Abstract
NF-κB has been well documented to play a critical role in signaling cell stress reactions. The extracellular signal-regulated kinase (ERK) regulates cell proliferation and survival. GADD45β is a primary cell cycle element responsive to NF-κB activation in anti-apoptotic responses. The present study provides evidence demonstrating that NK-κB, ERK and GADD45β are co-activated by ionizing radiation (IR) in a pattern of mutually dependence to increase cell survival. Stress conditions generated in human breast cancer MCF-7 cells by the administration of a single exposure of 5 Gy IR resulted in the activation of ERK but not p38 or JNK, along with an enhancement of the NF-κB transactivation and GADD45β expression. Overexpression of dominant negative Erk (DN-Erk) or pre-exposure to ERK inhibitor PD98059 inhibited NF-κB. Transfection of dominant negative mutant IκB that blocks NF-κB nuclear translocation, inhibited ERK activity and GADD45β expression and increased cell radiosensitivity. Interaction of p65 and ERK was visualized in living MCF-7 cells by bimolecular fluorescence complementation analysis. Anti-sense inhibition of GADD45β strikingly blocked IR-induced NF-κB and ERK but not p38 and JNK. Overall, these results demonstrate a possibility that NF-κB, ERK, and GADD45β are able to coordinate in a loop-like signaling network to defend cells against the cytotoxicity induced by ionizing radiation.
Protein kinase activities play a pivotal role in responses to a variety of stress conditions, e.g. UV light, ionizing radiation (IR),1 chemotherapy drugs, virus infection, osmotic shock, heat, nitric oxide, lipopolysaccharide, arsenic oxide, and inflammatory cytokines. A predominant feature of molecular responses under such stress conditions is the activation of early responsive transcription factors and signaling elements resulting in a temporary cell cycle arrest and anti-apoptotic responses leading to an increased cell survival (1–3). Numerous gene products are shown to be inducible in mammalian cells by the stress of IR (4–6). However, only the key signaling proteins that control cell cycle, DNA repair, and cell growth regulation are believed to be decisive in cell death or survival with the situation of lethal insults (5–7). Accumulating data have demonstrated that activation of the transcription factor NF-κB is critical in defending cells from stress-induced damages and influencing the cell survival rate following different cytotoxic conditions (8–10).
NF-κB, with five Rel family members, i.e. p65/RelA, p50, p52, c-Rel, and Rel B, comprise a stress-sensitive gene regulator. The NF-κB heterodimers (mostly p65/p50) are normally sequestered by binding to the inhibitor IκB and quickly activated by a stress that causes the phosphorylation and proteolysis of IκB (11–13). Being associated with the regulation of more than 150 effecter genes, NF-κB functions in an array of signaling pathways, including cell transformation and anti-apoptotic responses (14, 15). One of the most characterized features of NF-κB activation is to antagonize the signaling network of apoptotic response (16) and to increase cell resistance to radiation and chemotherapy (17). IR-treated HeLa cells demonstrate a transient NF-κB activation following IκB phosphorylation (18). In breast carcinoma MCF-7 cells (5) and Papillomavirus-transformed human keratinocytes (19), the basal and IR-induced NF-κB is paralleled with a reduced cell killing by IR. Interestingly, NF-κB is also associated with the mitochondrial stress resulting in the activation of calcineurin and mitochondrial antioxidant enzyme Mn-SOD (20, 21). Gene clustering analyses further demonstrate that NF-κB is responsible for at least a fraction of genes induced by IR, including Mn-SOD, indicating that the mitochondrial redox imbalance is important for cells to defend IR cytotoxicity (22). Blocking NF-κB inhibits the expression of Mn-SOD (22) as well as other IR-induced genes, which enhances IR-mediated cell death (19, 22). These findings illustrate a possibility that a group of stress-sensitive proteins are required for cell survival. The context of the IR-induced NF-κB and other signaling elements needs to be elucidated.
Accumulating results suggest that the group of mitogen-activated protein kinases (MAPKs), i.e. ERK, JNK, p38, functions to signal responses to IR (23–25) and other stress conditions (26–28). For instance, ERK is strongly induced by high doses of IR that can activate the membrane-associated tyrosine kinase (25, 29). ERK that is originally associated with mitotic response (30, 31) has been found to be a key mediator for tyrosine phosphorylation in growth factors and Ras activation (30–33). Therefore, ERK activation appears to be a necessary event in signaling cell proliferation and survival (26, 27, 34). In contrast, although JNK, a key member of MAPKs, induces NF-κB in anti-apoptotic responses (35), NF-κB-targeted genes are shown to inhibit JNK required for initiating tumor necrosis factor-α-induced apoptosis (8). Recent data further indicate that ERK-mediated anti-apoptotic response is dependent upon its cellular locations and interaction with NF-κB·IκB complexes (36). It remains unclear whether ERK and MAPKs are associated with NF-κB-mediated radioprotection.
GADD45β (growth arrest and DNA damage-inducible β, also named MyD118) is key member of nuclear proteins inducible by DNA-damaging stresses and has been shown to play an active role in cell cycle adjustment that affects cell survival (37). It has been long observed that IR-induced GADD45β expression (38) is linked to p53 pathways (39, 40). Experiments of antisense transfections find that GADD45β interacts with cyclin B1 complex during S and G2/M checkpoints following UV irradiation (41). GADD45β is also required in T cell receptor-induced responses and ERK, p38, and JNK activation are all substantially suppressed in GADD45β-deficient CD4(+) T cells (37), suggesting a tight link between GADD45β and MAPKs pathways. NF-κB is shown to induce Gadd45β promoter activity (42) and GADD45β induced by NF-κB can down-regulate the pro-apoptotic JNK (43, 44).
To characterize the signaling network of NF-κB activation that appears to play a key role in cell death or survival after high dose IR, we have examined the hypothesis that NF-κB, ERK, and GADD45β co-operate in radiation response. Inhibition of either factor by transfection of dominant negative mutant or pre-treatment with inhibitor compounds blocks the activity of other two components in a loop-like connection. Because antisense blocking gadd45β inhibited NF-κB and ERK but not other MAPKs, i.e. p38 and JNK, ERK, and gadd45β are likely to be specifically required for signaling NF-κB-mediated cell protection. The present study thus reveals a novel connection of NF-κB with activation of ERK and GADD45β in defending cell survival after IR insults.
EXPERIMENTAL PROCEDURES
Cell Culture and Exposure to Ionizing Radiation
MCF-7 cells were obtained from the American Type Culture Collection (passage number: 126) and maintained in Dulbecco’s minimum essential media (Cellgro, Herndon, VA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT), 1% L-glutamine, penicillin (100 units/ml), and streptomycin (100 μg/ml) in a humidified incubator (5% CO2) at 37 °C. Exponentially growing cells with 60–80% confluence were exposed to IR at room temperature with a Cs-137 mark I irradiator (dose rate, 436 cGy/min, J. L. Shepherd & Associates). Cells sheltered from IR source were used as the sham-IR control. After IR, cells were cultured at 37 °C incubator for further experiments.
