8 gene silencing
MiRNA acts as gene regulators by binding to the 3′ untranslated region of targeted mRNA. One miRNA regulates a network of genes because the targeting region is in the untranslated region of the mRNA, is present on many mRNA, and does not require perfect base pairing. Tumor suppressor genes, PDCD4 and SERPINB5, have been reported as being miR-21 suppressed genes [21, 33]. They are transcriptionally regulated by miR-21, specifically, when miR-21 is present, the mRNA of PDCD4 and SERPINB5 degraded. Indirect detection of miRNA activity was chosen due to limited degradation of miRNAs when anti-miRNAs are used [37]. The anti-miRNAs bind the miRNAs, which are stabilized, but unable to bind the mRNA target [41].
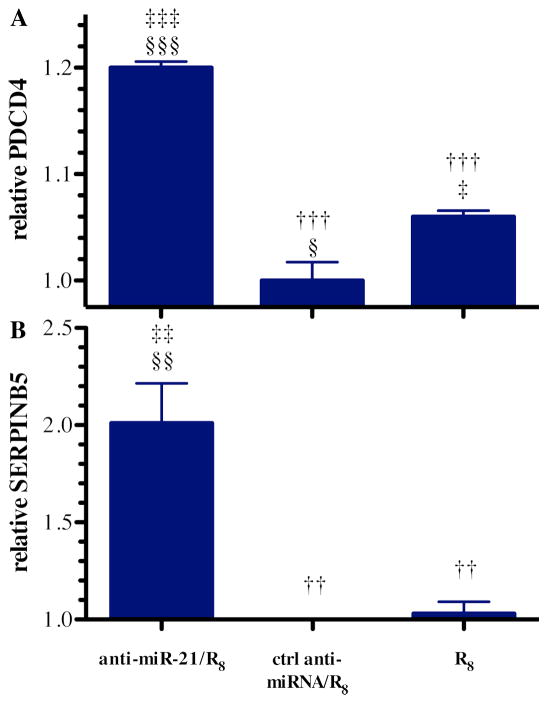
Based on our results with cell association and internalization, we observed the best miR-21 knockdown effect when preparing the complexes at charge ratio of 50 to 1 (+/−). As such, formula complexes at this charge ratio were examined. Both PDCD4 and SERPINB5 mRNA levels were significantly increased after anti-miR-21/R8 transfection comparing with the control groups, indicating successful anti-miR-21 intracellular delivery mediated by R8 (Figure 4). The relationship between PDCD4 and SERPINB5 mRNA level and miR-21 were validated by measuring their mRNA levels after anti-miR-21/Lipofectamine transfection (Figure S1).
Figure 4. Indirect measurement of anti-miRNA activity by detecting the mRNA of downstream miR-21 targets.
(A) PDCD4 and (B) SERPINB5 mRNA levels relative to GAPDH mRNA in cells treated with the R8 peptide, control (ctrl) anti-miRNA/R8 complexes, and anti-miRNA-21/R8 complexes where the RNA concentration was 55 nM and mixed with R8 at a charge ratio of 50 (mean±S.E.M.; n=3). Statistical significance compared to the anti- miRNA-21/R8 group (†), ctrl anti-miRNA/R8 group (‡), and R8 treatment group (§) is presented as one (0.01 < p < 0.05), two (0.001 < p < 0.01), or three (p < 0.001) symbols.
The delivery of oligonucleotides in many circumstances is associated with cytotoxicity. While examining the relative (per cell) mRNA expression, we also evaluated cytotoxicity at the concentration tested (Figure S3). None of the complexes showed significant toxicity at the concentrations examined. We, therefore, inferred that the change in mRNA levels were due to the inhibition of miRNA activity and not a result of cell death by some other mechanism. This was further confirmed by the use of the anti-miRNA control, which did not show a change in mRNA level.
