Abstract
The first and highly conserved step in glutathione (GSH) biosynthesis is formation of γ-glutamyl cysteine by the enzyme glutamate-cysteine ligase (GshA). However bioinformatic analysis revealed many prokaryotes encoding GSH-dependent proteins lack this enzyme. To understand how bacteria cope without gshA, we isolated E. coli ΔgshA multigenic suppressors that accumulated physiological levels of GSH. Mutations in both ProB and ProA, the first two genes in l-Pro biosynthesis, constituted a novel pathway of γ-glutamyl cysteine formation via the selective interception of ProB-bound γ-glutamyl phosphate by amino acid thiols, likely through an S→N acyl shift mechanism. Bioinformatic analysis suggested that the l-Pro biosynthetic pathway may have a second role in γ-glutamyl cysteine formation in prokaryotes. Also, we showed that this mechanism could be exploited to generate cytoplasmic redox buffers bio-orthogonal to GSH.
Glutathione, γ-l-glutamyl-l-cysteinyl-glycine (GSH; 1), is a highly conserved low molecular weight thiol found in many organisms including cyanobacteria, proteobacteria, a few strains of gram-positive bacteria, and in all eukaryotes having mitochondria and chloroplasts1. Glutathione is synthesized by the action of two enzymes: glutamate-cysteine ligase (gshA) and glutathione synthetase (gshB). Glutamate-cysteine ligase catalyzes the reaction between glutamic acid and cysteine to form γ-glutamyl cysteine (2), which is subsequently conjugated to glycine by glutathione synthetase to form GSH.
While GSH is essential for growth in eukaryotes, it is dispensable in bacteria grown under laboratory conditions. Nevertheless, GSH is known to play a central role in protecting bacteria from stressful conditions such as low pH, oxidative and osmotic stresses, toxic electrophiles and xenobiotics1. In addition, GSH provides electrons to glutaredoxins for the reduction of aberrant, catalytic or regulatory disulfide bonds formed in cytoplasmic proteins and also in pathogenicity1,2. Despite these important functions, the majority of aerobic bacteria do not encode a gshA homologue, suggesting that they lack the ability to produce GSH. While subset of these organisms instead synthesize other low molecular weight thiols such as mycothiol, or bacillithiol3,4, there are numerous aerobic bacteria which are not known to make GSH or other low molecular weight thiols3,5.
To address how bacteria lacking gshA might be able to carry out biological processes that normally require GSH, we sought to isolate suppressor mutations in E. coli ΔgshA cells that will confer arsenate (AsO43-) resistance. In E. coli, the ability to grow in the presence of elevated concentrations of AsO43- requires the presence of GSH, as arsenate reductase (arsC gene product) which is essential for growth on AsO43-, depends on GSH for its activity6. Mutagenesis of E. coli ΔgshA followed by selection for growth on AsO43- led to isolation of suppressor mutations in both proB and proA, the first two genes in the proline biosynthetic pathway. The suppressor mutations in proB and proA resulted in the formation of γ-glutamyl cysteine which was then converted to GSH by gshB, conferring growth on AsO43-. Genetic analysis revealed that ProA(supp) and ProB(supp) (supp indicates a suppressor) are the only two proteins required for GSH biosynthesis. Biochemical studies with purified proteins indicated that amino acid substitutions in ProB(supp) reduce the Km for l-Glu and relieve l-Pro mediated feedback inhibition of ProB, whereas those in ProA(supp) abolish NADPH oxidase activity, and together they mediate the highly specific condensation of γ-glutamyl phosphate with cysteine to form γ-glutamyl cysteine. Bioinformatic analysis revealed the existence of several proB and proA homologues containing the amino acids found in the E.coli proB(supp) proA(supp) alleles.
RESULTS
Bioinformatic analysis of GSH biosynthesis in bacteria
A large number of bacterial genomes do not encode gshA homologues (Supplementary Fig. 1 online). Among them there are a few species which synthesize other low molecular weight (L.M.W) thiols, such as species in the actinobacteria phylum that are known to produce mycothiol3. However for most other aerobic bacteria, there is little information on whether they employ L.M.W. thiols in electron transfer. More interestingly, among the bacteria that do not contain gshA, we identified 202 different strains that have genes coding for proteins which may require GSH for their activity (Supplementary Fig. 1 online). From this data, it appears that these bacteria may produce a GSH analogue or may have an alternate pathway for biosynthesis of GSH.
Mutations that confer growth of E. coli ΔgshA on AsO43-
E. coli WP758 (DHB4 ΔgshA) does not form colonies on MOPScasein minimal medium agar plates with 0.1 mM AsO43-, whereas its isogenic parent, DHB4 plates at 100% efficiency in the same medium containing 1 mM AsO43- (Supplementary Fig. 2 online). Following chemical mutagenesis of WP758 cells with 1-methyl-3-nitro-1-nitrosoguanidine (MNNG), several clones that could form colonies on MOPScasein minimal medium agar plates containing 0.1 mM AsO43- were obtained after one day at 37°C. The three fastest growing clones were designated CM1, CM8, and CM15.