Preparation of Nuclear Extracts and Gel Shift Analyses
Subsequent to IR exposure, cells were rinsed with phosphate-buffered saline containing 1 mM EDTA, collected by centrifugation, and resuspended in ice-cold hypotonic lysis buffer supplemented with protease inhibitors (10 mM Hepes, pH 7.9, 0.1 mM EDTA, 0.1 mM EGTA, 10 mM KCl, 0.3% Nonidet P-40, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 1 μg/ml pepstatin A). Following 15 min of incubation on ice, the cell lysates were vortexed and centrifuged at 15,000 × g for 1 min to obtain the nuclear pellet and the cytosolic protein-containing supernatant. The nuclear pellets were washed using the aforementioned buffer with the exclusion of Nonidet P-40. Then the nuclei were resuspended in the ice-cold solution (20 mM Hepes, pH 7.9, 420 mM NaCl, 1 mM EDTA, 0.1 mM EGTA, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 1 μg/ml pepstatin A). After 30 min of incubation on ice, the samples were centrifuged at 15,000 × g for 5 min, and the resulting supernatant was saved as nuclear extracts stored in the −80 °C freezer until use.
Gel shift analysis was performed with 3 μg of nuclear proteins incubated on ice for 10 min in a total 20 μl of DNA binding buffer (10 mM Tris-HCl, pH 7.5, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 5% glycerol, and 50 μg/ml polydeoxyinosinnicdeoxycytidylic acid strand DNA) and then incubated for 20 min at room temperature with 5 × 104 cpm 32P-labeled NF-κB oligonucleotides probe. The DNA·protein complexes were resolved on 10% native polyacrylamide gel electrophoresis and visualized in x-ray film. For supershift assays nuclear extracts were incubated with antibodies p65 (sc-7151) or p50 (sc-7178) at room temperature for 20 min prior to the addition of the radioactive-labeled DNA oligonucleotides.
Immunoblotting Analysis
Cellular extracts were fractionated using a 10% SDS-polyacrylamide gel and transferred by way of a semi-dry apparatus (Bio-Rad, Hercules, CA) to polyvinylidene difluoride membranes (Bio-Rad). Immunoblot analysis was carried out using antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) and visualized using the horseradish peroxidase-coupled mouse anti-rabbit immunoglobulins followed by the ECL Western blotting detection system (Amersham Biosciences).
Transient Co-transfection of NF-κB Reporters with Dominant Negative Mutant IκB or Dominant Mutant Erk
Transfection of NF-κB luciferase reporter with dominant negative mutant IκB-α vector (22) or with dominant negative Erk vector (45) were performed as previously detailed (22). Cells in 12-well plates were co-transfected with 0.3 μg of NF-κB and 0.2 μg of β-galactosidase reporters for 3 h, and luciferase activity was measured 24 h following exposure to sham or IR. Luciferase activity was measured using an illuminometer (Turner Designs, Sunnyvale, CA). An aliquot of the same cell lysate was used for measurement of β-galactosidase activity to normalize luciferase results.
Establishment of Stable Transfectants MCF-7/mIκB and MCF-7/DN-Erk
MCF-7 cells were stably transfected with dominant negative mutant mIκB or dominant negative Erk (DN-Erk) plasmids, and stable transfectants were obtained using Lipofectamine reagent (Invitrogen) as previously described (22). MCF-7 cells (5 × 106 in 100-mm cell culture dishes) were transfected with 15 μg of mIκB or DN-Erk plasmid, 2 μg of G418 marker DNA pcDNA3, and 40 μg of Lipofectamine in 6 ml of serum-reduced OPTI-EMEM (Invitrogen). pcDNA3 only was transfected into MCF-7 cells as a vector control. Cells were transfected with transfection solution for 72 h and recovered for 24 h in complete medium, trypsinized, and cultured in the selecting medium for 14–21 days. The selected mutant and vector control clones were maintained in Dulbecco’s modified Eagle’s medium, and all transfectants were cultured for at least two passages in G418-free medium before experiments.
Clonogenic Survival Assay
Prior to conducting clonogenic assays, the plating efficiency of the wild type MCF-7, mutant gene transfectants MCF-7/mIκB and MCF-7/DN-Erk, and vector control transfectants MCF-7/V were determined. Contingent upon these phycoerythrin findings, the number of cells seeded was calculated to give an identical number of clones for the controls. The number of cells seeded ranged from 1,000 to 200,000 according to the radiation doses 2–12 Gy. 14–18 days after irradiation, the plates were stained and colonies with more than 50 cells were counted and normalized to the plating efficiency of control and transfected cell lines.
Cell Proliferation Assay
Exponentially growing MCF-7 cells were digested with trypsin, and different cell numbers were plated into multiple well plates for 18–24 h. Cells were then treated with PD98059 or Gadd45β antisense oligonucleotides followed by an exposure to a single dose 5 Gy IR. Cell proliferation was measured by cell numbers calculated with hemocytometers or by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay (Promega, Madison, MI).
Protein Kinase Assays
Cell lysates were prepared 24 h after radiation with the lysis buffer provided in the Bio-Rad Cell Lysis Kit by following the user manual. Protein concentration was measured with a DC™ protein assay kit (Bio-Rad Laboratories), and protein concentration was adjusted using the cell lysis buffer. p-JNK, p-p38, and p-ERK1/2 were measured using the Bio-Plex™ phosphoprotein assay (Bio-Rad Laboratories). Briefly, the filter plate was rinsed with 100 μl of phosphoprotein wash buffer, followed by adding 50 μl of antibody-conjugated beads, 25 μl of each cell lysate, and 25 μl of phosphoprotein assay buffer. The plate was shaken overnight and incubated with biotinylated anti-phospho-p44/p42 MAPK (ERK1/ERK2; Santa Cruz Biotechnology) antibody, incubated with 50 μl of streptavidin-labeled phycoerythrin, and read on the Bio-Plex™ system.
PD98059 and IR Treatment
PD98059 (Sigma) was dissolved in Me2SO and MCF-7 cells were incubated with control Me2SO or Me2SO with a range of concentrations of PD98059 for 2 h. Treatments were terminated by replacing with complete medium without PD98059, and cell pellets were collected 24 h after exposure to a single dose of IR. Proteins were purified from control and PD98059/IR-treated cells, and ERK kinase activity was measured by using the Bio-Plex™ phospho-protein assay (Bio-Rad) and Western blotting. In a parallel experiment, cells were plated into multiple-well plates, and cell growth was determined after PD98059 and IR treatments.
Antisense Preparation and Treatments
Phosphorothioate-modified antisense oligonucleotides of Gadd45β (5′-GAAGTTGCGGAAACCAACGGT-3′) were synthesized with the gene sequence obtained from GenBank™ (AF533019; 1836–1856). MCF-7 cells grown in a T-75 tissue culture flask were transfected with a specific range of antisense concentrations and Lipofectamine reagent (Invitrogen). Cells were exposed to antisense oligonucleotides for 24 h and then replaced with the complete medium and treated with or without IR. At different time intervals cell pellets were collected for analysis of MAPK activity. In a parallel experiment, cells were plated into multiple well plates, and cell proliferation was determined after antisense and IR treatments.
Imaging of ERK/p65 Interactions in Living Cells
Plasmids with full-length sequence encoding human NF-κB p65, ERK1, and ERK2 were fused to N- and C-terminal fragments of enhanced yellow fluorescent protein. The EEK1, ERK2, p65 coding regions were connected by linker sequences as described (46). These expression vectors encoding p65-YC156, ERK1-YN173, and ERK2-YN173 were kind gifts from Dr. C. D. Hu, and co-transfection was performed as previously described (22). Briefly, MCF-7 cells were maintained in complete Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (HyClone, Logan, UT), and cells in 24-well plates were co-transfected with the expression vectors indicated in each experiment (a total of 0.4 μg of DNA in each well) using Lipofectamine 2000 (Invitrogen). The fluorescence emissions were observed in living cells 14–16 h after transfection using a Nikon TE300 inverted fluorescence microscope with a cooled charge-coupled device camera.