Tumor Cell Migration Inhibition
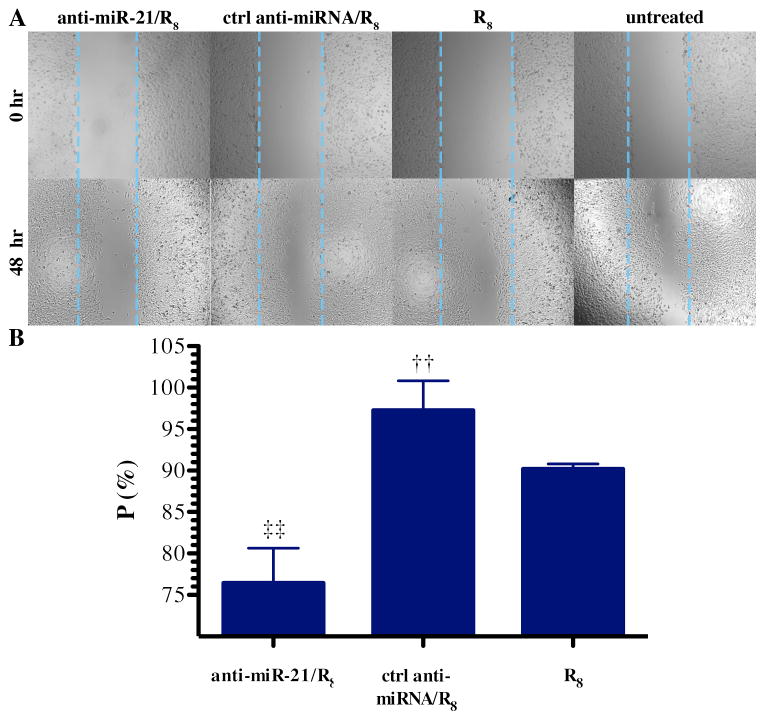
The two selected miRNA-21 targeted genes, PDCD4 and SERPINB5, have also been reported to play inhibitory roles in cancer cell invasion and migration [2, 13, 36]. To examine the influence of knockdown of miR-21 by anti-miR-21/R8 complexes on cell migration, we used a wound-healing assay. In monolayer cell culture, the recovery of wound area was compared at 72-hour post transfection with no measureable reduction in cell survival (Figure S4). The 72-hour time point was chosen based upon preliminary experiments to give sufficient time for anti-miR-21 exerts effects on target mRNAs and subsequent proteins expression. Anti-miR-21/R8 complexes significantly inhibited cell migration by 25% compared with the negative control treated group (Figure 5), indicating the amount of anti-miR-21 transfected is biological active. The inhibition of cell migration by anti-miRNA-21 on U251 cells was also tested by Lipofectamine transfection as positive control (Figure S2).
Figure 5. Inhibition of U251 cell migration after anti-miR-21/R8 transfection.
(A) Micrographs of U251 cells immediately after (0 hr) and 48 hours after wounding. (B) Wound recovery (P) measured 72 hours post transfection following treatment with R8 peptide, ctrl anti-miRNA/R8 complexes, or anti-miRNA-21/R8 complexes where the RNA concentration was 55 nM and mixed with R8 at a charge ratio of 50 (mean±S.E.M.; n=3) The wound recovery (P) of the three groups were normalized to the cell control group underwent wounding assay but no treatment. Statistical significance compared to the anti-miRNA-21/R8 group (†) and ctrl anti-miRNA/R8 group (‡) treatment group is presented (0.001 < p < 0.01).
Discussion
Within the last decade, the involvement of miRNA in human cancer oncogenesis and progression has become apparent [20]. Cancer is intimidating because of its pharmacological complexity and constant development of resistance against therapy. MiRNAs act in accordance with our current understanding of cancer as a “pathway disease” that presumably can only be successfully treated when simultaneously intervening with multiple oncogenic pathways [3].
Even though double stranded siRNA and single stranded anti-miRNA are both seen as RNAi technology, their mechanisms are different. RNAi mediated by double stranded siRNA aims to target one gene at a time by knocking down a specific mRNA. The nascent siRNA associates with Dicer, TRBP, and Argonaute 2 (Ago2) to form the RNA-Induced Silencing Complex (RISC) [11]. Once in RISC, one strand of the siRNA (the passenger strand) is degraded or discarded while the other strand (the guide strand) remains to guide the cleavage of the target mRNA by Ago2, a ribonuclease. Anti-miRNAs sterically block miRNA function by hybridizing and repressing the activity of a mature miRNA whether it exists in single-stranded form, double-stranded form with the natural passenger strand or bound to an Argonaute protein in the miRNA induced silencing complex (miRISC) [27].
Due to this difference in mechanisms and differential effect on a given mRNA, it would be quite difficult and less meaningful to directly compare the gene knockdown efficiency of anti-miRNA/R8 with siRNA/R8. Thus, we examined the ability of the peptide to condense anti-miRNA and how the complexes interact with cells, but the knockdown efficiency was not directly compared. Arginine-rich CPPs have been used to deliver double stranded siRNA, but the delivery of single stranded anti-miRNA via arginine-rich CPP in noncovalent manner has never been described to our knowledge. Therefore, our successful delivery of single stranded anti-miRNAs achieving migration repression and gene knockdown is significant and shows therapeutic potential.