Genetic mapping experiments were used to identify the chromosomal alleles responsible for AsO43- resistance in CM1. Several transposon insertions displaying 45%-99% linkage (Fig. 1) were isolated. Sequencing of 5 kb DNA flanking the 99%-linked transposon revealed a transition mutation in both the proB and proA genes leading to the amino acid substitutions ProB(A117V) and ProA(S362F). Similarly, the proB and proA genes in CM8 and CM15 also had one transition mutation each [CM8: ProB(T120I), ProA(S362F); CM15: ProB(E143K), ProA(R143C)] (designated as proB(supp) and proA(supp)). ProB, the first enzyme of the l-Pro biosynthetic pathway, is a γ-glutamyl kinase whose reaction product, γ-glutamyl phosphate, is reduced by the γ-glutamyl phosphate reductase, ProA, to glutamic acid 5-semialdehyde7 (Fig. 2).
Figure 1.
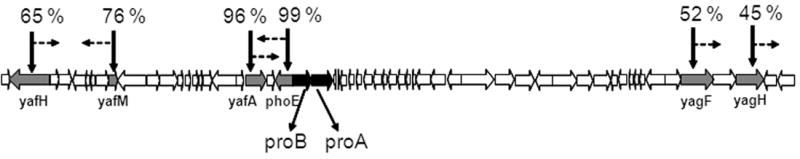
Suppressor mutations in the proBA operon are responsible for the growth of CM1 on AsO43-. The open reading frames that were disrupted by the transposon insertions are shown in gray and the proBA locus is shown in black. The percentages represent the P1 linkage between the transposon and the mutation. Dashed arrows indicate the orientation of the transposon.
Figure 2.
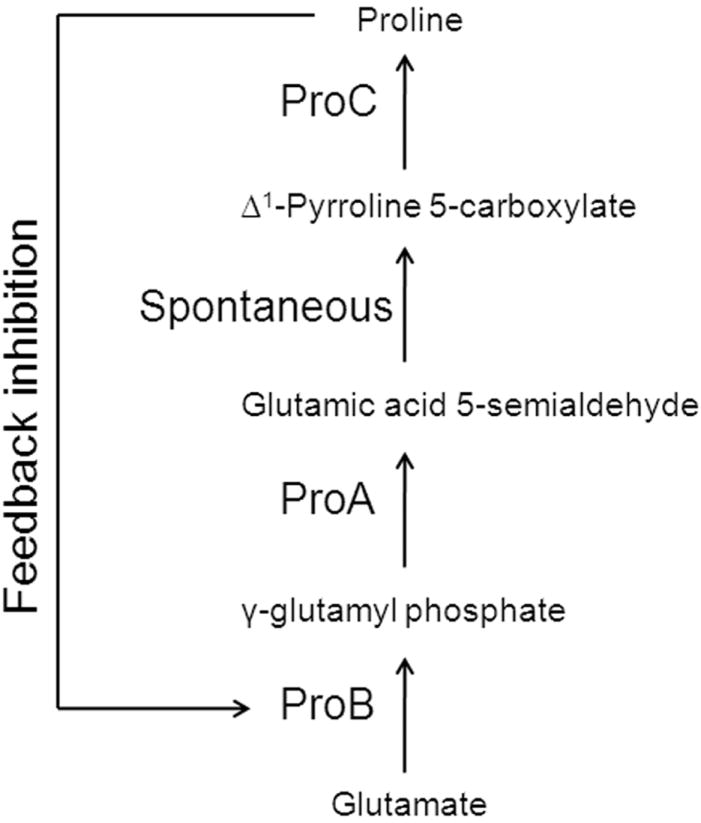
Proline biosynthetic pathway in E. coli. The enzyme ProB converts glutamate to γ-glutamyl phosphate which is then reduced to glutamic acid 5-semialdehyde by ProA. Glutamic acid 5-semialdehyde spontaneously rearranges to form Δ1-Pyrroline 5-carboxylate which is converted by ProC, to proline, a known inhibitor of ProB.
The chromosomal region containing the proBA(supp) (proB and proA genes belong to the operon proBA) alleles in CM1, CM8, and CM15 was transferred to the parent WP758 cells by P1 transduction and the respective strains (WM1, WM8, and WM15) were found to be resistant to AsO43- indicating that this phenotype is likely conferred by the proB(supp) and proA(supp) mutations alone (Supplementary Fig. 2 online). Expression of proBA(supp) from a plasmid conferred AsO43- resistance to WP758 ΔproBA while the wild-type proBA genes could not. Only plasmids carrying mutations in both the proB and proA genes conferred good growth on AsO43- (Table 1). In addition, chromosomal deletions of either the proB(supp) or proA(supp) gene alone in WM1, WM8, or WM15 abolished colony formation in 0.1 mM AsO43-. These results clearly indicate mutations in both proB and proA are required for suppression.
Table 1. Suppression of Growth Defect of WP758 ΔproBA cells expressing proB and proA variants from the plasmid pBAD24.
Single colonies were streaked on MOPScasein minimal medium agar plates containing 0.2 % arabinose (to induce expression from araBAD promoter in pBAD24 plasmid), ampicillin, and 0.1 mM AsO43-. Growth was examined after 15 hours. – indicates no growth, + indicates good growth, and +/- forms tiny colonies.