RESULTS
IR Activates NF-κB and ERK but Not p38 or JNK
Accumulating results in literature (47, 48) and our previous reports (5, 22) have demonstrated that ionizing radiation enhances NF-κB DNA binding and transcriptional activities that are associated with reduced cell radiosensitivity. To get insight of the network of NF-κB-mediated radioprotection, the activity of a group of MAPKs was measured in irradiated breast carcinoma MCF-7 cells. Agreeing with NF-κB activation (Fig. 1A), a single exposure to 5 Gy IR significantly induced the kinase activity of p-ERK (75% increase compared with the sham-IR controls; Fig. 1B). In contrast, no difference was detected in the basal and IR-induced kinase activities of p-p38 and p-JNK. Thus, ERK kinase activity appears to be specifically paralleled with IR-induced NF-κB activation.
Fig. 1. NF-κB and ERK but not p38 or JNK was activated by IR.
A, NF-κB activation by 5 Gy IR. MCF-7 cells were co-transfected with NF-κB luciferase and β-galactosidase reporters for 6 h, and luciferase activity was measured 24 h after exposure to sham-IR (−) or a single dose of 5 Gy IR (+). Luciferase reporter activity was normalized to β-galactosidase (mean ± S.E., n = 3). B, p-ERK but not p-JNK or p-p38 was induced by IR. MCF-7 cells treated with sham-IR (0 Gy) and IR (5 Gy) were collected 24 h after irradiation. p-ERK, p-JNK, and p-p38 antibodies were incubated with 20 μg of whole cell lysates, and the immunocomplex was labeled with streptavidin-phycoerythrin and detected by the Bio-Plex (Bio-Rad laboratory) protein array system as described under “Experimental Procedures” (mean ± S.E., n = 3; **, p < 0.01).
Overexpression of mIκB Blocked IR-induced ERK Activity and GADD45β Expression
To test the hypothesis that IR-induced NF-κB and ERK activation is mutually dependent, IR-induced ERK activity was measured with the inhibition of NF-κB. The dose dependence of NF-κB inhibition by overexpression of the dominant negative mutant IκB is shown in Fig. 2A. The mutant form of IκBα that has been shown to inhibit NF-κB activity in MCF-7 cells (22) dose-dependently inhibited the basal (MCF-7, Fig. 2A, left panel) and IR-induced (MCF-7 plus 5Gy, Fig. 2A, right panel) NF-κB-controlled luciferase activities. IR-mediated NF-κB DNA binding activity was assayed by gel shift analysis in stable transfectants (MCF-7/mIκB, with vector control transfectants MCF-7/V as a control). Compared with the wild type MCF-7 and MCF-7/V cells, a striking reduction in IR-induced NF-κB DNA binding was induced in MCF-7/mIκB transfectants (Fig. 2B). Both NF-κB subunits p65 and p50 were found to be present in the IR-induced NF-κB·DNA binding complexes (Fig. 2C), suggesting that p65 and p50 are the major components of IR-induced NF-κB complexes. ERK activity measured by substrate Elk-1 phosphorylation (Fig. 2D) and GADD45β measured by Western blot (Fig. 2E) were significantly enhanced by IR in the wild type (MCF-7) and vector control (MCF-7/V) cells. In contrast, ERK activity and GADD45β expression were not enhanced by IR in MCF-7 cells overexpressing mIκB (MCF-7/mIκB).
Fig. 2. Overexpression of mIκB inhibited NF-κB activity and Elk-1 phosphorylation.
A, basal and radiation-induced NF-κB activity was dose-dependently inhibited by transfection of dominate negative mutant IκB (mIκB). NF-κB luciferase reporters were co-transfected with the indicated amounts of mIκB plasmids into MCF-7 cells, and luciferase activity was measured 24 h after treatment with or without 5 Gy IR. B, IR-induced NF-κB DNA binding activity was inhibited in stable transfectants of mIκB. Wild type MFC-7 and stable transfectants of mIκB (MCF-7/mIκB) or empty vector (MCF-7/V) were treated with or without 5 Gy IR. NF-κB DNA binding activity was assayed with DNA retardation gel analysis. MCF-7/V+cold oligonucleotide was included as a negative control with the presence of unlabeled NF-κB nucleotides. C, NF-κB subunits p65 and p50 were detected in the NF-κB·DNA complexes induced by IR. Nuclear extracts from 5 Gy IR-treated MCF-7 cells were assayed with DNA retardation gel with or without antibody to NF-κB subunit p65 and p50 as indicated. NF-κB and the antibody super-shifted bands are indicated with arrowheads. D, expression of mIκB inhibited IR-induced ERK kinase activity. Wild type MFC-7, empty vector control transfectants (MCF-7/V) and mIκB MCF-7 transfectants (MCF-7/mIκB) were irradiated with 0 (−) or 5 Gy (+) IR. Phos-pho-Elk-1 (sc-355; Santa Cruz Biotechnology) was detected by Western blotting as a downstream ERK kinase substrate. E, expression of mIκB inhibited IR-induced GADD45β. Wild type MFC-7, empty vector control transfectants (MCF-7/V), and mI3B MCF-7 transfectants (MCF-7/mIκB) were irradiated with 0 (−) or 5 Gy (+) IR. GADD45β was detected by Western blot (antibody sc-8775; Santa Cruz Biotechnology).
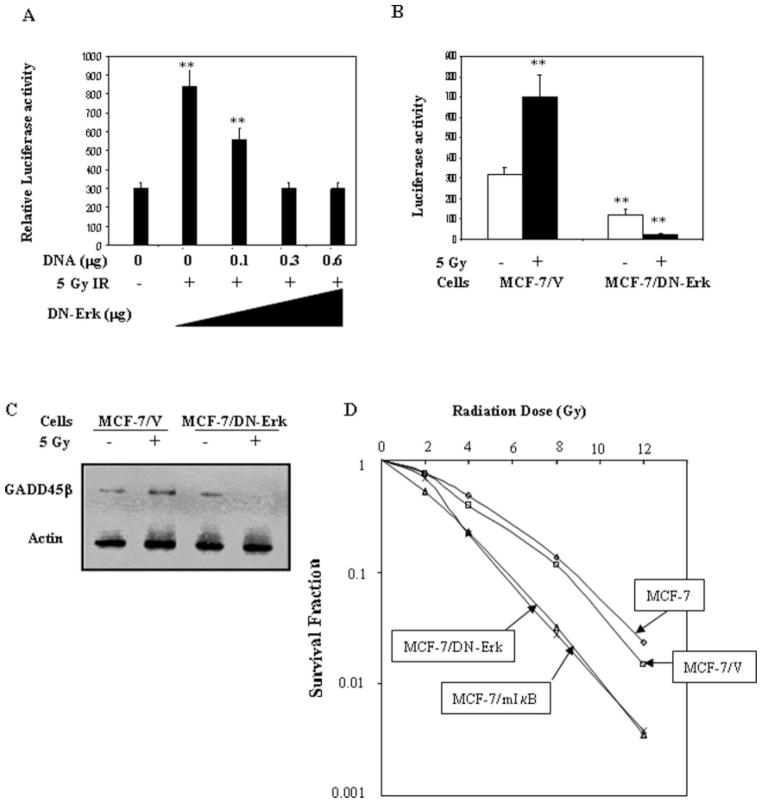
Overexpression of Dominant Negative ERK (DN-Erk) Blocked IR-induced NF-κB Activity and GADD45β Expression and Enhanced Cell Radiosensitivity
To determine if IR-induced ERK is associated with NF-κB activation, we analyzed the dose-dependence of NF-κB inhibition by overexpression of the dominant negative mutant Erk gene that, when overexpressed, inhibits ERK kinase activity (45). IR-induced NF-κB luciferase activity was found to be dose-dependently down-regulated by DN-Erk DNA transfection (Fig. 3A). Transfection of 0.3 and 0.6 μg of DN-Erk DNA reduced NF-κB transcriptional activity to the basal level of wild type MCF-7 cells without IR exposure (Fig. 3A). In addition, DN-Erk stable transfectants (MCF-7/DN-Erk) demonstrated a predominant reduction in basal and IR-induced NF-κB activation (Fig. 3B). Similar to NF-κB inhibition, GADD45β expression was totally blocked in MCF-7/DN-Erk transfectants (Fig. 3C), suggesting that both NF-κB and GADD45β can be downstream elements of IR-induced ERK activation. Clonogenic survival of MCF-7/DN-Erk and MCF-7/mIκB transfectants was found to be significantly decreased after the exposure to an array of IR doses (0–12 Gy; Fig. 3D). Compared to the wild type MCF-7 cells the dose-modifying factors at 10% isosurvival level were 1.43 and 1.39, respectively, for MCF-7/DN-Erk and MCF-7/mIκB cells. These results strongly indicate that IR-induced ERK activity is able to induce NF-κB and GADD45β and increase cell clonogenic survival after IR.