Single stranded anti-miRNA is a flexible oligonucleotide that could have many different conformations while double stranded siRNA exists in A-form double helix, the most stable form of RNA secondary structures [42]. Thermodynamically speaking, it will cost more energy to stabilize a system composed of single stranded anti-miRNA than double stranded siRNA. On the other hand, the rigid structure of double stranded siRNA functions as skeleton and forces R8 to interact alongside the double helix conformation. Interestingly, this discrepancy leads to different cell association and endosome escape efficiency. It has been reported that the polyplexes formed by circular plasmid DNA/CPPs and siRNA/CPPs differ significantly in terms of physicochemical properties, cellular uptake and endosome escape due to their molecular weight and structure differences [35]. The dramatic difference between anti-miRNA/R8 and siRNA/R8 reported here advanced our understanding about the interaction between CPPs and oligonucleotides. Special consideration should be given to the application of single stranded anti-miRNAs transfection with CPPs.
In addition, oligoarginines can be tailored for the establishment of a wide variety of miRNAs therapeutics delivery system since they can be easily encapsulated into or conjugated with other polymeric carrier. Our group has previously reported a matrix metalloproteinases-2 responsive hydrogel drug delivery system aiming for glioblastoma, a great option to combine with the result reported here [40, 48]. Although we have not proceed to in vivo animal experiment yet, Kim et al has shown that HER-2-specific siRNA/R15 complexes resulted in a marked reduction of SKOV-3 xenograft tumor growth in nude mice in vivo [24]. Further study is warranted to investigate the miRNA-21 silencing efficiency mediated by anti-miR-21/R8 complexes in vivo.
Conclusion
For the first time, the difference between single stranded anti-miRNA/R8 and double stranded siRNA/R8 was understood by comparing their physicochemical property, cell association and endosome escape efficiency. Single stranded anti-miRNAs pose greater challenge for intracellular delivery via CPPs. In addition, effective in vitro miRNA-21 interference was achieved via anti-miR-21/R8 complexes, resulting in significant inhibition on U251 cell migration, which warrants further exploration of oligoarginine as intracellular anti-miRNA carrier.
Supplementary Material
Highlights.
R8-associated anti-miRNA enters cells less efficiently than R8-associated siRNA.
R8-associated anti-miR elicited efficient downstream gene upregulation.
Glioma cell migration was inhibited compared to the negative control group.
This is the first demonstration of miRNA modulation with anti-miR-21/R8 complexes.
Acknowledgments
The authors thank Dr. Hayat Onyuksel for generous use of instrumentation. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR15482 from the National Center for Research Resources, NIH and funded by the University of Illinois at Chicago Center for Clinical and Translational Science (CCTS) award UL1 RR029879. Yu Zhang was supported by University Fellowship from University of Illinois at Chicago.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Akita H, Ito R, Khalil IA, Futaki S, Harashima H. Quantitative three-dimensional analysis of the intracellular trafficking of plasmid DNA transfected by a nonviral gene delivery system using confocal laser scanning microscopy. Mol Ther. 2004;9:443–51. doi: 10.1016/j.ymthe.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 3.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–30. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Bhardwaj A, Singh S, Singh AP. MicroRNA-based Cancer Therapeutics: Big Hope from Small RNAs. Mol Cell Pharmacol. 2010;2:213–9. doi: 10.4255/mcpharmacol.10.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–32. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 8.Buhrman JS, Cook LC, Rayahin JE, Federle MJ, Gemeinhart RA. Proteolytically activated anti-bacterial hydrogel microspheres. J Control Release. 2013;171:288–95. doi: 10.1016/j.jconrel.2013.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 11.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi YJ, Kang SJ, Kim YJ, Lim YB, Chung HW. Comparative studies on the genotoxicity and cytotoxicity of polymeric gene carriers polyethylenimine (PEI) and polyamidoamine (PAMAM) dendrimer in Jurkat T-cells. Drug and chemical toxicology. 2010;33:357–66. doi: 10.3109/01480540903493507. [DOI] [PubMed] [Google Scholar]
