Abstract
Many pathogens depend on nitric oxide (NO•) detoxification and repair to establish an infection, and inhibitors of these systems are under investigation as next-generation antibiotics. Due to the broad reactivity of NO• and its derivatives with biomolecules, a deep understanding of how pathogens sense and respond to NO•, as an integrated system, has been elusive. Quantitative kinetic modeling has been proposed as a method to enhance analysis and understanding of NO• stress at the systems-level. Here we review the motivation for, current state of, and future prospects of quantitative modeling of NO• stress in bacteria, and suggest that such mathematical approaches would prove equally useful in the study of other broadly reactive antimicrobials, such as hydrogen peroxide (H2O2).
Introduction
NO• is a potent antimicrobial produced by immune cells to combat pathogens [1,2]. The importance of NO• to immunity is evidenced by the many pathogens, including Mycobacterium tuberculosis, Neisseria meningitides, Vibrio cholerae, Salmonella enterica serovar Typhimurium, Pseudomonas aeruginosa, and enterohemorrhagic Escherichia coli (EHEC), whose virulence depends on NO• detoxification and repair systems (Table 1) [3–8]. Collectively, these studies suggest that knowledge of how pathogens sense and respond to NO• could illuminate antibacterial strategies that synergize with host immunity. Research on NO• stress has continued for over two decades, and the cumulative picture that has emerged is immensely complex [1,9–12]. This derives from the broad reactivity of NO• and its reactive intermediates (reactive nitrogen species: RNS) with biomolecules [1,9,12]. Depending on the environment, dosage, and delivery rate, NO• will destroy iron-sulfur (Fe-S) clusters, reversibly bind heme, directly react with O2 and superoxide (O2•−), and/or be enzymatically detoxified, whereas its derivatives (NO2•, N2O3, N2O4, HNO, and ONOO−) damage thiols, tyrosine residues, and DNA bases (Figure 1) [1,9,12–14]. This systems-level stress becomes even further complicated when one considers that Fe-S clusters and thiols are used for a broad range of enzymatic and regulatory functions throughout the cellular network [15–19]. To decipher this response and understand how bacteria, as an integrated system, sense and respond to NO•, a quantitative understanding of intracellular NO• reactivity is required. NO• has many available reaction paths upon entering a cell, and the biological outcome of NO• exposure, whether it is continued growth, bacteriostasis, expression of virulence factors, transition to an antibiotic-tolerant state and/or cell death [17, 20– 22], is governed by a complex, kinetic competition. Quantitative knowledge of this competition and the factors that control it will reveal novel targets within the NO• response network for the discovery and development of therapeutics that synergize with host-derived NO•.
Table 1.
Pathogens for which NO• detoxification or repair has been identified as a virulence factor.
| Pathogen | Gene(s) | Description | Ref. |
|---|---|---|---|
|
E. coli (enterohemorrhagic) |
norV | Strains harboring an inactive norV gene (norVs) exhibited reduced survival in murine macrophages. |
[4] |
| E. coli (uropathogenic) | hmp | Isolates from patients with urinary tract infection had increased hmp expression, and Δhmp mutants were outcompeted by the wild-type in a mouse infection model. |
[78] |
| M. tuberculosis |
mpa, pafA uvrB, dlaT |
Mutants deficient in proteasome components (mpa or pafA) [3] or nucleotide excision repair (uvrB) [31] exhibited attenuated virulence in mice. |
[3,31] |
| N. meningitides | cycP, norB | Mutants lacking cytochrome c′ (cycP) or NO• reductase (norB) exhibited reduced survival in human macrophages and human nasopharyngeal mucosa organ cultures. |
[6] |
| P. aeruginosa | norCBD | A mutant deficient in NO• reductase (norCBD) exhibited reduced viability in murine macrophages. |
[8] |
| S. Typhimurium |
hmp, xth, nfo, ytfE, STM1808 |
Mutants lacking hmp exhibited reduced survival in human macrophages [25] and attenuated virulence in mice [26]. Mutations in base excision repair (xth and nfo) [32], Fe-S assembly (ytfE) [7], or previously-unidentified STM1808 [7] each caused attenuated virulence in mice. |
[7,25,26, 32] |
| S. aureus | hmp | Mutants deficient in hmp exhibited attenuated virulence in mice. |
[79] |
| V. cholerae | hmpA | Mutants lacking hmpA were outcompeted in a mouse intestine colonization assay. |
[5] |
| Y. pestis | hmp | A mutation in hmp resulted in longer incubation times and attenuated virulence in rats. |
[30] |
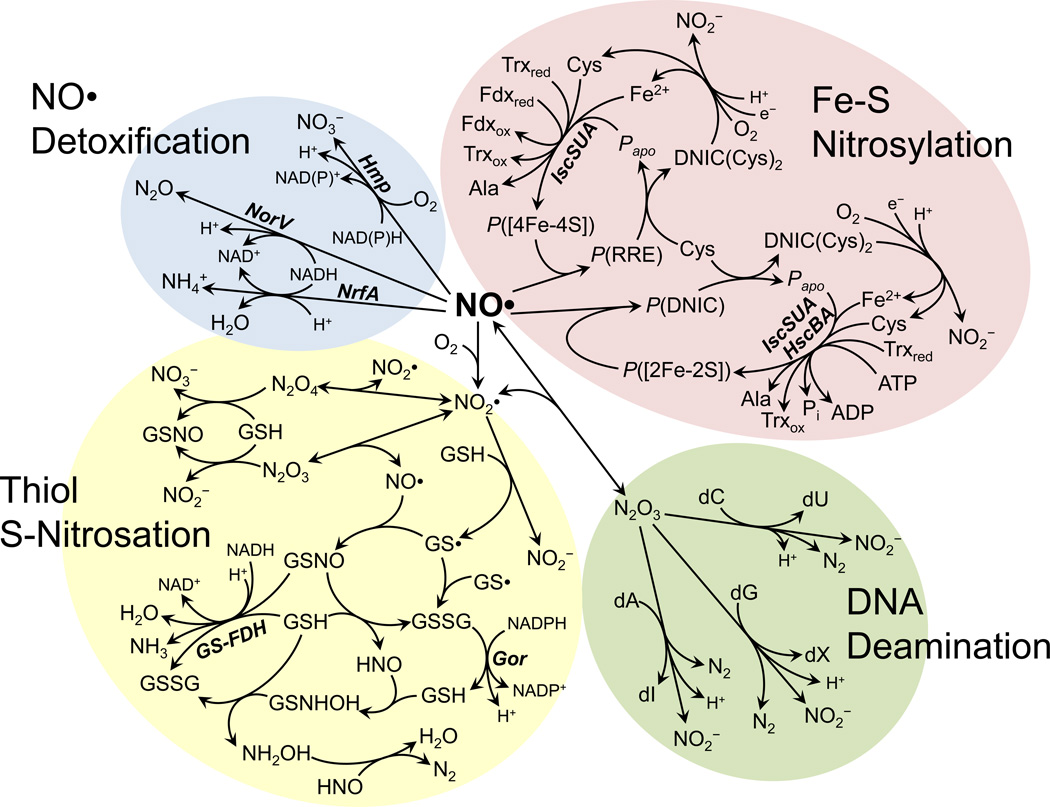
Figure 1.