| Plasmid | Growth of WP758 ΔproBA on AsO43- |
|---|---|
| pBAD24-proB proA | - |
| pBAD24-proB(A117V) proA(S362F) | + |
| pBAD24-proB(T120I) proA(S362F) | + |
| pBAD24-proB(E143K) proA(R143C) | + |
| pBAD24-proB(A117V) proA | +/- |
| pBAD24-proB proA(S362F) | - |
| pBAD24-proB(T120I) proA | +/- |
| pBAD24-proB(E143K) proA | +/- |
| pBAD24-proB proA(R143C) | - |
To evaluate whether other chromosomal genes normally present in WP758 might be required for suppression we carried out a genetic search for Tn insertions that confer AsO43- sensitivity to WM15. Out of a total of 7000 colonies screened, 16 were found to be sensitive to AsO43-. The majority of the Tn insertions disrupted arsC or arsB, genes which are known to be involved directly in AsO43- resistance. Tn insertions in ppk, encoding a polyphosphate kinase and in betT which encodes a choline transporter were also found but given the known biochemical roles of these genes, it is unlikely that they are involved directly in AsO43- resistance. While an earlier report had suggested that the carboxylate amine ligase YbdK displays some glutamate-cysteine ligase activity in vitro8, deletion of ybdk in the proBA(supp) strains did not affect AsO43- resistance. Collectively, these results suggest that unexpectedly, suppression of AsO43- sensitivity in gshA cells is a multigenic trait in that mutations in proB and proA are necessary and sufficient to allow growth under selective conditions.
Suppression mechanism involves biosynthesis of GSH
In an earlier report a Saccharomyces cerevisiae Δgsh1 (gshA homologue) pro2 mutant (proA homologue) was found to accumulate minute amounts of GSH (approx. 0.5% of wild-type level) by an unknown mechanism9. Using standard growth medium, the E. coli ΔgshA proBA(supp) strain WM15 accumulated 1.68 ± 0.12 mM of GSH corresponding to 25% of the level produced by the gshA+ strain DHB4 (Fig. 3a, Supplementary Fig. 3a online). Addition of 5 mM l-Cys to the growth medium increased the levels of GSH in WM15 to 97% of the wild-type levels indicating that the proBA(supp) pathway for GSH biosynthesis is very effective and also that it is limited by the intracellular level of cysteine (Fig. 3a, Supplementary Fig. 4a online). Interestingly, E.coli WM15 cells growing in medium with 5 mM l-Cys accumulated only ~0.3 mM GSH during exponential phase (Supplementary Fig. 4b online). Unlike GSH, l-Cys is much more efficient in reducing Fe3+ and therefore l-Cys in the cell is maintained at low levels to reduce oxidative damage via the Fenton reaction10. GSH biosynthesis was dependent on gshB gene in vivo as proBA(supp) cells lacking gshB accumulated γ-glutamyl cysteine (Supplementary Fig. 3b online).
Figure 3.
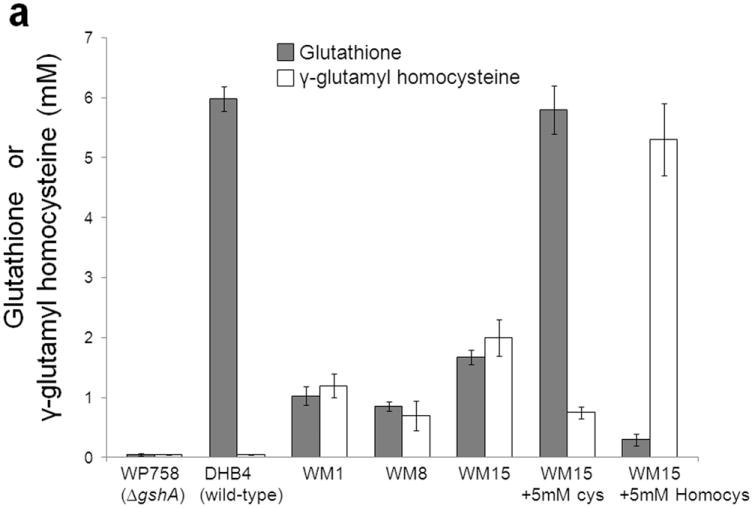
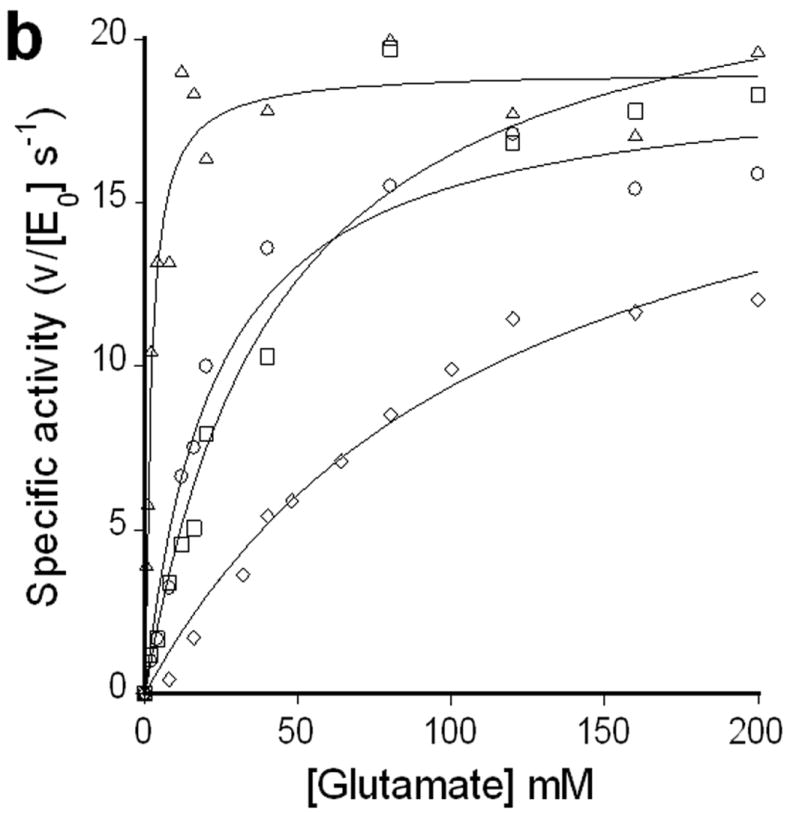
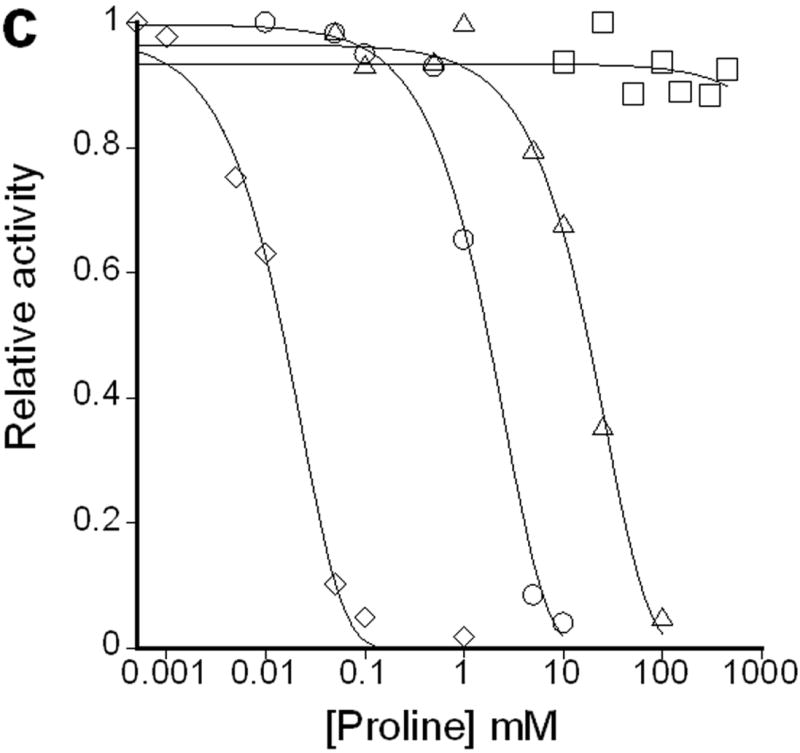