- 13.Chou RH, Wen HC, Liang WG, Lin SC, Yuan HW, Wu CW, et al. Suppression of the invasion and migration of cancer cells by SERPINB family genes and their derived peptides. Oncol Rep. 2012;27:238–45. doi: 10.3892/or.2011.1497. [DOI] [PubMed] [Google Scholar]
- 14.Darefsky AS, King JT, Jr, Dubrow R. Adult glioblastoma multiforme survival in the temozolomide era: a population-based analysis of Surveillance, Epidemiology, and End Results registries. Cancer. 2012;118:2163–72. doi: 10.1002/cncr.26494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Sayed A, Khalil IA, Kogure K, Futaki S, Harashima H. Octaarginine- and octalysine-modified nanoparticles have different modes of endosomal escape. J Biol Chem. 2008;283:23450–61. doi: 10.1074/jbc.M709387200. [DOI] [PubMed] [Google Scholar]
- 16.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nature reviews Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 17.Farkhani SM, Valizadeh A, Karami H, Mohammadi S, Sohrabi N, Badrzadeh F. Cell penetrating peptides: Efficient vectors for delivery of nanoparticles, nanocarriers, therapeutic and diagnostic molecules. Peptides. 2014;57C:78–94. doi: 10.1016/j.peptides.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–84. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Molecular and cellular biology. 2008;28:5369–80. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaur AB, Holbeck SL, Colburn NH, Israel MA. Downregulation of Pdcd4 by mir-21 facilitates glioblastoma proliferation in vivo. Neuro-oncology. 2011;13:580–90. doi: 10.1093/neuonc/nor033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geihe EI, Cooley CB, Simon JR, Kiesewetter MK, Edward JA, Hickerson RP, et al. Designed guanidinium-rich amphipathic oligocarbonate molecular transporters complex, deliver and release siRNA in cells. Proc Natl Acad Sci U S A. 2012;109:13171–6. doi: 10.1073/pnas.1211361109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermansen SK, Dahlrot RH, Nielsen BS, Hansen S, Kristensen BW. MiR-21 expression in the tumor cell compartment holds unfavorable prognostic value in gliomas. J Neuro-Oncol. 2013;111:71–81. doi: 10.1007/s11060-012-0992-3. [DOI] [PubMed] [Google Scholar]
- 24.Kim SW, Kim NY, Choi YB, Park SH, Yang JM, Shin S. RNA interference in vitro and in vivo using an arginine peptide/siRNA complex system. Journal of controlled release: official journal of the Controlled Release Society. 2010;143:335–43. doi: 10.1016/j.jconrel.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Kollmer M, Keskar V, Hauk TG, Collins JM, Russell B, Gemeinhart RA. Stem cell-derived extracellular matrix enables survival and multilineage differentiation within superporous hydrogels. Biomacromolecules. 2012;13:963–73. doi: 10.1021/bm300332w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lampugnani MG. Cell migration into a wounded area in vitro. doi: 10.1385/1-59259-258-9:177. [DOI] [PubMed] [Google Scholar]
- 27.Lennox KA, Owczarzy R, Thomas DM, Walder JA, Behlke MA. Improved Performance of Anti-miRNA Oligonucleotides Using a Novel Non-Nucleotide Modifier. Molecular therapy Nucleic acids. 2013;2:e117. doi: 10.1038/mtna.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindberg S, Munoz-Alarcon A, Helmfors H, Mosqueira D, Gyllborg D, Tudoran O, et al. PepFect15, a novel endosomolytic cell-penetrating peptide for oligonucleotide delivery via scavenger receptors. Int J Pharm. 2013;441:242–7. doi: 10.1016/j.ijpharm.2012.11.037. [DOI] [PubMed] [Google Scholar]
- 29.Moore LM, Zhang W. Targeting miR-21 in glioma: a small RNA with big potential. Expert Opin Ther Targets. 2010;14:1247–57. doi: 10.1517/14728222.2010.527334. [DOI] [PubMed] [Google Scholar]