Biochemical reaction network of NO• in E. coli. The diagram illustrates several key pathways involved in NO• metabolism. Reactions are grouped into categories of NO• detoxification, Fe-S nitrosylation and repair, DNA deamination, and thiol S-nitrosation and denitrosation. Enzymes catalyzing a reaction are shown in bold italics next to the reaction arrow. Enzyme abbreviations: NorV, NO• reductase; Hmp, NO• dioxygenase; NrfA, formate-dependent nitrite reductase; GS-FDH, glutathione-dependent formaldehyde dehydrogenase; Gor, glutathione reductase; IscSUA, Fe-S cluster assembly system; HscBA, Fe-S assembly chaperone. Metabolite abbreviations: GSH, glutathione; GS•, glutathionyl radical; GSNO, S-nitrosoglutathione; GSSG, glutathione disulfide; dA, deoxyadenosine; dG, deoxyguanosine; dC, deoxycytidine; dI, deoxyinosine; dX, deoxyxanthosine; dU, deoxyuridine; P([2Fe-2S]), protein-bound [2Fe-2S] cluster; P([4Fe-4S]), protein-bound [4Fe-4S] cluster; P(DNIC), protein-bound dinitrosyl-iron complex; P(RRE), protein-bound Roussins’ red ester; DNIC(Cys)2, L-cysteine-bound dinitrosyl-iron complex; Papo, apo-protein (lacking Fe-S cluster); Trxred, reduced thioredoxin; Trxox, oxidized thioredoxin; Fdxred, reduced ferredoxin; Fdxox, oxidized ferredoxin.
Due to the complexity of the competition for NO• among biomolecules, mathematical models are required to quantitatively analyze and understand data from NO•-stressed environments [13,14,23,24]. Beyond data interpretation, these models enable identification of emergent properties of the NO• response network and formulation of testable predictions concerning the impact of genetic and environmental perturbations. Here, we summarize evidence that suggests quantitative modeling will facilitate the discovery and development of NO•-based antibiotics, review the current state of NO• models along with their contributions to the present understanding of NO• stress, and reflect upon the future prospects of quantitative modeling to enhance the study of systems-level stresses from not only NO•, but other broadly reactive antimicrobials as well, such as H2O2.
NO• detoxification and repair systems are prevalent virulence factors
The ability to withstand NO• stress has been linked to the virulence of an impressive number of pathogens, several of which are presented in Table 1. Notably, in S. Typhimurium, Stevanin and colleagues found that a mutant defective in NO• dioxygenase (Δhmp) exhibited reduced survival in human macrophages, and that the effect was eliminated upon treatment with an inhibitor of inducible nitric oxide synthase (iNOS) [25]. More recently, this effect was corroborated in vivo, where Δhmp S. Typhimurium had attenuated virulence in mice, and iNOS inhibition restored virulence [26]. In EHEC, a genomic study of clinical isolates found that the presence of a functional norV gene, which encodes NO• reductase, correlated with an increased frequency of hemolytic-uremic syndrome (HUS) [27]. This connection was substantiated by a study demonstrating that EHEC possessing an inactive norV gene exhibited reduced survival in mouse macrophages compared to those with an active norV [4]. Recently, the genome of the EHEC strain responsible for the 2011 outbreak in Germany, which resulted in the highest incidence of HUS on record [28], was found to contain a functional norV [29], lending even further support for the previous genomic study. For Yersinia pestis, a microarray analysis of a model rat infection identified hmp expression to be significantly up-regulated, and subsequent experiments revealed that a Δhmp mutant exhibited attenuated virulence [30]. Beyond NO• detoxification, microbial repair systems have also been found to be important for resisting NO• stress and were shown to contribute to virulence. A transposon screen in M. tuberculosis found that mutations in proteasome components (mpa and pafA) and a nucleotide excision repair gene (uvrB) increased killing by NO• produced from acidified nitrite in vitro, and reduced virulence in mouse infection models [3,31]. In S. Typhimurium, Richardson and colleagues found that base excision repair mutants (ΔxthΔnfo) had attenuated virulence in mice, which was fully restored upon administration of an iNOS inhibitor [32]. These and related studies support a role for NO• and its derivatives as key mediators of host defense, and suggest that targeting the NO• defenses of pathogens may be an effective way to inhibit infection [33].