(a) Glutathione (grey bars) and γ-glutamyl homocysteine (white bars) levels in stationary phase cells. WM15+5mM cys or WM15+5mM Homocys indicates that this strain is grown in medium supplemented with 5 mM cysteine or homocysteine respectively (The optimal concentration of cysteine or homocysteine was found to be 5 mM for maximum GSH or γ-glutamyl homocysteine accumulation respectively. See Supplementary Fig. 3a online). The bimane adducts of GSH (m/z~498) and γ-glutamyl homocysteine (m/z~455) were confirmed by LC-MS. The data was obtained by averaging three independent experiments ± s.d.. Influence of l-Glu (b) and l-Pro (c) on the activities of the enzymes ProB(wt) (◇), ProB(A117V) (Δ), ProB(T120I) (○), and ProB(E143K) (□). In plot b, v denotes the rate of oxidation of NADPH (concentration/time), and [E0] is the concentration of ProB variant used in the coupled assay. In plot c, the relative activity on the y-axis represents the fraction of activity of the enzyme in the absence of l-Pro.
Biochemical analysis of ProB(supp) and ProA(supp)
AsO43- resistant mutants (WM1, WM8, and WM15) were found to be proline auxotrophs because they could not grow in MOPS minimal medium without casein but supplemented with all amino acids (50 μg/ml) except proline. This could be because of either (i) ProB(supp) or/and ProA(supp) are loss of function mutations; or (ii) the mutations affect the integrity of the ProB:ProA bienzymatic complex whose formation is required for efficient channeling of the highly unstable intermediate, γ-glutamyl phosphate11.
A coupled in vitro assay, which measures NADPH oxidation by ProA(wt), was used to determine the reaction kinetics of purified samples of ProB(wt) and ProB(supp) [ProB(A117V), ProB(T120I), and ProB(E143K)]. Compared to the wild-type ProB, all ProB(supp) enzymes displayed substantially lower Km values for l-Glu (~40-fold lower for ProB(A117V) with no effect on kcat) (Fig. 3b, Supplementary Table 2 online) when analyzed in the presence of ProA(wt). Additionally, the ProB(supp) enzymes showed dramatically reduced inhibition by l-Pro with at least 100-fold higher IC50. In the case of ProB(E143K), no inhibition was observed even at 500 mM l-Pro (Fig. 3c, Supplementary Table 2 online). The latter result is consistent with earlier reports on the effect of amino acid substitutions in E143 on inhibition by l-Pro12, 13, 14. Remarkably, the ProB(supp) variants exhibited both a substantially higher IC50 for l-Pro and a lower Km for l-Glu even though the binding sites overlap for l-Glu and l-Pro in the ProB crystal structure 15.
In contrast to ProA(wt), no activity in the coupled assay was detected with complexes of ProA(supp) with either ProB(wt) or ProB(supp). On the other hand, ProB(supp)+ProA(supp) or ProB(supp)+ProA(wt) catalyzed the formation of γ-glutamyl hydroxamate from l-Glu and hydroxyl amine, a reaction that has previously been reported to take place with inactive ProA as long as it can still form a complex with ProB7,16 (Supplementary Fig. 5 online). Collectively, these data support the notion that ProA(supp) are loss of function mutations, consistent with the finding that the AsO43- resistant mutants are proline auxotrophs.