Fig. 3. Expression of dominant negative Erk, DN-Erk, or mIκB in MCF-7 cells blocked NF-κB activity and increased cell sensitivity to radiation.
A, NF-κB activity was dose-dependently inhibited by transfection of DN-Erk. NF-κB luciferase reporters were co-transfected with the indicated amounts of DN-Erk into MCF-7 cells, and luciferase activity was measured 24 h after treatment with or without 5 Gy IR (mean ± S.E., n = 3, p < 0.01) compared with the control of MCF-7 (without IR). B, basal and IR-induced NF-κB activities were inhibited in MCF-7/DN-Erk transfectants. NF-κB luciferase reporters were transfected into parental MCF-7 and DN-Erk transfectants (MCF-7/DNErk), and luciferase activity was measured 24 h after exposure to 5 Gy IR. C, IR-induced GADD45β was inhibited in MCF-7/DN-Erk transfectants. GADD45β was measured with Western blotting in MCF-7/V and MCF-7/DN-Erk transfectants 24 h after exposure to 5 Gy IR. D, radiosensitivity was increased in MCF-7/DN-Erk and MCF-7/mIκB transfectants. Wild type MCF-7, vector control MCF-7/V, MCF-7/mIκB, and MCF-7/DN-Erk transfectants were irradiated with a range of IR doses (0–12 Gy), and clonogenic survival was determined 18 days after IR. Colonies with more than 50 cells were counted, and survival fractions of each cell line were normalized to the plating efficiency.
PD98059 Inhibited IR-induced ERK and NF-κB Activation and Increased Cell Radiosensitivity
PD98059 has been shown to specifically inhibit MAPK activity (49, 50) leading to IR-induced apoptosis with a decreased clonogenic survival in MCF-7 cells (51). To investigate if PD98059-mediated radio-sensitivity is caused by the inhibition of both NF-κB and pERK, Fig. 4A shows that IR-induced p-ERK levels (p-MAPK, p44/42) were dose-dependently inhibited by the pre-treatment of cells to PD98059. IR-induced NF-κB activity measured by luciferase reporter was inhibited with a similar dose-dependent pattern (Fig. 4B). Radiosensitivity measured by cell proliferation showed that cells pre-exposed to PD98059 increased IR-induced growth inhibition by 24–40% during the time 48–72 h after radiation. Without IR, PD98059 did not induce significant growth inhibition (Fig. 4C).
Fig. 4. PD98059 inhibited p-ERK and NF-κB activity and increased MCF-7 cell radiosensitivity.
A, ERK inhibitor PD98059 dose-dependently reduced IR-induced p-ERK. MCF-7 cells treated with the indicated concentrations of PD98059 for 2 h before exposure to 5 Gy IR and p-ERK activity was determined by Western blotting with antibody of p-ERK (sc-7383; sc-93 for ERK). B, PD98059 dose-dependently inhibited NF-κB luciferase reporter activity. MCF-7 cells transfected with NF-κB luciferase reporters were exposed to the indicated concentrations of PD98059 for 2 h, and luciferase activity was determined 24 h after IR. C, PD98059-increased IR-induced cytotoxicity. MCF-7 cells cultured in 6-well plates were treated with PD98059 by the indicated concentrations of PD98059 for 2 h before 5 Gy IR. Cell proliferation was determined by cell numbers counted 24, 48, and 72 h after radiation (mean ± S.E., n = 5; **, p < 0.01).
Antisense-blocking Gadd45β Inhibited ERK but Not p38 or JNK
GADD45 proteins have been shown to play important roles in maintaining cell genomic stability and regulating the cell cycle (52, 53). The GADD45β-induced anti-apoptotic pathway is believed to occur via NF-κB activation (43). To test if GADD45β is required for IR induction of ERK and NF-κB, p-ERK, p-p38, and p-JNK activities were measured in MCF-7 cells transfected with the antisense of Gadd45β (antisense sequences are described under “Experimental Procedures”). Fig. 5A shows that pre-treatment with antisense Gadd45β significantly blocked IR-induced ERK activation without affecting the activity of p38 and JNK. These results are consistent with the observation shown in Fig. 1 indicating that ERK is specifically induced in MCF-7 cells by IR. Analyses with Western blotting shown in Fig. 5 (B–D) demonstrated that both basal and IR-induced p-ERK were inhibited by pre-treatment with Gadd45β antisense compared to cells without antisense transfection. No detectable difference was induced in p-p38 and p-JNK activities by antisense pre-treatment with or without following IR exposure.
Fig. 5. Transfection of antisense Gadd45β reduced IR-induced p-ERK but not p-p38 and p-JNK.
A, MCF-7 cells were incubated with antisense (AS) Gadd45β oligonucleotides 0.2 μM for 24 h, and p-ERK, p-JNK, and p-p38 kinase activities were measured 24 h after 5 Gy IR with a Bio-Plex kit (**, p < 0.01, n = 3). B–D, antisense Gadd45β inhibited p-ERK. 40 μg of whole cell lysate was analyzed by Western blotting with antibodies of p-ERK (sc-7383; sc-93 for ERK as control; B), p-p38 (sc-7975-R, sc-728 for p-38 as control; C), and p-JNK (sc-6254; sc-4061 for JNK1 as control; D); each blot was re-hybridized with the control antibody; results are representative of three blots.
Antisense-blocking GADD45β Inhibited IR-induced NF-κB Activation Causing Decreased Cell Survival
To determine if GADD45β is required for enhancing cell survival after IR insults, the inhibited protein levels of GADD45β was verified in irradiated MCF-7 cells transfected with antisense Gadd45β (Fig. 6A). Equivalent with GADD45β inhibition, IR-induced NF-κB luciferase transcription was totally abolished by anti-sense transfection (Fig. 6B). Without antisense transfection, MCF-7 cells showed ~4-fold NF-κB luciferase activity induced by IR. Cell growth showed that pre-treatment with Gadd45β antisense completely eliminated cell proliferation after IR in contrast to the cell growth treated with 2 Gy IR alone (Fig. 6C). Thus, GADD45β expression levels appear to be a sensitive factor in mediating IR-induced ERK and NF-κB activation and cell survival.