Therapeutic potential of NO•-based antibiotics
Several studies have found chemical inhibitors of the NO• response network that increase the sensitivity of pathogens to NO• [3,33,34]. Two chemical inhibitors were shown to block activity of the M. tuberculosis proteasome, and successfully reproduced the NO•-sensitive phenotype of proteasome-deficient mutants [3]. Helmick and colleagues found that imidazoles could inhibit NO• dioxygenase in vitro, and whole-cell NO• detoxification in Staphylococcus aureus and E. coli cultures, though the effects were far less pronounced in E. coli due to the poor Gram-negative membrane permeability of imidazoles [33]. By performing a screen to identify inhibitors of DlaT, an enzyme important for M. tuberculosis to tolerate NO•-stress, Bryk and colleagues discovered that rhodanines enhance killing of non-replicating M. tuberculosis treated with NO• by several orders of magnitude [34]. Further, D157070 (DlaT inhibitor) reduced M. tuberculosis viability in murine bone-marrow macrophages. These studies demonstrate the potential of targeting the NO• response network for the discovery of novel antibiotics, and suggest that a deeper understanding of NO• stress will reveal additional therapeutic strategies for investigation, since all targets are not equally accessible, as demonstrated with imidazoles and E. coli [33]. It is also worth noting that, in addition to potentiating immune-derived NO•, chemicals that target the NO• response network would prove useful for therapies that directly administer exogenous NO• to infection sites. Direct administration techniques have been garnering attention in recent years, due to the ability of NO• to eliminate antibiotic resistant pathogens [35–38], and numerous delivery mechanisms, including nanoparticles [35,39], probiotic patches [37], and dendrimers [38], have been explored. Several excellent reviews on the topic have recently been published [40–42], so here we will only highlight an important design constraint. Specifically, the delivery method must achieve NO• concentrations high enough to be antibacterial but low enough to remain non-toxic to eukaryotic cells. This therapeutic window can be as small as 5-fold [41], thereby presenting a significant challenge for direct delivery methods. One way to relieve this constraint is to couple direct delivery with agents that increase the sensitivity of pathogens to NO•, effectively expanding the therapeutic window.
NO• elicits a complex, systems-level stress response
A comprehensive, quantitative understanding of NO• stress has been elusive due to the reactivity of NO• and its derivatives with a wide range of biomolecules [1,9,10,12], and the depth to which these perturbations propagate through cellular networks. This complexity is best illustrated by the findings of transcriptomic [7,17,19,43–47], proteomic [15,18], and metabolomic [48,49] studies, which have collectively demonstrated diverse, systems-wide responses to NO•. Hyduke and colleagues conducted a DNA microarray analysis of NO•-stressed E. coli, and found that 709 genes were significantly perturbed, affecting a diverse range of cellular functions, including branched-chain amino acid synthesis, respiration, Fe-S assembly, and energy metabolism [17]. This study also included a chemoinformatic analysis of the E. coli proteome, wherein 554 proteins were identified as potential RNS targets based on the presence of possible NO•-reactive features (such as Fe-S clusters, heme groups, or reactive thiol motifs). A transcriptomic study of anaerobically-grown E. coli found that approximately 4% of the genome (173 genes) exhibited a significant change in expression upon NO• treatment [43]. Again, the perturbed genes spanned a diversity of functions beyond NO• detoxification, including DNA metabolism, cofactor and prosthetic group synthesis, fatty acid metabolism, cell structure maintenance, metal ion and multidrug transport, purine and pyrimidine synthesis, and energy metabolism. The transcriptome of oral pathogen Porphyromonas gingivalis was measured following NO• exposure, and it too demonstrated an expansive, systems-level response where expression of approximately 19% of its genome (380 genes) was significantly perturbed [44]. Although the majority of the affected genes were of unknown function, those associated with energy metabolism, translation, and regulatory functions were found to be altered by NO•. A recent metabolomic study of NO•-treated V. cholerae found extensive metabolic distress, as demonstrated by an accumulation of upper glycolytic metabolites, impairment of arginine synthesis, and accumulation of citrate, thereby suggesting TCA cycle dysfunction [48]. Similarly, Auger and colleagues studied NO•-stressed Pseudomonas fluorescens and observed obstruction of the TCA cycle [49]. Proteomic analyses have also revealed systems-level perturbations by NO•; for example, Rhee and colleagues measured S-nitrosation of M. tuberculosis proteins by acidified nitrite- or macrophage-derived NO•, and found 29 enzymes whose functions included amino acid biosynthesis, energy metabolism, antioxidant defense, heat shock response, RNA polymerase, and lipid metabolism to be nitrosation targets [15]. Analysis of S-nitrosation in E. coli revealed some similarities, as well as novel targets, including a subunit of pyruvate dehydrogenase [18]. These studies demonstrate the extent to which NO• perturbs numerous facets of bacterial physiology, and highlight the challenges associated with gaining a complete understanding of how a pathogen, as an integrated system, responds to NO•.
Quantitative modeling of NO• stress
On a molecular level, the complex bacterial responses to NO• all originate from the NO• biochemical reaction network. Therefore, a deeper understanding of NO• stress must begin with a quantitative examination of intracellular NO• dynamics. The broad reactivity of NO• and derived RNS, culminating in the breadth of physiological perturbations identified by -omics studies, underscores the need for a quantitative, mathematical approach to deconvolute the effects of NO• stress and the ensuing microbial response. Furthermore, the dynamics of these processes span multiple time scales, ranging from fractions of a second (spontaneous chemical reactions) to minutes (regulatory responses), thus requiring a computational approach for their integration. The need for a model-based approach to quantitatively study the complex reaction network of NO• in biological systems has motivated the construction of kinetic models to simulate the NO• biochemical network and gain insight into its biological roles.
Initial models to examine NO• stress in biological systems
Initial attempts to model NO• biochemical reaction networks were largely motivated by a desire to understand its role in mammalian signaling. Lancaster constructed a kinetic model of NO• chemistry accounting for the oxidation, nitration, and nitrosation reaction types governing the fate of NO• and its reactive intermediates, and was able to make predictions regarding the relative importance of each pathway under different biological conditions (e.g., inflammatory and non-inflammatory levels of NO• production) [13]. This model was reformulated by Lim and colleagues to describe intracellular NO• chemistry of inflamed tissue at steady-state, and was expanded to include additional antioxidants, amino acids, and lipid-phase reactions [14]. Simulations provided valuable information on approximate concentrations of RNS that are generally too unstable and/or scarce to measure experimentally, as well as their major intracellular sinks [14]. Bagci and colleagues used mathematical modeling to explore the participation of NO• in apoptosis by integrating an NO• chemical network with a mitochondrial apoptotic signaling network, thus providing quantitative insight into the pro- and anti-apoptotic activity of NO• [23]. Though these methods laid the foundation for the quantitative study of NO• in biological systems, little consideration was given to cellular responses to NO•, such as repair of damaged biomolecules and regulatory responses, which we have recently found to be critical in simulating the dynamics of NO• stress in microbes [24].