These results suggest that the l-Pro resistant ProB(supp) and loss of function ProA(supp) proteins mediate the accumulation of the γ-glutamyl phosphate intermediate in the cell, which reacts with free l-Cys generating γ-glutamyl cysteine. The γ-glutamyl cysteine, in turn, is converted to GSH by the gshB gene product (Fig. 4). γ-glutamyl phosphate is extremely unstable in solution and undergoes cyclization to form 5-oxoproline. To prevent this unwanted reaction, it is predicted to be tightly bound to ProB15. Presumably, binding of γ-glutamyl phosphate into the ProB active site serves to neutralize the phosphate charge facilitating attack by the l-Cys thiolate, an event expected to be disfavored in solution due to electrostatic effects. Hence, as expected no 5-oxoproline could be detected in proB(supp)proA(supp) cells.
Figure 4.
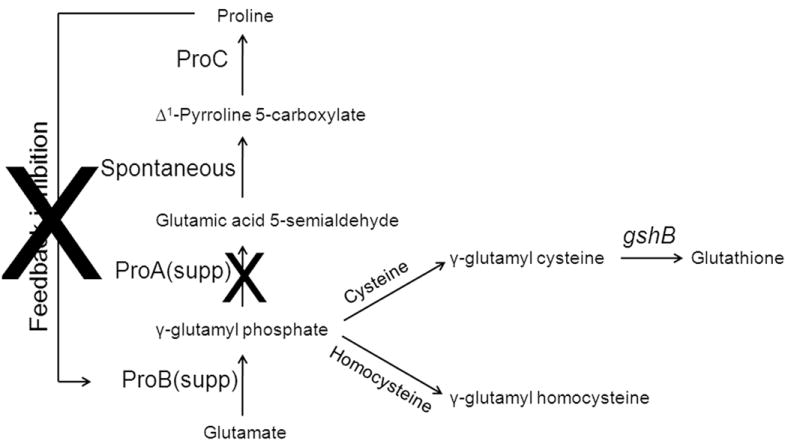
De novo pathway for GSH biosynthesis. The combined effect of ProB(supp) (higher IC50 for l-Pro) and ProA(supp) (loss of function) leads to accumulation of γ-glutamyl phosphate which reacts with free cysteine or homocysteine in the cell to form γ-glutamyl cysteine or γ-glutamyl homocysteine. The enzyme GshB adds glycine to γ-glutamyl cysteine to form GSH.
In vitro, ProB(wt)+ProA(wt) did not catalyze the formation of γ-glutamyl cysteine in the presence of NADPH, l-Pro and l-Cys (Supplementary Table 3 online). However, in the absence of l-Pro, ProB(wt) is not inhibited and therefore the γ-glutamyl phosphate that formed can react with l-Cys to give rise to a small amount of γ-glutamyl cysteine as long as NADPH is not present (Supplementary Table 3 online). Under the same reaction conditions, ProB(supp)+ProA(supp) gave high yields of γ-glutamyl cysteine irrespective of whether NADPH or l-Pro was present, consistent with the finding that the ProB(supp) have a reduced feedback inhibition by l-Pro and ProA(supp) does not display the γ-glutamyl phosphate reductase activity. The formation of γ-glutamyl cysteine from ProB(supp)+ProA(wt) was favored even in the presence of NADPH indicating that at high concentrations of l-Cys, reaction of γ-glutamyl phosphate with l-Cys outcompetes reduction of γ-glutamyl phosphate by ProA. However since both ProB(supp) and ProA(supp) are essential for suppression in vivo, in the intracellular environment where l-Cys is present in μM concentrations, the reductase activity of ProA must be shut off to allow γ-glutamyl cysteine formation.
Biosynthesis of unnatural γ-glutamyl cysteine analogues
We found that γ-glutamyl phosphate (synthesized in vitro by ProB(supp)) reacts selectively with thiol containing amino acids l-Cys and l-Hcys (homocysteine), but not with l-Ser or any amino acid lacking a thiol group (Supplementary Table 3 online). In addition, no reaction was observed when the thiol in l-Cys was blocked by using 2-iodoacetamide. These results reveal that the S atom is required for reaction, which leads to an intriguing mechanistic possibility involving an S→N acyl shift reaction. In this scheme, the deprotonated thiolate of l-Cys attacks the γ-carboxyl, leading to loss of the phosphate moiety as a leaving group and generation of a thioester intermediate (Scheme 1). This is followed by rearrangement to give the stronger amide bond, facilitated by proximity in the thioester intermediate bearing similarities to native chemical ligation17.
Scheme 1.