Fig. 6. Antisense blocking GADD45β inhibited NF-κB causing increased radiosensitivity.
A, MCF-7 cells were incubated with 5 ml of transfection mixture containing 0.2 μM antisense (AS) Gadd45β oligonucleotides for 6 h and further transfected with 0.1 μM antisense Gadd45β oligonucleotides for 24 h before IR with 5 Gy. Cell lysate was prepared 24 h after IR, and Western blotting was performed to confirm GADD45β inhibition. B, antisense Gadd45β inhibited IR-induced NF-κB. MCF-7 cells transfected with NF-κB luciferase reporters were treated with or without antisense transfection and exposed to IR. Lu-ciferase activity was determined 24 h after IR (**, p < 0.01, n = 3). C, MCF-7 cells in multiple-well plates were treated with (+) or without (−) antisense Gadd45β oligonucleotides for 24 h followed by 2 Gy IR. Cell proliferation was calculated at different time intervals after IR (data are the combined mean values of three experiments).
Visualization of Interaction between ERK and NF-κB p65
bimolecular fluorescence complementation analysis (46) provides the image of protein interactions in living cells. To confirm the direct interactions of NF-κB p65 and ERK in MCF-7 cells, expression vectors encoding p65-YC156, Erk1-YN173, and Erk2-YN173 were co-transfected, and protein-protein interaction was analyzed under fluorescence microscopy (46). Fluorescence was observed within 14–16 h after co-transfection of p65-YC165 with EEK1-YN173 or ERK2-TN173 (Fig. 7, C and E). In the control experiments, expression of p65-YC156, ERK1-YN173, or ERK2-YN173 alone did not produce significant fluorescence (Fig. 7, A, B, and D). Protein interactions appeared to occur mainly in the nucleus of MCF-7 cells (Fig. 7, C and E), indicating a possibility that NF-κB and ERK form complexes to regulate specific genes to increase cell survival. Dynamic analysis of IR-induced protein interactions between NF-κB subunits, ERK, and GADD45β and nuclear transportation is underway.
Fig. 7. Visualization of p65/ERK interactions in living MCF-7 cells.

Using bimolecular fluorescence complementation analysis, fluorescence images of MCF-7 cells expressing the fusion proteins indicated in each panel were acquired 14–16 h after co-transfection. A, p65-YC156 alone; B, Erk1-YN173 alone; C, p65-YC156 with Erk1-YN173; D, Erk2-YN173 alone; E, p65-YC156 with Erk2-YN173.
IR-Induced G2/M Delay Was Reduced by Blocking ERK/NF-κB/GADD45β
We then tested the hypothesis that the increased radiosensitivity, upon inhibiting the ERK/NF-κB/GADD45β pathway is caused by alterations in cell cycle adjustment. Cell cycle distribution of wild type MCF-7 cells pre-exposed to the corresponding inhibitors was calculated after exposure to a single dose of 5 Gy (Table I). In contrast to a remarkable G2/M delay (3.32-fold) induced in wild type MCF-7 cells by IR, none of MCF-7/DN-Erk transfectants or MCF-7 cells pre-treated with PD98059 and Gadd45β antisense showed a significant delay in G2/M border. The basal cell accumulation in G2/M border without IR insult was slightly increased in cells with the inhibition of the ERK/NF-κB/GADD45β pathway suggesting an opposite influence of blocking ERK/NF-κB/GADD45β pathway versus IR. In addition, compared with the wild type MCF-7 cells, no significant alteration in G0/G1 and S phases was induced by inhibition of ERK/NF-κB/GADD45β. These results strongly imply that a shortage in IR-induced G2/M delay causes the radiosensitization induced by inhibition of ERK/NF-κB/GADD45β pathways.
Table I. Cell cycle distribution of MCF-7/DN-Erk and MCF-7/PD98039 cells.
The control MCF-7, MCF-7/DN-Erk, and MCF-7 cells treated with PD98039 or antisense GADD45β were treated with (+) or without (−) a single dose of 5-Gy radiation. Cells were collected 48 h after radiation and fixed in 85% ethanol and stained with propidium iodide (PI). PI-stained nuclear DNA content, indicating the distribution of cells in G0/G1, S, and G2, was determined with FACScan plus software (BD Biosciences), and the results of 1 × 104 counted cells were analyzed using Multicycle software.
| Cell line | 5 Gy | G0/G1 | S | G2/M |
|---|---|---|---|---|
| MCF-7 | − | 69.15 | 27.76 | 3.09a |
| MCF-7 | + | 70.79 | 18.95 | 10.26a |
| MCF-7/DN-Erk | − | 65.07 | 25.26 | 9.67 |
| MCF-7/DN-Erk | + | 65.24 | 20.53 | 14.23 |
| MCF-7/PD98039 | − | 69.42 | 25.50 | 5.94 |
| MCF-7/PD98039 | + | 71.19 | 23.03 | 7.59 |
| MCF-7/anti-GADD | − | 67.22 | 23.50 | 8.77 |
| MCF-7/anti-GADD | + | 69.39 | 19.44 | 11.11 |
p < 0.01 compared to sham-IR control cells.
DISCUSSION
The present study provides evidence suggesting a novel connection of ERK, NF-κB, and GADD45β in enhancing cell survival after IR exposure. These factors were found to be co-activated and mutually dependent in radiation responses leading to a protective cell phenotype. Inhibition of ERK by transfection of dominant negative Erk (MCF-7/DN-Erk) or treated with ERK inhibitor PD98059 inhibited NF-κB activity and increased cell radiosensitivity. Inhibition of NF-κB with transfection of dominant negative IκB (MCF-7/mIκB) showed a similar pattern of inhibition and radiosensitization. Additionally, antisense-blocking GADD45β inhibited NF-κB and ERK but not p38 or JNK, causing enhanced IR cytotoxicity. These results illustrate a loop-like connection in IR-induced transcription factors and cell cycle elements required for increasing cell survival.
Identifying the signaling network associated with cell death or survival after IR stress is an essential issue in the research of radioprotection and anticancer radiotherapy. The present study suggests that two key stress-sensitive signaling elements ERK and GADD45β are closely related to NF-κB activity, forming a loop-like connection to increase cell survival. This is evidenced by the fact that blocking each factor causes the inhibition of the other two and significantly changes cell radiosensitivity. Although the exact mechanisms underlying the formation of the protective network remain to be elucidated, accumulating evidence strongly implicates that a large group of gene regulators and gene products are required for signaling cell death or survival (21, 54–58). The challenge therefore is to identify the specific signaling pattern that plays a decisive role in increasing cell survival. Among all the stress-responsive transcription factors, NF-κB has been tightly linked with cell resistance against IR-mediated cytotoxicity and thus have been tested for a possible application in anticancer therapy (22, 58–60). Our prior data showed that NF-κB is causally related to radiosensitivity of human cancer or virus-transformed cells (19, 24, 56). However, inhibition of NF-κB does not always increase cell radiosensitivity. For instance, inhibition of NF-κB by overexpressing IκB induces little effect in the radiosensitivity of PC3 prostate cancer cells, nor in HD-MyZ Hodgkin’s lymphoma cells (61). This apparent paradox strongly implicates that IR-induced signaling elements interact with each other to form a network that provides a by-pass to keep cell survival. The loop-like connection of ERK/NF-κB/GADD45β found in the present study illustrates a typical interaction of such network.