Recent advances in model-driven analyses of microbial NO• stress
Recent work in our laboratory has demonstrated the feasibility and utility of a model-based approach in studying NO• stress in a microbial system [24]. Drawing upon existing kinetic models and the available body of literature, a comprehensive kinetic model of NO• biochemistry in E. coli was constructed and experimentally validated. The model encompassed processes such as damage and repair of Fe-S clusters, DNA, and thiols, as well as enzymatic NO• detoxification, autoxidation, and reversible inhibition of respiratory cytochromes. Model simulations exhibited excellent predictive accuracy with regard to major system perturbations, such as the deletion of hmp, which encodes the primary aerobic NO• detoxification system in E. coli. In addition, parametric analysis identified the rate of NO• delivery to the system as a control parameter having strong influence on the distribution of NO• consumption, which was confirmed experimentally. Finally, the model was found to accurately capture NO• dynamics under microaerobic O2 concentrations, and successfully predict the importance of the major aerobic (Hmp) and anaerobic (NorV) detoxification systems in this medically important regime.
Potential of quantitative modeling to transform the study of NO• stress in bacteria
As discussed above, bacteria mount a systems-level response to NO• that spans energy metabolism, amino acid biosynthesis, translation, transcription, respiration, DNA metabolism, protein cofactor synthesis, and direct detoxification [1]. Quantitative modeling offers a means to interpret NO• responses and investigate their underlying architecture. For example, network analysis techniques, such as parameter variation [24] or metabolic control analysis [50], can be used to identify species, pathways, or other network components that significantly alter the NO• distribution upon perturbation. In this way, emergent systems properties of the NO• stress response can be discovered, painting a more complete picture of the complex network, and offering novel therapeutic targets. In addition, quantitative models provide an excellent framework to integrate diverse types of data, such as metabolite, transcript, and protein levels, since explicit variables for concentrations of cellular components are used. Further, a rigorously-constructed model represents the current knowledgebase, and observed phenomena that disagree with simulations represent a knowledge gap to be filled. For example, a screen may identify a novel gene or chemical that modifies how a bacterium processes NO•, resulting in an unexplained NO• dynamic. To understand the basis of such novel phenotypes, analyses can be performed to identify parameters whose modulation reconciles simulations with experiments. These model adjustments provide readily testable hypotheses, such as altered gene expression or protein degradation, to explain the mechanism by which a mutation or chemical perturbs the NO• response network.
Challenges facing quantitative modeling of NO• stress
Although model-based approaches to study NO• stress offer numerous benefits, they are all inherently limited by the availability of kinetic data and knowledge of the system architecture, such as the repertoire of NO• detoxification enzymes present. For microbes where this information is scarce, coarse-grained models, where reaction pathways and parameters have been lumped together, can be trained on experimental data and used for quantitative analyses, until more detailed information becomes available. Another challenge is related to rapid quantification of unstable, short-lived intermediates that often exist in trace quantities, such as N2O3 and NO2• [14]. The lack of precise measurements of these species prevents direct validation of those components in the model, and therefore conclusions based on simulation of those species should be handled with caution. One approach to reconcile the lack of a direct measurement, however, is to use a reliable proxy, such as a stable and measureable downstream product. Overall, these challenges limit the accuracy of quantitative models of NO• stress, but it is important to note that failure of a rigorously-constructed model to capture a phenotypic response represents an opportunity to discover novel biology not contained within the available knowledgebase.
Beyond NO•
The broad reactivity of NO• makes quantitative modeling an attractive tool for studying its systems-level effects on bacteria. This quality of NO• is mirrored in other immune antimicrobials, such as H2O2 [51]. The importance of H2O2 to immunity has also been supported by the many pathogens that require H2O2 detoxification systems to establish or sustain an infection, such as S. Typhimurium [52], M. tuberculosis [53,54], S. aureus [55], Helicobacter pylori [56], Streptococcus pyogenes [57], and Enterococcus faecalis [58] (Table 2). In phagocytic cells, H2O2 is derived from the dismutation of O2•− that is produced by NADPH oxidase [59], and readily diffuses into bacterial cells [60] to react with cysteine [61] and methionine [62,63] residues, Fe-S clusters [64], transition metals [65], and α-keto acids [66,67], or undergo enzymatic detoxification by catalases [68], and hydroperoxidases [69] (Figure 2). H2O2 can also be reduced by Fe2+ to form the stronger oxidant, HO•, which reacts with most biomolecules at diffusion-limited rates [70]. Given this broad reactivity, it is not surprising that transcriptomic studies have shown that H2O2 treatment results in systems-level changes in the expression of genes involved in DNA repair, virulence, membrane function, metabolism, and peroxide detoxification [71–74].
Table 2.
Pathogens for which H2O2 detoxification has been identified as a virulence factor.
| Pathogen | Gene(s) | Description | Ref. |
|---|---|---|---|
| E. faecalis | tpx, npr, ahp | Mutants lacking thiol peroxidase (tpx) have attenuated virulence in a mouse peritonitis model. Triple mutants lacking Tpx, NADH peroxidase (npr), and alkyl hydroperoxide reductase (ahp) were more significantly attenuated. |
[58] |
| H. pylori | katA, kapA | Mutants lacking katA and kapA were less able to sustain long-term infection in mice. |
[56] |
| M. tuberculosis | katG | Mutations in katG decreased persistence within infected mice [53] and human monocytes in vitro [54]. |
[53,54] |
| S. Typhimurium |
katE, katG katN, ahpC, tsaA |
Inactivation of all five catalase and hydroperoxidase genes resulted in high sensitivity to H2O2 and decreased survival within murine macrophages. |
[52] |
| S. aureus | katA, ahpC | Mutations in katA and ahpC decreased ability to colonize the nasal cavities of cotton rats. |
[55] |
| S. pyogenes | gpoA |
S. pyogenes requires glutathione peroxidase (GpoA) for virulence in several mouse models. |
[57] |
Figure 2.