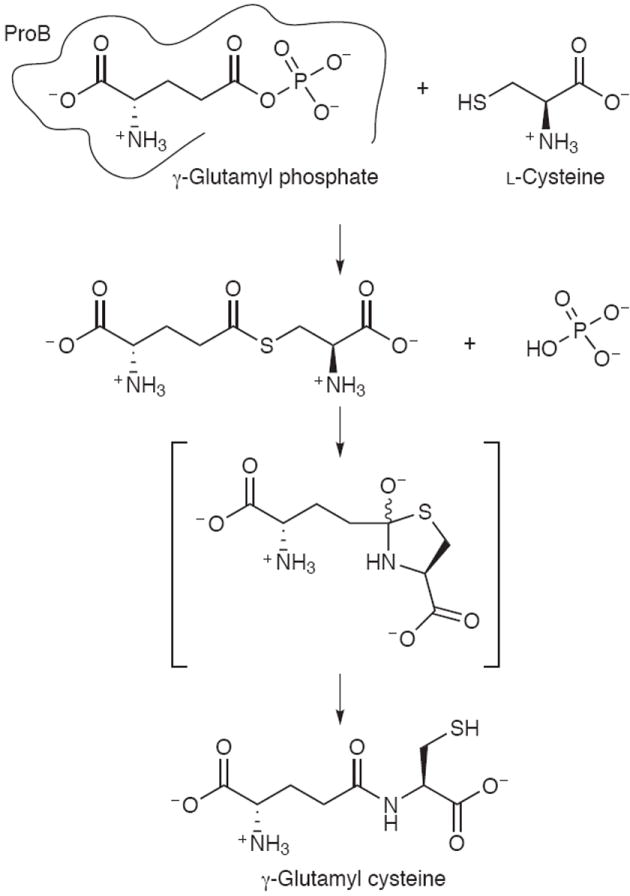
Proposed scheme for reaction between cysteine and γ-glutamyl phosphate.
This reaction is not limited to γ-carboxyl acids as we found for example that aspartokinase III (LysC) can be employed to synthesize β-aspartyl cysteine (3) from β-aspartyl phosphate (Supplementary Fig. 6 online). The enzyme LysC from E. coli was used to convert l-Asp to β-aspartyl phosphate which in the presence of l-Cys formed β-aspartyl cysteine in vitro. LC-MS was used to confirm the formation of β-aspartyl cysteine. Alternatively, growth of ProB(supp)+ProA(supp) mutants in medium containing l-Hcys can be exploited to engineer cells that accumulate the unnatural anti-oxidant γ-glutamyl homocysteine (4) at concentrations comparable to those of GSH in wild-type E. coli strain (Fig. 3a, Fig. 4). Analogous to GSH, γ-glutamyl homocysteine is overwhelmingly present in reduced form (Supplementary Fig. 7 online).
ProB(supp) variants are found in many bacterial species
From a physiology standpoint, bioinformatic analysis revealed that homologues of ProB containing Val at the position corresponding to Ala117 in the E. coli enzyme are found among bacterial strains that do not encode gshA but nonetheless make GSH-dependent proteins such as glutaredoxins (Supplementary Table 4 online). For example, all Streptococcus pyogenes genomes in the database lack gshA but make GSH-dependent proteins including glutathione peroxidase (gpoA) which is important for virulence2. Some of these organisms may be able to accumulate γ-glutamyl cysteine and also produce l-Pro either through regulation of ProA activity or possibly by encoding more than one ProA proteins. Along these lines we note that a few organisms (3/28 in Table S4) encoding ProB(supp) homologues contain two copies of the ProBA operon. Additionally we found that the E. coli mutant WM1 with ProB(A117V)+ProA(wt) on an F’ plasmid accumulated both l-Pro (4.3 ± 0.6 mM) and GSH (2.3 ± 0.3 mM) (see Supplementary methods online for intracellular free proline measurements) indicating that when synthesis of γ-glutamyl phosphate is deregulated, about 35% of the flux is diverted towards the biosynthesis of GSH.
DISCUSSION
GSH and other L.M.W. thiols primarily function as antioxidants and help maintain redox homeostasis but in addition they take part in a variety of other cellular functions. Surprisingly, among bacterial strains that do not have a gshA homologue, there are quite a few species which contain genes for GSH-dependent enzymes such as glutathione S-transferases or glutaredoxins. To understand how cells, lacking the canonical gshA-dependent GSH biosynthetic pathway, can perform the functions that are normally carried out by GSH, we resorted to a genetic approach in E.coli ΔgshA cells. Resistance to AsO43- in E. coli requires the reduction of AsO43- to AsO2-, a reaction catalyzed by ArsC with electrons provided by glutaredoxins and GSH. Since in other bacteria, ArsC is recycled by thioredoxin, we had expected that suppression of the arsenate sensitivity of E.coli ΔgshA mutants might involve mutations that affect the electron transfer of the enzyme. Instead, to our surprise we found that suppression was mediated by pairs of single transition mutations in the genes proB and proA. We showed that mutations in both proB and proA are both necessary and sufficient for conferring growth of E.coli ΔgshA cells on 0.1 mM AsO43- in minimal medium.