An important feature in cell response to ionizing radiation is the generation of redox imbalance. Most signaling proteins are known to be sensitive to redox alterations (62, 63), and several redox-sensitive transcription factors have been shown to induce the mitochondrial antioxidant enzyme manganese-containing superoxide dismutase (Mn-SOD) that is believed to play a central role in redox regulation (22, 54, 64). NF-κB is actively involved in regulation of Mn-SOD expression (20, 65). Notably, both pro- and anti-apoptotic pathways can be activated by NF-κB via expression of Mn-SOD (66). We have previously observed that IR-induced mitochondrial Mn-SOD is via NF-κB activation and blocking NF-κB or Mn-SOD expression down-regulates a group of IR-induced genes and increases cell radiosensitivity (22). Shonai et al. (67) have found that ERK-mediated anti-apoptotic signals are activated through inhibition of mitochondrial related caspase-8 activity. Especially, ERK can selectively inhibit IR-induced loss of mitochondrial membrane potential and subsequent cell death. Therefore, IR-induced mitochondrial signaling pathways require ERK activation. The bimolecular fluorescence complementation analysis (46) is applied in the present study to detect the interaction between NF-κB subunits and ERK. Interaction of p65 and ERK is detected in living MCF-7 cells after co-transfection (Fig. 7, C and E). These results and data reported by others (68) indicate but could not prove that activation of ERK/NF-κB/GADD45β is involved in regulation of Mn-SOD and other specific genes that in turn affect mitochondria-mediated cell death and overall cell survival.
GADD45β in the loop of ERK/NF-κB/GADD45β may provide a specific function for signaling radioprotection. GADD45β, a key nuclear protein responsive to DNA damage, plays an active role in cell cycle adjustment (37). GADD45 proteins interact with the complex of Cdk1 and CyclinB1, both of which are required for IR-induced cell cycle regulation and radioresistance (5, 69). IR strongly induces GADD45 expression with the dependence of p53 activation (39, 70, 71). IR with a dose as low as 0.5 Gy induces a rapid induction of GADD45β (70). NF-κB subunit p65 is shown to be sufficient to activate GADD45β expression (42). Induction of Gadd45β gene expression by p65 is shown to be dependent on three κB elements of the Gadd45β promoter region (42). Each of these sites is able to bind to NF-κB complexes and required for optimal promoter transactivation. Antisense study suggests that all GADD45 proteins are likely to cooperate in the activation of S and G2/M check-points following exposure to UV irradiation (41). Our present data support that IR-mediated regulation of Gadd45β is due to NF-κB activation and GADD45β further enhances NF-κB and ERK activation. A connection of GADD45β with activation of ERK and NF-κB may be a necessary step to efficiently adjust cell cycle distribution altered by IR stress. Because inhibition of ERK, NF-κB, or GADD45β significantly reduces G2/M delay that is necessary for repairing damaged DNA as to increase cell survival (Table I), the result of forming the loop of ERK/NF-κB/GADD45β appears to enhance the efficiency of repairing the damaged DNA.
Papa et al. (44) have analyzed the mechanisms by which GADD45β inhibits the pathways initiated by activation of JNK. In this case, GADD45β is shown to bind to MKK7/JNKK2, a specific activator of JNK, and block the catalytic activity of MKK7/JNKK2 (44). Interestingly, GADD45β is able to inhibit tumor necrosis factor-α-induced cytotoxicity. Disrupting GADD45β/MKK7 interaction also blocks GADD45β and NF-κB and suppresses tumor necrosis factor-α-induced cytotoxicity. Both results suggest that GADD45β interacts with JNK pathways. However, in our present study, a single dose of IR exposure induced little activity of JNK and p38 (Fig. 1) and antisense-blocking GADD45β did not induce any changes in p38 and JNK activities (Fig. 5). These findings establish a basis for the hypothesis that the connection of NF-κB with GADD45β and ERK is a unique network for increasing cell survival under IR stress. JNK and p38 appear not to be directly linked with NF-κB-mediated radioresistance.
PD98059 has been shown to inhibit IR-induced ERK (72). Notably, the results reported by Suzuki et al. (72) have demonstrated that IR of a very low dose range, i.e. between 2 and 5 cGy, is able to stimulate the proliferation of normal human diploid cells and tumor cells. Irradiation with less than 1 Gy induces the phosphorylation of ERK, which is decreased if the radiation dose is reduced to 0.5 Gy (72), providing solid evidence that ERK is very sensitive to IR-mediated cytotoxicity. The activated ERK is shown to augment the phosphorylation of Elk-1 protein. As shown in Fig. 2, our present study indicates that Elk-1 phosphorylation is induced by IR and inhibited by overexpression of mIκB. The phosphorylation of the ERK is also reduced by pre-treatment with PD98059 (Fig. 4), which inhibits ERK phosphorylation inducible under the stress of IR with 2 cGy or 6 Gy of x-rays (72). In addition, overexpression of ERK in NCI-H1299 human lung carcinoma cells has demonstrated an enhanced proliferation; antisense-blocking ERK abrogates IR-induced protective responses (72). Overall, ERK phosphorylation appears to be an essential step in signaling cell survival under the stress of IR, which obviously requires co-activation of NF-κB and GADD45β.
In summary, the present study demonstrates that two key stress signaling elements, ERK and GADD45β, are co-activated with NF-κB activation in MCF-7 cells exposed to a single dose of ionizing radiation. Blocking NF-κB by overexpressing dominant negative mutant IκB inhibits ERK activation and decreases cell survival. Inhibition of ERK by overexpression of dominant negative ERK blocks NF-κB activation and GADD45β expression. Interaction between p65 and ERK proteins was visualized in the nuclei of living MCF-7 cells. Anti-sense-blocking GADD45β inhibits NF-κB and ERK but not p38 and JNK. These results suggest that NF-κB, ERK, and GADD45β are able to coordinate to increase cell survival after the lethal damage induced by ionizing radiation.
Acknowledgments
We thank L. M. Lee at Laboratory of Molecular Technology, NCI, National Institutes of Health for the assistance of designing Gadd45β antisense oligonucleotides and Dr. C. D. Hu, at Purdue University School of Pharmacy, for the kind gifts of p65-YC156, Erk1-YN173, and Erk2-YN173 plasmids.
Footnotes
This work was supported in part by National Institutes of Health RO1 Grant CA101990 and by the Office of Science, U. S. Department of Energy Grant DE-FG02-05ER63945 (to J. J. L.).
The abbreviations used are: IR, ionizing radiation; ERK, extracellular signal-regulated kinase; FBS, fetal bovine serum; FIR, fractionated ionizing radiation; JNK, c-Jun N-terminal kinase; NF-κB, nuclear factor-κB; ROI, reactive oxygen intermediates; MAPK, mitogen-activated protein kinase; SOD, superoxide dismutase; DN-Erk, dominant negative Erk; GADD45β, growth arrest and DNA damage-inducible β; mIκB, mutant IκB.