Biochemical reaction network of H2O2 in E. coli. The diagram highlights the complexity of the intracellular H2O2 reaction network. Reactions are grouped into categories of antioxidant enzymes, antioxidant metabolites, oxidative DNA damage, and oxidative protein damage. Enzyme abbreviations: Cat, catalase; Ahp, alkyl hydroperoxidase; Msr, methionine sulfoxide reductase; TrxR, thioredoxin reductase; Gor, glutathione reductase. Metabolite abbreviations: R-COCOOH, generic α-keto acid; R-COOH, carboxylic acid; Metsox, L-methionine-S-oxide; SSB, DNA single strand break; DSB, DNA double strand break; dG, deoxyguanosine; 8-OHdG, 8-hydroxy-2-deoxyguanosine; 8-oxodG, 8-oxo-7,8-dihydro-2’-deoxyguanosine; Trxred, reduced thioredoxin; Trxox, oxidized thioredoxin; GSH, reduced glutathione; GSSG, glutathione disulfide; P([4Fe-4S]), protein with [4Fe-4S] cluster; P([3Fe-4S]), damaged protein with [3Fe-4S] cluster; P(Cys-CH2-SH), protein-bound L-cysteine; -SOH, sulfenic acid; -SO2H, sulfinic acid; -SO3H, sulfonic acid; P(Pro), protein-bound L-proline; P(Arg), protein-bound L-arginine; P(Lys), protein-bound L-lysine; P(glut-semi), protein-bound glutamic semialdehyde; P(glut-amino), protein-bound aminoadipic semialdehyde.
The complexity of the H2O2 biochemical reaction network suggests that quantitative modeling could provide a deeper understanding of how bacteria sense and respond to H2O2 as an integrated system. Currently, the best models of H2O2 biochemistry are specific to mammalian systems due to its importance as a cellular signaling molecule and implication in a number of diseases [75–77]. These models have included H2O2 elimination by the antioxidants glutathione and thioredoxin, enzymes catalase, glutathione peroxidase, glutathione reductase, glutaredoxin, and peroxiredoxin, as well as processing of oxidized protein thiols [75–77], but are incomplete due to the lack of reactions describing damage and repair of many biomolecules and the exclusion of transcriptional regulation. Analogous to NO• stress, quantitative modeling has the potential to provide a deeper understanding of H2O2 stress, and thereby illuminate therapeutic targets to sensitize pathogens to oxidative immune attack.
Conclusion
NO• is an important immune antimicrobial that produces a systems-level stress that is difficult to understand quantitatively without the use of mathematical models. These models offer utilities far beyond data interpretation, such as a platform to investigate systems-level control of NO• metabolism, and an ability to mechanistically dissect novel phenotypes of the NO• response network. Recent advances in this area include a detailed model of NO• stress in E. coli [24], which led to the identification of an emergent property of the NO• response network, and increased understanding of NO• stress under the physiologically relevant microaerobic regime. While important knowledge can be gained through analysis and expansion of this model, it can also serve as a template to develop models for less well-characterized bacteria, which is a necessary step to transform quantitative modeling into a common practice for investigations of stress caused by NO• and other broadly-reactive antimicrobials, such as H2O2.
Highlights.
NO• detoxification and repair have been linked to virulence in many pathogens.
NO• and its derivatives are broadly reactive and elicit a systems-level response.
Quantitative models can enhance understanding of the NO• response network.
Quantitative models can improve study of other broadly-reactive antimicrobials.
Acknowledgements
This work was supported in part by the National Science Foundation Graduate Research Fellowship under Grant No. DGE 1148900, and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R21AI105342.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* Of special interest
** Of outstanding interest
- 1. Bowman LAH, McLean S, Poole RK, Fukuto JM. The Diversity of Microbial Responses to Nitric Oxide and Agents of Nitrosative Stress: Close Cousins but Not Identical Twins. Adv Microb Physiol. 2011;59:135–219. doi: 10.1016/B978-0-12-387661-4.00006-9. This is an excellent review describing the chemistry and biochemistry of NO• and its derivatives, as well as the associated microbial response. Also discusses various experimental approaches used to study nitrogen oxide chemistry.
- 2.Fang FC. Antimicrobial reactive oxygen and nitrogen species: Concepts and controversies. Nat Rev Microbiol. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 3. Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. The authors found an NO•-protective role for the proteasome of M. tuberculosis. An infection study showed that the proteasome contributes to M. tuberculosis virulence in mice, and increased NO• sensitivity could be achieved using chemical inhibitors of proteasome activity.
- 4.Shimizu T, Tsutsuki H, Matsumoto A, Nakaya H, Noda M. The nitric oxide reductase of enterohaemorrhagic Escherichia coli plays an important role for the survival within macrophages. Mol Microbiol. 2012;85:492–512. doi: 10.1111/j.1365-2958.2012.08122.x. [DOI] [PubMed] [Google Scholar]
- 5.Stern AM, Hay AJ, Liu Z, Desland FA, Zhang J, Zhong ZT, Zhu J. The NorR Regulon Is Critical for Vibrio cholerae Resistance to Nitric Oxide and Sustained Colonization of the Intestines. mBio. 2012:3. doi: 10.1128/mBio.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevanin TM, Moir JWB, Read RC. Nitric oxide detoxification systems enhance survival of Neisseria meningitidis in human macrophages and in nasopharyngeal mucosa. Infect Immun. 2005;73:3322–3329. doi: 10.1128/IAI.73.6.3322-3329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karlinsey JE, Bang IS, Becker LA, Frawley ER, Porwollik S, Robbins HF, Thomas VC, Urbano R, McClelland M, Fang FC. The NsrR regulon in nitrosative stress resistance of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2012;85:1179–1193. doi: 10.1111/j.1365-2958.2012.08167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kakishima K, Shiratsuchi A, Taoka A, Nakanishi Y, Fukumori Y. Participation of nitric oxide reductase in survival of Pseudomonas aeruginosa in LPS-activated macrophages. Biochem Bioph Res Co. 2007;355:587–591. doi: 10.1016/j.bbrc.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 9. Toledo JC, Augusto O. Connecting the Chemical and Biological Properties of Nitric Oxide. Chem Res Toxicol. 2012;25:975–989. doi: 10.1021/tx300042g. This is a detailed review on NO• chemistry and its interaction with a variety of cellular targets, emphasizing the importance of connecting reactivity with biological activity.