- 30.Nakase I, Niwa M, Takeuchi T, Sonomura K, Kawabata N, Koike Y, et al. Cellular uptake of arginine-rich peptides: roles for macropinocytosis and actin rearrangement. Mol Ther. 2004;10:1011–22. doi: 10.1016/j.ymthe.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 31.Nakase I, Takeuchi T, Tanaka G, Futaki S. Methodological and cellular aspects that govern the internalization mechanisms of arginine-rich cell-penetrating peptides. Adv Drug Deliv Rev. 2008;60:598–607. doi: 10.1016/j.addr.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. Jama. 2013;310:1842–50. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 33.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–72. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 34.Ross AE, Tang MY, Gemeinhart RA. Effects of Molecular Weight and Loading on Matrix Metalloproteinase-2 Mediated Release from Poly(Ethylene Glycol) Diacrylate Hydrogels. AAPS J. 2012;14:482–90. doi: 10.1208/s12248-012-9356-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scholz C, Wagner E. Therapeutic plasmid DNA versus siRNA delivery: common and different tasks for synthetic carriers. J Control Release. 2012;161:554–65. doi: 10.1016/j.jconrel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 36.Shi HY, Stafford LJ, Liu Z, Liu M, Zhang M. Maspin controls mammary tumor cell migration through inhibiting Rac1 and Cdc42, but not the RhoA GTPase. Cell Motil Cytoskeleton. 2007;64:338–46. doi: 10.1002/cm.20187. [DOI] [PubMed] [Google Scholar]
- 37.Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3:1. doi: 10.1186/1758-907X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suh JS, Lee JY, Choi YS, Chong PC, Park YJ. Peptide-mediated intracellular delivery of miRNA-29b for osteogenic stem cell differentiation. Biomaterials. 2013;34:4347–59. doi: 10.1016/j.biomaterials.2013.02.039. [DOI] [PubMed] [Google Scholar]
- 39.Tauro JR, Gemeinhart RA. Matrix metalloprotease triggered local delivery of cancer chemotherapeutics. Bioconjugate Chem. 2005;16:1133–9. doi: 10.1021/bc0501303. [DOI] [PubMed] [Google Scholar]
- 40.Tauro JR, Lee BS, Lateef SS, Gemeinhart RA. Matrix Metalloprotease Selective Peptide Substrates Cleavage within Hydrogel Matrices for Cancer Chemotherapy Activation. Peptides. 2008;29:1965–73. doi: 10.1016/j.peptides.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomson DW, Bracken CP, Szubert JM, Goodall GJ. On measuring miRNAs after transient transfection of mimics or antisense inhibitors. PLoS One. 2013;8:e55214. doi: 10.1371/journal.pone.0055214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tinoco I, Jr, Bustamante C. How RNA folds. J Mol Biol. 1999;293:271–81. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 43.van Asbeck AH, Beyerle A, McNeill H, Bovee-Geurts PH, Lindberg S, Verdurmen WP, et al. Molecular parameters of siRNA--cell penetrating peptide nanocomplexes for efficient cellular delivery. ACS Nano. 2013;7:3797–807. doi: 10.1021/nn305754c. [DOI] [PubMed] [Google Scholar]
- 44.Vartak D, Gemeinhart RA. Matrix metalloproteases: Underutilized Targets for Drug Delivery. J Drug Target. 2007;15:1–21. doi: 10.1080/10611860600968967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vives E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem. 1997;272:16010–7. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 46.Wagstaff KM, Jans DA. Protein transduction: cell penetrating peptides and their therapeutic applications. Curr Med Chem. 2006;13:1371–87. doi: 10.2174/092986706776872871. [DOI] [PubMed] [Google Scholar]
- 47.Zadran S, Remacle F, Levine RD. miRNA and mRNA cancer signatures determined by analysis of expression levels in large cohorts of patients. Proc Natl Acad Sci U S A. 2013;110:19160–5. doi: 10.1073/pnas.1316991110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Gemeinhart RA. Improving matrix metalloproteinase-2 specific response of a hydrogel system using electrophoresis. Int J Pharm. 2012;429:31–7. doi: 10.1016/j.ijpharm.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Wang Z, Gemeinhart RA. Progress in microRNA delivery. Journal of controlled release: official journal of the Controlled Release Society. 2013;172:962–74. doi: 10.1016/j.jconrel.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Wang ZJ, Gemeinhart RA. Progress in microRNA Delivery. J Control Release. 2013;172:962–74. doi: 10.1016/j.jconrel.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng M, Pavan GM, Neeb M, Schaper AK, Danani A, Klebe G, et al. Targeting the blind spot of polycationic nanocarrier-based siRNA delivery. ACS nano. 2012;6:9447–54. doi: 10.1021/nn301966r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu H, Acquaviva J, Ramachandran P, Boskovitz A, Woolfenden S, Pfannl R, et al. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc Natl Acad Sci U S A. 2009;106:2712–6. doi: 10.1073/pnas.0813314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.