References
- 1.Hartwell LH, Kastan MB. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 2.Fornace AJ, Jr, Amundson SA, Bittner M, Myers TG, Meltzer P, Weinsten JN, Trent J. Gene Expr. 1999;7:387–400. [PMC free article] [PubMed] [Google Scholar]
- 3.Bebien M, Salinas S, Becamel C, Richard V, Linares L, Hipskind RA. Oncogene. 2003;22:1836–1847. doi: 10.1038/sj.onc.1206334. [DOI] [PubMed] [Google Scholar]
- 4.Amundson SA, Bittner M, Chen Y, Trent J, Meltzer P, Fornace AJ., Jr Oncogene. 1999;18:3666–3672. doi: 10.1038/sj.onc.1202676. [DOI] [PubMed] [Google Scholar]
- 5.Li Z, Xia L, Lee ML, Khaletskiy A, Wang J, Wong JYC, Li JJ. Radiat Res. 2001;155:543–553. doi: 10.1667/0033-7587(2001)155[0543:egaimh]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 6.Sreekumar A, Nyati MK, Varambally S, Barrette TR, Ghosh D, Lawrence TS, Chinnaiyan AM. Cancer Res. 2001;61:7585–7593. [PubMed] [Google Scholar]
- 7.Amundson SA, Bittner M, Meltzer P, Trent J, Fornace AJ., Jr Radiat Res. 2001;156:657–661. doi: 10.1667/0033-7587(2001)156[0657:iogeaa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 8.Tang G, Minemoto Y, Dibling B, Purcell NH, Li Z, Karin M, Lin A. Nature. 2001;414:313–317. doi: 10.1038/35104568. [DOI] [PubMed] [Google Scholar]
- 9.Kataoka Y, Murley JS, Khodarev NN, Weichselbaum RR, Grdina DJ. Int J Radiat Oncol Biol Phys. 2002;53:180–189. doi: 10.1016/s0360-3016(01)02820-6. [DOI] [PubMed] [Google Scholar]
- 10.Wang T, Zhang X, Li JJ. Int Immunopharmacol. 2002;2:1509–1520. doi: 10.1016/s1567-5769(02)00058-9. [DOI] [PubMed] [Google Scholar]
- 11.Granville DJ, Carthy CM, Jiang H, Levy JG, McManus BM, Matroule JY, Piette J, Hunt DW. Blood. 2000;95:256–262. [PubMed] [Google Scholar]
- 12.Ravi R, Bedi GC, Engstrom LW, Zeng Q, Mookerjee B, Gelinas C, Fuchs EJ, Bedi A. Nat Cell Biol. 2001;3:409–416. doi: 10.1038/35070096. [DOI] [PubMed] [Google Scholar]
- 13.Li Q, Verma IM. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 14.Barkett M, Gilmore TD. Oncogene. 1999;18:6910–6924. doi: 10.1038/sj.onc.1203238. [DOI] [PubMed] [Google Scholar]
- 15.Romashkova JA, Makarov SS. Nature. 1999;401:86–90. doi: 10.1038/43474. [DOI] [PubMed] [Google Scholar]
- 16.Wang CY, Mayo MW, Baldwin AS., Jr Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 17.Wang CY, Cusack JC, Jr, Liu R, Baldwin AS., Jr Nat Med. 1999;5:412–417. doi: 10.1038/7410. [DOI] [PubMed] [Google Scholar]
- 18.Curry HA, Clemens RA, Shah S, Bradbury CM, Botero A, Goswami P, Gius D. J Biol Chem. 1999;274:23061–23067. doi: 10.1074/jbc.274.33.23061. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, Shen B, Xia L, Khaletzkiy A, Chu D, Wong JY, Li JJ. Cancer Res. 2002;62:1213–1221. [PubMed] [Google Scholar]
- 20.Xu Y, Kiningham KK, Devalaraja MN, Yeh CC, Majima H, Kasarskis EJ, StClair DK. DNA Cell Biol. 1999;18:709–722. doi: 10.1089/104454999314999. [DOI] [PubMed] [Google Scholar]
- 21.Murley JS, Kataoka Y, Hallahan DE, Roberts JC, Grdina DJ. Free Radic Biol Med. 2001;30:1426–1439. doi: 10.1016/s0891-5849(01)00554-8. [DOI] [PubMed] [Google Scholar]
- 22.Guo G, Yan-Sanders Y, Lyn-Cook BD, Wang T, Tamae D, Ogi J, Khaletskiy A, Li Z, Weydert C, Longmate JA, Huang TT, Spitz DR, Oberley LW, Li JJ. Mol Cell Biol. 2003;23:2362–2378. doi: 10.1128/MCB.23.7.2362-2378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hara T, Namba H, Yang TT, Nagayama Y, Fukata S, Kuma K, Ishikawa N, Ito K, Yamashita S. Biochem Biophys Res Commun. 1998;244:41–44. doi: 10.1006/bbrc.1998.8210. [DOI] [PubMed] [Google Scholar]
- 24.Bradbury CM, Markovina S, Wei SJ, Rene LM, Zoberi I, Horikoshi N, Gius D. Cancer Res. 2001;61:7689–7696. [PubMed] [Google Scholar]
- 25.Dent P, Yacoub A, Fisher PB, Hagan MP, Grant S. Oncogene. 2003;22:5885–5896. doi: 10.1038/sj.onc.1206701. [DOI] [PubMed] [Google Scholar]
- 26.Meier KE, Gause KC, Wisehart-Johnson AE, Gore AC, Finley EL, Jones LG, Bradshaw CD, McNair AF, Ella KM. Cell Signal. 1998;10:415–426. doi: 10.1016/s0898-6568(97)00140-x. [DOI] [PubMed] [Google Scholar]
- 27.Kurada P, White K. Cell. 1998;95:319–329. doi: 10.1016/s0092-8674(00)81764-x. [DOI] [PubMed] [Google Scholar]
- 28.Limoli CL, Kaplan MI, Giedzinski E, Morgan WF. Free Radic Biol Med. 2001;31:10–19. doi: 10.1016/s0891-5849(01)00542-1. [DOI] [PubMed] [Google Scholar]
- 29.Kasid U, Suy S, Dent P, Ray S, Whiteside TL, Sturgill TW. Nature. 1996;382:813–816. doi: 10.1038/382813a0. [DOI] [PubMed] [Google Scholar]
- 30.Cook SJ, McCormick F. Science. 1993;262:1069–1072. doi: 10.1126/science.7694367. [DOI] [PubMed] [Google Scholar]
- 31.Macdonald SG, Crews CM, Wu L, Driller J, Clark R, Erikson RL, McCormick F. Mol Cell Biol. 1993;13:6615–6620. doi: 10.1128/mcb.13.11.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moodie SA, Paris M, Villafranca E, Kirshmeier P, Willumsen BM, Wolfman A. Oncogene. 1995;11:447–454. [PubMed] [Google Scholar]
- 33.Anders M, Christian C, McMahon M, McCormick F, Korn WM. Cancer Res. 2003;63:2088–2095. [PubMed] [Google Scholar]
- 34.Kolch W. Biochem J. 2000;351:289–305. [PMC free article] [PubMed] [Google Scholar]
- 35.Javelaud D, Besancon F. Oncogene. 2001;20:4365–4372. doi: 10.1038/sj.onc.1204570. [DOI] [PubMed] [Google Scholar]
- 36.Ajenjo N, Canon E, Sanchez-Perez I, Matallanas D, Leon J, Perona R, Crespo P. J Biol Chem. 2004;279:32813–32823. doi: 10.1074/jbc.M313656200. [DOI] [PubMed] [Google Scholar]