- 10.Reiter TA. NO• chemistry: a diversity of targets in the cell. Redox Rep. 2006;11:194–206. doi: 10.1179/135100006X116718. [DOI] [PubMed] [Google Scholar]
- 11.Vine CE, Cole JA. Unresolved sources, sinks, and pathways for the recovery of enteric bacteria from nitrosative stress. FEMS Microbiol Lett. 2011;325:99–107. doi: 10.1111/j.1574-6968.2011.02425.x. [DOI] [PubMed] [Google Scholar]
- 12.Stern AM, Zhu J. An Introduction to Nitric Oxide Sensing and Response in Bacteria. Adv Appl Microbiol. 2014;87:187–220. doi: 10.1016/B978-0-12-800261-2.00005-0. [DOI] [PubMed] [Google Scholar]
- 13.Lancaster JR. Nitroxidative, nitrosative, and nitrative stress: Kinetic predictions of reactive nitrogen species chemistry under biological conditions. Chem Res Toxicol. 2006;19:1160–1174. doi: 10.1021/tx060061w. [DOI] [PubMed] [Google Scholar]
- 14.Lim CH, Dedon PC, Deen WA. Kinetic Analysis of Intracellular Concentrations of Reactive Nitrogen Species. Chem Res Toxicol. 2008;21:2134–2147. doi: 10.1021/tx800213b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhee KY, Erdjument-Bromage H, Tempst P, Nathan CF. S-nitroso proteome of Mycobacterium tuberculosis: Enzymes of intermediary metabolism and antioxidant defense. Proc Natl Acad Sci U S A. 2005;102:467–472. doi: 10.1073/pnas.0406133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richardson AR, Payne EC, Younger N, Karlinsey JE, Thomas VC, Becker LA, Navarre WW, Castor ME, Libby SJ, Fang FC. Multiple Targets of Nitric Oxide in the Tricarboxylic Acid Cycle of Salmonella enterica Serovar Typhimurium. Cell Host Microbe. 2011;10:33–43. doi: 10.1016/j.chom.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hyduke DR, Jarboe LR, Tran LM, Chou KJY, Liao JC. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli . Proc Natl Acad Sci U S A. 2007;104:8484–8489. doi: 10.1073/pnas.0610888104. The authors used transcriptomics to elucidate the NO• response network of E. coli and to identify participating regulators, and attributed the bacteriostatic effect of NO• to its inhibition of branched chain amino acid synthesis.
- 18.Brandes N, Rinck A, Leichert LI, Jakob U. Nitrosative stress treatment of E. coli targets distinct set of thiol-containing proteins. Mol Microbiol. 2007;66:901–914. doi: 10.1111/j.1365-2958.2007.05964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pullan ST, Gidley MD, Jones RA, Barrett J, Stevanin TA, Read RC, Green J, Poole RK. Nitric oxide in chemostat-cultured Escherichia coli is sensed by Fnr and other global regulators: Unaltered methionine biosynthesis indicates lack of S nitrosation. J Bacteriol. 2007;189:1845–1855. doi: 10.1128/JB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009;325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vareille M, de Sablet T, Hindre T, Martin C, Gobert AP. Nitric oxide inhibits Shiga-toxin synthesis by enterohemorrhagic Escherichia coli . Proc Natl Acad Sci U S A. 2007;104:10199–10204. doi: 10.1073/pnas.0702589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones-Carson J, Laughlin JR, Stewart AL, Voskuil MI, Vazquez-Torres A. Nitric oxide-dependent killing of aerobic, anaerobic and persistent Burkholderia pseudomallei . Nitric Oxide. 2012;27:25–31. doi: 10.1016/j.niox.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bagci EZ, Vodovotz Y, Billiar TR, Ermentrout B, Bahar I. Computational Insights on the Competing Effects of Nitric Oxide in Regulating Apoptosis. PLoS One. 2008;3:e2249. doi: 10.1371/journal.pone.0002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robinson JL, Brynildsen MP. A Kinetic Platform to Determine the Fate of Nitric Oxide in Escherichia coli . PLoS Comput Biol. 2013;9:e1003049. doi: 10.1371/journal.pcbi.1003049. This study presents the construction and experimental validation of the first comprehensive model of NO• biochemistry in bacteria, and demonstrates its utility in predicting novel network features.
- 25.Stevanin TM, Poole RK, Demoncheaux EAG, Read RC. Flavohemoglobin Hmp Protects Salmonella enterica Serovar Typhimurium from Nitric Oxide-Related Killing by Human Macrophages. Infect Immun. 2002;70:4399–4405. doi: 10.1128/IAI.70.8.4399-4405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bang IS, Liu LM, Vazquez-Torres A, Crouch ML, Stamler JS, Fang FC. Maintenance of Nitric Oxide and Redox Homeostasis by the Salmonella Flavohemoglobin Hmp. J Biol Chem. 2006;281:28039–28047. doi: 10.1074/jbc.M605174200. [DOI] [PubMed] [Google Scholar]
- 27.Kulasekara BR, Jacobs M, Zhou Y, Wu ZN, Sims E, Saenphimmachak C, Rohmer L, Ritchie JM, Radey M, McKevitt M, et al. Analysis of the Genome of the Escherichia coli O157:H7 2006 Spinach-Associated Outbreak Isolate Indicates Candidate Genes That May Enhance Virulence. Infect Immun. 2009;77:3713–3721. doi: 10.1128/IAI.00198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frank C, Werber D, Cramer JP, Askar M, Faber M, an der Heiden M, Bernard H, Fruth A, Prager R, Spode A, et al. Epidemic Profile of Shiga-Toxin-Producing Escherichia coli O104:H4 Outbreak in Germany. N Engl J Med. 2011;365:1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 29.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, et al. Prospective Genomic Characterization of the German Enterohemorrhagic Escherichia coli O104:H4 Outbreak by Rapid Next Generation Sequencing Technology. PloS One. 2011;6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sebbane F, Lemaitre N, Sturdevant DE, Rebeil R, Virtaneva K, Porcella SF, Hinnebusch BJ. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc Natl Acad Sci U S A. 2006;103:11766–11771. doi: 10.1073/pnas.0601182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darwin KH, Nathan CF. Role for Nucleotide Excision Repair in Virulence of Mycobacterium tuberculosis . Infect Immun. 2005;73:4581–4587. doi: 10.1128/IAI.73.8.4581-4587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Richardson AR, Soliven KC, Castor ME, Barnes PD, Libby SJ, Fang FC. The Base Excision Repair System of Salmonella enterica serovar Typhimurium Counteracts DNA Damage by Host Nitric Oxide. PLoS Pathog. 2009;5:e1000451. doi: 10.1371/journal.ppat.1000451. The authors found that an S. Typhimurium mutant deficient in base excision repair was more sensitive to NO•, and this effect contributed to attenuated virulence in mice.