Biochemical experiments revealed that the mutations A117V, T120I, or E143K in γ-glutamyl kinase (ProB) abolished or substantially reduced feedback inhibition by proline. The residues A117 and T120 are far away (>15 A°) from the glutamate and proline binding sites15 in E. coli ProB and hence it is difficult to provide a basis for the effect of mutations A117V and T120I on kinetic properties of the enzyme. Interestingly, the mutation A117V in ProB from S. marcescens (corresponds to A117V in E. coli), was shown to result in 700 fold decrease in proline mediated feedback inhibition compared to wild-type ProB13. Amino acid residues 136-148 in E. coli ProB, which belong to the β4-αE loop, are known to be important for proline binding15. Proline binding is predicted to trigger the movement of the β4-αE loop which results in amino acids such as the D148 and E143 in this loop facing inward towards the proline binding site. For example, the amino acid D148 in this mobile loop forms a salt bridge with imino group of proline and hence a change D148N or D148A greatly reduced feedback inhibition by proline18. Similarly the E143K mutation in ProB might affect proline binding resulting in loss of feedback inhibition. Consistent with these results, the mutation E143A in E. coli ProB hampers feedback inhibition by proline12. Interestingly ProB enzymes from plants have a positively charged amino acid (Lys or Arg) at this position (corresponding to 143 in E. coli) in agreement with the observation that the ProB enzymes from plants have reduced inhibition by proline relative to their bacterial counterparts14. It is predicted that this lower sensitivity to feedback inhibition by proline helps plants to accumulate high intracellular proline levels during osmotic stress which plants frequently encounter. Consistent with these observations, γ-glutamyl kinase encoded by proJ in B. subtilis which is induced during osmotic stress contains a basic amino acid arginine at this position (corresponding to position 143 in E. coli) resulting in high intracellular levels of proline during osmotic stress14.
GSH plays a role in many cellular processes by providing a means of shuttling electrons to different enzymatic systems. As a result, thiol-dependent redox metabolic processes are highly coupled and the effects of gshA deletions are pleiotropic. We show here that unnatural GSH analogues such as γ-glutamyl homocysteine can be synthesized at high levels in E. coli ΔgshA proB(supp)proA(supp) cells grown in medium with homocysteine. Deploying an engineered enzyme that can use an unnatural GSH analogue, such as γ-glutamyl homocysteine, in a ΔgshA proB(supp) proA(supp) cell can provide an opportunity to biochemically isolate this pathway from the network of other GSH-dependent redox reactions. In other words, γ-glutamyl homocysteine can serve as a bio-orthogonal electron transfer system, a system to shuttle electrons in parallel but largely independent of GSH. Such a bio-orthogonal electron transfer system may be used to selectively direct electron equivalents to biotransformation reactions of commercial importance.
METHODS
Chemical Mutagenesis and Mutant Selection
Chemical mutagenesis using 1-methyl-3-nitro-1-nitrosoguanidine (MNNG) was performed as described19 except that cells were grown to an A600~1 in MOPScasein minimal medium before harvesting. Selection for growth was performed on MOPScasein minimal medium agar plates containing 0.1 mM AsO43-.
Arsenate resistance assays
Overnight cultures were diluted 100 fold into MOPScasein minimal medium and grown to an A600~0.5-0.7. Then cells were diluted 106 fold and plated on MOPScasein minimal medium agar plates containing different concentrations of AsO43-. Colonies were counted after incubating the plates at 37°C for 36 hours. % survival denotes, (#colonies observed/#colonies observed on plates containing no AsO43-)*100.
Genetic Analysis Using Replica Spotting assays
A library of random transposon insertions using EZ∷TN<DHFR> transposome was constructed in the mutant strain WM15 also carrying ProB(E143K) and ProA(R143C) on an F plasmid. Single colonies from this transposon library were grown till stationary phase in 96 well plates containing MOPScasein minimal medium. Around 2 μl of stationary phase cultures of each colony were spotted on master and replica plates maintaining the same spatial pattern on both the plates. The master plate contains MOPScasein minimal agar medium and the replica plate contains MOPScasein minimal agar medium + 0.1 mM AsO43-. Culture spots were allowed to grow for 16-24 hours and the spots that appeared on master plate but not on the replica plate were selected for further analysis. The spots that appeared only on the master plate were colony purified and single colonies were picked and the genomic DNA was sequenced using transposon specific primers to identify the location of the transposon.
Bioinformatic analyses
Hidden Markov models for the identification of gshA, gshB, and GSH-dependent proteins were obtained from Pfam (http://pfam.sanger.ac.uk//family). The Pfam models used for gshA are: PF04107, PF04262, and PF08886. The Pfam models used for gshB are: PF02951, PF02955, PF03917, and PF03199. Fusion proteins containing activities of both GshA and GshB are also included in the analysis and have hits in the Pfam model PF04262. The Pfam models used for GSH-dependent proteins are: PF00462 (Glutaredoxin), PF02798 (Glutathione transferase N-terminal), PF00043 (Glutathione transferase C-terminal), PF03738 (Glutathionyl spermidine synthase), PF04828 (Glutathione dependent formaldehyde activating enzyme), PF00255 (Glutathione peroxidase), and PF04399 (Glutaredoxin 2).
Hmmer (http://hmmer.janelia.org), and the seed alignments obtained from TIGR (http://cmr.jcvi.org/tigr-scripts), were used to create Markov models for ProA and ProB.
Hmmer was used to search a set of 817 Bacterial and Archaeal genomes that was downloaded on Jan. 1, 2009 from the NCBI ftp site (ftp://ftp.ncbi.nih.gov/genbank/genomes/) with each model. ProA and ProB homologues were aligned to the corresponding E. coli sequence using the alignments to the models in the Hmmer output. The amino acids at positions corresponding to 117, 120, and 143 in ProB and at positions 143, and 362 in ProA were identified from the alignments.
Other Methods
Protocols for mutation mapping and linkage analysis19, enzyme purification and kinetics, in vitro assays, intracellular glutathione20, γ-glutamyl homocysteine and proline measurements are described in Supplementary Methods online.
Supplementary Material
Acknowledgments
We thank E. Stone and M. Faulkner for discussions. This work was supported by NIH grant GMO41883 to J.B. and NIH grant GM55090 to G.G.