- 37.Lu B, Ferrandino AF, Flavell RA. Nat Immunol. 2004;5:38–44. doi: 10.1038/ni1020. [DOI] [PubMed] [Google Scholar]
- 38.Papathanasiou MA, Kerr NC, Robbins JH, McBride OW, Alamo I, Jr, Barrett SF, Hickson ID, Fornace AJ., Jr Mol Cell Biol. 1991;11:1009–1016. doi: 10.1128/mcb.11.2.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kastan MB, Zhan Q, El-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ., Jr Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 40.Chin PL, Momand J, Pfeifer GP. Oncogene. 1997;15:87–99. doi: 10.1038/sj.onc.1201161. [DOI] [PubMed] [Google Scholar]
- 41.Vairapandi M, Balliet AG, Hoffman B, Liebermann DA. J Cell Physiol. 2002;192:327–338. doi: 10.1002/jcp.10140. [DOI] [PubMed] [Google Scholar]
- 42.Jin R, De Smaele E, Zazzeroni F, Nguyen DU, Papa S, Jones J, Cox C, Gelinas C, Franzoso G. DNA Cell Biol. 2002;21:491–503. doi: 10.1089/104454902320219059. [DOI] [PubMed] [Google Scholar]
- 43.De Smaele E, Zazzeroni F, Papa S, Nguyen DU, Jin R, Jones J, Cong R, Franzoso G. Nature. 2001;414:308–313. doi: 10.1038/35104560. [DOI] [PubMed] [Google Scholar]
- 44.Papa S, Zazzeroni F, Bubici C, Jayawardena S, Alvarez K, Matsuda S, Nguyen DU, Pham CG, Nelsbach AH, Melis T, Smaele ED, Tang WJ, D’Adamio L, Franzoso G. Nat Cell Biol. 2004;6:146–153. doi: 10.1038/ncb1093. [DOI] [PubMed] [Google Scholar]
- 45.Huang C, Ma WY, Li J, Goranson A, Dong Z. J Biol Chem. 1999;274:14595–14601. doi: 10.1074/jbc.274.21.14595. [DOI] [PubMed] [Google Scholar]
- 46.Hu CD, Chinenov Y, Kerppola TK. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 47.Gius D, Botero A, Shah S, Curry HA. Toxicol Lett. 1999;106:93–106. doi: 10.1016/s0378-4274(99)00024-7. [DOI] [PubMed] [Google Scholar]
- 48.Abeyama K, Eng W, Jester JV, Vink AA, Edelbaum D, Cockerell CJ, Bergstresser PR, Takashima A. J Clin Invest. 2000;105:1751–1759. doi: 10.1172/JCI9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He H, Wang X, Gorospe M, Holbrook NJ, Trush MA. Cell Growth Differ. 1999;10:307–315. [PubMed] [Google Scholar]
- 50.Zelivianski S, Spellman M, Kellerman M, Kakitelashvilli V, Zhou XW, Lugo E, Lee MS, Taylor R, Davis TL, Hauke R, Lin MF. Int J Cancer. 2003;107:478–485. doi: 10.1002/ijc.11413. [DOI] [PubMed] [Google Scholar]
- 51.Liang K, Lu Y, Jin W, Ang KK, Milas L, Fan Z. Mol Cancer Ther. 2003;2:1113–1120. [PubMed] [Google Scholar]
- 52.Yang Q, Manicone A, Coursen JD, Linke SP, Nagashima M, Forgues M, Wang XW. J Biol Chem. 2000;275:36892–36898. doi: 10.1074/jbc.M005319200. [DOI] [PubMed] [Google Scholar]
- 53.Chung HK, Yi YW, Jung NC, Kim D, Suh JM, Kim H, Park KC, Song JH, Kim DW, Hwang ES, Yoon SH, Bae YS, Kim JM, Bae I, Shong M. J Biol Chem. 2003;278:28079–28088. doi: 10.1074/jbc.M212835200. [DOI] [PubMed] [Google Scholar]
- 54.Kiningham KK, StClair DK. Cancer Res. 1997;57:5265–5271. [PubMed] [Google Scholar]
- 55.Zhao W, Spitz DR, Oberley LW, Robbins ME. Cancer Res. 2001;61:5537–5543. [PubMed] [Google Scholar]
- 56.Li Z, Khaletskiy A, Wang J, Wong JY, Oberley LW, Li JJ. Free Radic Biol Med. 2001;30:260–267. doi: 10.1016/s0891-5849(00)00468-8. [DOI] [PubMed] [Google Scholar]
- 57.Xia L, Paik A, Li JJ. Cancer Res. 2004;64:221–228. doi: 10.1158/0008-5472.can-03-0969. [DOI] [PubMed] [Google Scholar]
- 58.Spitz DR, Azzam EI, Li JJ, Gius D. Cancer Metastasis Rev. 2004;23:311–322. doi: 10.1023/B:CANC.0000031769.14728.bc. [DOI] [PubMed] [Google Scholar]
- 59.Baldwin AS., Jr Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 60.Duffey DC, Chen Z, Dong G, Ondrey FG, Wolf JS, Brown K, Siebenlist U, Van Waes C. Cancer Res. 1999;59:3468–3474. [PubMed] [Google Scholar]
- 61.Pajonk F, Pajonk K, McBride WH. J Natl Cancer Inst. 1999;91:1956–1960. doi: 10.1093/jnci/91.22.1956. [DOI] [PubMed] [Google Scholar]
- 62.Kamata H, Hirata H. Cell Signal. 1999;11:1–14. doi: 10.1016/s0898-6568(98)00037-0. [DOI] [PubMed] [Google Scholar]
- 63.Harris AL. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 64.Manna SK, Zhang HJ, Yan T, Oberley LW, Aggarwal BB. J Biol Chem. 1998;273:13245–13254. doi: 10.1074/jbc.273.21.13245. [DOI] [PubMed] [Google Scholar]
- 65.Jones PL, Ping D, Boss JM. Mol Cell Biol. 1997;17:6970–6981. doi: 10.1128/mcb.17.12.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bernard D, Monte D, Vandenbunder B, Abbadie C. Oncogene. 2002;21:4392–4402. doi: 10.1038/sj.onc.1205536. [DOI] [PubMed] [Google Scholar]
- 67.Shonai T, Adachi M, Sakata K, Takekawa M, Endo T, Imai K, Hareyama M. Cell Death Differ. 2002;9:963–971. doi: 10.1038/sj.cdd.4401050. [DOI] [PubMed] [Google Scholar]
- 68.Basu S, Rosenzweig KR, Youmell M, Price BD. Biochem Biophys Res Commun. 1998;247:79–83. doi: 10.1006/bbrc.1998.8741. [DOI] [PubMed] [Google Scholar]
- 69.Hassan KA, Ang KK, El-Naggar AK, Story MD, Lee JI, Liu D, Hong WK, Mao L. Cancer Res. 2002;62:6414–6417. [PubMed] [Google Scholar]
- 70.Zhan Q, Bae I, Kastan MB, Fornace AJ., Jr Cancer Res. 1994;54:2755–2760. [PubMed] [Google Scholar]
- 71.Smith ML, Ford JM, Hollander MC, Bortnick RA, Amundson SA, Seo YR, Deng CX, Hanawalt PC, Fornace AJ., Jr Mol Cell Biol. 2000;20:3705–3714. doi: 10.1128/mcb.20.10.3705-3714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suzuki K, Kodama S, Watanabe M. Cancer Res. 2001;61:5396–5401. [PubMed] [Google Scholar]