- 33.Helmick RA, Fletcher AE, Gardner AM, Gessner CR, Hvitved AN, Gustin MC, Gardner PR. Imidazole Antibiotics Inhibit the Nitric Oxide Dioxygenase Function of Microbial Flavohemoglobin. Antimicrob Agents Chemother. 2005;49:1837–1843. doi: 10.1128/AAC.49.5.1837-1843.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bryk R, Gold B, Venugopal A, Singh J, Samy R, Pupek K, Cao H, Popescu C, Gurney M, Hotha S, et al. Selective Killing of Nonreplicating Mycobacteria. Cell Host Microbe. 2008;3:137–145. doi: 10.1016/j.chom.2008.02.003. This paper identified chemical inhibitors of DlaT that enhanced NO•-mediated killing of non-replicating M. tuberculosis, demonstrating the potential of targeting the NO• response network in designing novel antibiotics
- 35.Friedman A, Blecher K, Sanchez D, Tuckman-Vernon C, Gialanella P, Friedman JM, Martinez LR, Nosanchuk JD. Susceptibility of Gram-positive and -negative bacteria to novel nitric oxide-releasing nanoparticle technology. Virulence. 2011;2:217–221. doi: 10.4161/viru.2.3.16161. [DOI] [PubMed] [Google Scholar]
- 36.Heilman BJ, St John J, Oliver SRJ, Mascharak PK. Light-Triggered Eradication of Acinetobacter baumannii by Means of NO Delivery from a Porous Material with an Entrapped Metal Nitrosyl. J Am Chem Soc. 2012;134:11573–11582. doi: 10.1021/ja3022736. [DOI] [PubMed] [Google Scholar]
- 37.Sulemankhil I, Ganopolsky JG, Dieni CA, Dan AF, Jones ML, Prakash S. Prevention and Treatment of Virulent Bacterial Biofilms with an Enzymatic Nitric Oxide-Releasing Dressing. Antimicrob Agents Chemother. 2012;56:6095–6103. doi: 10.1128/AAC.01173-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun B, Slomberg DL, Chudasama SL, Lu Y, Schoenfisch MH. Nitric Oxide-Releasing Dendrimers as Antibacterial Agents. Biomacromolecules. 2012;13:3343–3354. doi: 10.1021/bm301109c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedman AJ, Blecher K, Schairer D, Tuckman-Vernon C, Nacharaju P, Sanchez D, Gialanella P, Martinez LR, Friedman JM, Nosanchuk JD. Improved antimicrobial efficacy with nitric oxide releasing nanoparticle generated S-nitrosoglutathione. Nitric Oxide. 2011;25:381–386. doi: 10.1016/j.niox.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 40. Schairer DO, Chouake JS, Nosanchuk JD, Friedman AJ. The potential of nitric oxide releasing therapies as antimicrobial agents. Virulence. 2012;3:271–279. doi: 10.4161/viru.20328. The authors review the various NO• delivery methods that have been developed for use as antimicrobial therapies. The efficacy and feasibility of each treatment in the context of clinical application is discussed.
- 41.Jones ML, Ganopolsky JG, Labbe A, Wahl C, Prakash S. Antimicrobial properties of nitric oxide and its application in antimicrobial formulations and medical devices. Appl Microbiol Biot. 2010;88:401–407. doi: 10.1007/s00253-010-2733-x. [DOI] [PubMed] [Google Scholar]
- 42.Riccio DA, Schoenfisch MH. Nitric oxide release: Part I. Macromolecular scaffolds. Chem Soc Rev. 2012;41:3731–3741. doi: 10.1039/c2cs15272j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Justino MC, Vicente JB, Teixeira M, Saraiva LM. New Genes Implicated in the Protection of Anaerobically Grown Escherichia coli against Nitric Oxide. J Biol Chem. 2005;280:2636–2643. doi: 10.1074/jbc.M411070200. [DOI] [PubMed] [Google Scholar]
- 44.Boutrin MC, Wang C, Aruni W, Li XJ, Fletcher HM. Nitric Oxide Stress Resistance in Porphyromonas gingivalis Is Mediated by a Putative Hydroxylamine Reductase. J Bacteriol. 2012;194:1582–1592. doi: 10.1128/JB.06457-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore CM, Nakano MM, Wang T, Ye RW, Helmann JD. Response of Bacillus subtilis to Nitric Oxide and the Nitrosating Agent Sodium Nitroprusside. J Bacteriol. 2004;186:4655–4664. doi: 10.1128/JB.186.14.4655-4664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukhopadhyay P, Zheng M, Bedzyk LA, LaRossa RA, Storz G. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc Natl Acad Sci U S A. 2004;101:745–750. doi: 10.1073/pnas.0307741100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, Schoolnik GK. Inhibition of Respiration by Nitric Oxide Induces a Mycobacterium +tuberculosis Dormancy Program. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stern AM, Liu BB, Bakken LR, Shapleigh JP, Zhu J. A Novel Protein Protects Bacterial Iron-Dependent Metabolism from Nitric Oxide. J Bacteriol. 2013;195:4702–4708. doi: 10.1128/JB.00836-13. This is the first metabolomic study quantifying the change in a broad range of metabolite concentrations as a result of NO• stress. Also from their data, the authors determine that NnrS provides NO• resistance to V. cholerae through protection of the cellular iron pool from nitrosylation.