Footnotes
AUTHOR CONTRIBUTIONS
K.V. and G.G. designed research; K.V., and D.B. performed research; K.V., D.B., B.I., J.B., and G.G. analyzed data; K.V., D.B., B.I., J.B., and G.G. wrote the paper.
COMPETING FINANCIAL INTERESTS STATEMENT
The authors declare no competing financial interests.
References
- 1.Masip L, Veeravalli K, Georgiou G. The many faces of glutathione in bacteria. Antioxid Redox Signal. 2006;8:753–62. doi: 10.1089/ars.2006.8.753. [DOI] [PubMed] [Google Scholar]
- 2.Brenot A, King KY, Janowiak B, Griffith O, Caparon MG. Contribution of glutathione peroxidase to the virulence of Streptococcus pyogenes. Infect Immun. 2004;72:408–13. doi: 10.1128/IAI.72.1.408-413.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newton GL, et al. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J Bacteriol. 1996;178:1990–5. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newton GL, et al. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat Chem Biol. 2009;5:625–627. doi: 10.1038/nchembio.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahey RC, Brown WC, Adams WB, Worsham MB. Occurrence of glutathione in bacteria. J Bacteriol. 1978;133:1126–9. doi: 10.1128/jb.133.3.1126-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oden KL, Gladysheva TB, Rosen BP. Arsenate reduction mediated by the plasmid-encoded ArsC protein is coupled to glutathione. Mol Microbiol. 1994;12:301–6. doi: 10.1111/j.1365-2958.1994.tb01018.x. [DOI] [PubMed] [Google Scholar]
- 7.Smith CJ, Deutch AH, Rushlow KE. Purification and characteristics of a gamma-glutamyl kinase involved in Escherichia coli proline biosynthesis. J Bacteriol. 1984;157:545–51. doi: 10.1128/jb.157.2.545-551.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehmann C, et al. YbdK is a carboxylate-amine ligase with a gamma-glutamyl:Cysteine ligase activity: crystal structure and enzymatic assays. Proteins. 2004;56:376–83. doi: 10.1002/prot.20103. [DOI] [PubMed] [Google Scholar]
- 9.Spector D, Labarre J, Toledano MB. A genetic investigation of the essential role of glutathione: mutations in the proline biosynthesis pathway are the only suppressors of glutathione auxotrophy in yeast. J Biol Chem. 2001;276:7011–6. doi: 10.1074/jbc.M009814200. [DOI] [PubMed] [Google Scholar]
- 10.Park S, Imlay JA. High levels of intracellular cysteine promote oxidative DNA damage by driving the fenton reaction. J Bacteriol. 2003;185:1942–50. doi: 10.1128/JB.185.6.1942-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baich A. Proline synthesis in Escherichia coli. A proline-inhibitable glutamic acid kinase. Biochim Biophys Acta. 1969;192:462–7. doi: 10.1016/0304-4165(69)90395-x. [DOI] [PubMed] [Google Scholar]
- 12.Rushlow KE, Deutch AH, Smith CJ. Identification of a mutation that relieves gamma-glutamyl kinase from allosteric feedback inhibition by proline. Gene. 1985;39:109–12. doi: 10.1016/0378-1119(85)90115-5. [DOI] [PubMed] [Google Scholar]
- 13.Omori K, Suzuki S, Imai Y, Komatsubara S. Analysis of the mutant proBA operon from a proline-producing strain of Serratia marcescens. J Gen Microbiol. 1992;138:693–9. doi: 10.1099/00221287-138-4-693. [DOI] [PubMed] [Google Scholar]
- 14.Fujita T, et al. Identification of regions of the tomato gamma-glutamyl kinase that are involved in allosteric regulation by proline. J Biol Chem. 2003;278:14203–10. doi: 10.1074/jbc.M212177200. [DOI] [PubMed] [Google Scholar]
- 15.Marco-Marin C, et al. A novel two-domain architecture within the amino acid kinase enzyme family revealed by the crystal structure of Escherichia coli glutamate 5-kinase. J Mol Biol. 2007;367:1431–46. doi: 10.1016/j.jmb.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 16.Hayzer DJ, Leisinger T. The gene-enzyme relationships of proline biosynthesis in Escherichia coli. J Gen Microbiol. 1980;118:287–93. doi: 10.1099/00221287-118-2-287. [DOI] [PubMed] [Google Scholar]
- 17.Schnolzer M, Kent SB. Constructing proteins by dovetailing unprotected synthetic peptides: backbone-engineered HIV protease. Science. 1992;256:221–5. doi: 10.1126/science.1566069. [DOI] [PubMed] [Google Scholar]
- 18.Perez-Arellano I, Rubio V, Cervera J. Mapping active site residues in glutamate-5-kinase. The substrate glutamate and the feed-back inhibitor proline bind at overlapping sites. FEBS Lett. 2006;580:6247–53. doi: 10.1016/j.febslet.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 19.Zhan X, et al. Genetic analysis of disulfide isomerization in Escherichia coli: expression of DsbC is modulated by RNase E-dependent mRNA processing. J Bacteriol. 2004;186:654–60. doi: 10.1128/JB.186.3.654-660.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faulkner MJ, Veeravalli K, Gon S, Georgiou G, Beckwith J. Functional plasticity of a peroxidase allows evolution of diverse disulfide-reducing pathways. Proc Natl Acad Sci U S A. 2008;105:6735–40. doi: 10.1073/pnas.0801986105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