- 49.Auger C, Lemire J, Cecchini D, Bignucolo A, Appanna VD. The Metabolic Reprogramming Evoked by Nitrosative Stress Triggers the Anaerobic Utilization of Citrate in Pseudomonas fluorescens . PLoS One. 2011;6:e28469. doi: 10.1371/journal.pone.0028469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cintolesi A, Clomburg JM, Rigou V, Zygourakis K, Gonzalez R. Quantitative Analysis of the Fermentative Metabolism of Glycerol in Escherichia coli . Biotechnol Bioeng. 2012;109:187–198. doi: 10.1002/bit.23309. [DOI] [PubMed] [Google Scholar]
- 51.Flannagan RS, Cosio G, Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol. 2009;7:355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- 52.Hébrard M, Viala JPM, Meresse S, Barras F, Aussel L. Redundant Hydrogen Peroxide Scavengers Contribute to Salmonella Virulence and Oxidative Stress Resistance. J Bacteriol. 2009;191:4605–4614. doi: 10.1128/JB.00144-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li ZM, Kelley C, Collins F, Rouse D, Morris S. Expression of katG in Mycobacterium tuberculosis Is Associated with Its Growth and Persistence in Mice and Guinea Pigs. J Infect Dis. 1998;177:1030–1035. doi: 10.1086/515254. [DOI] [PubMed] [Google Scholar]
- 54.Manca C, Paul S, Barry CE, Freedman VH, Kaplan G. Mycobacterium tuberculosis Catalase and Peroxidase Activities and Resistance to Oxidative Killing in Human Monocytes In Vitro. Infect Immun. 1999;67:74–79. doi: 10.1128/iai.67.1.74-79.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cosgrove K, Coutts G, Jonsson IM, Tarkowski A, Kokai-Kun JF, Mond JJ, Foster SJ. Catalase (KatA) and Alkyl Hydroperoxide Reductase (AhpC) Have Compensatory Roles in Peroxide Stress Resistance and Are Required for Survival, Persistence, and Nasal Colonization in Staphylococcus aureus . J Bacteriol. 2007;189:1025–1035. doi: 10.1128/JB.01524-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harris AG, Wilson JE, Danon SJ, Dixon MF, Donegan K, Hazell SL. Catalase (KatA) and KatA-associated protein (KapA) are essential to persistent colonization in the Helicobacter pylori SS1 mouse model. Microbiology. 2003;149:665–672. doi: 10.1099/mic.0.26012-0. [DOI] [PubMed] [Google Scholar]
- 57.Brenot A, King KY, Janowiak B, Griffith O, Caparon MG. Contribution of Glutathione Peroxidase to the Virulence of Streptococcus pyogenes . Infect Immun. 2004;72:408–413. doi: 10.1128/IAI.72.1.408-413.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.La Carbona S, Sauvageot N, Giard JC, Benachour A, Posteraro B, Auffray Y, Sanguinetti M, Hartke A. Comparative study of the physiological roles of three peroxidases (NADH peroxidase, Alkyl hydroperoxide reductase and Thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis . Mol Microbiol. 2007;66:1148–1163. doi: 10.1111/j.1365-2958.2007.05987.x. [DOI] [PubMed] [Google Scholar]
- 59.Diacovich L, Gorvel JP. Bacterial manipulation of innate immunity to promote infection. Nat Rev Microbiol. 2010;8:117–128. doi: 10.1038/nrmicro2295. [DOI] [PubMed] [Google Scholar]
- 60.Karlsson A, Dahlgren C. Assembly and Activation of the Neutrophil NADPH Oxidase in Granule Membranes. Antioxid Redox Signal. 2002;4:49–60. doi: 10.1089/152308602753625852. [DOI] [PubMed] [Google Scholar]
- 61.Kim J-R, Yoon HW, Kwon K-S, Lee S-R, Rhee SG. Identification of Proteins Containing Cysteine Residues That Are Sensitive to Oxidation by Hydrogen Peroxide at Neutral pH. Anal Biochem. 2000;283:214–221. doi: 10.1006/abio.2000.4623. [DOI] [PubMed] [Google Scholar]
- 62.Davies MJ. The oxidative environment and protein damage. Biochim Biophys Acta. 2005;1703:93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 63.Luo S, Levine RL. Methionine in proteins defends against oxidative stress. FASEB J. 2009;23:464–472. doi: 10.1096/fj.08-118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jang SJ, Imlay JA. Micromolar Intracellular Hydrogen Peroxide Disrupts Metabolism by Damaging Iron-Sulfur Enzymes. J Biol Chem. 2007;282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stadtman ER, Levine RL. Protein Oxidation. Ann N Y Acad Sci. 2000;899:191–208. doi: 10.1111/j.1749-6632.2000.tb06187.x. [DOI] [PubMed] [Google Scholar]
- 66.Perera A, Parkes HG, Herz H, Haycock P, Blake DR, Grootveld MC. High Resolution 1H NMR Investigations of the Reactivities of alpha-Keto Acid Anions with Hydrogen Peroxide. Free Radic Res. 1997;26:145–157. doi: 10.3109/10715769709097793. [DOI] [PubMed] [Google Scholar]
- 67.Vlessis AA, Bartos D, Trunkey D. Importance of spontaneous alpha-ketoacid decarboxylation in experiments involving peroxide. Biochem Biophys Res Commun. 1990;170:1281–1287. doi: 10.1016/0006-291x(90)90532-r. [DOI] [PubMed] [Google Scholar]
- 68.Yoshpe-Purer Y, Henis Y. Factors Affecting Catalase Level and Sensitivity to Hydrogen Peroxide in Escherichia coli . Appl Environ Microbiol. 1976;32:465–469. doi: 10.1128/aem.32.4.465-469.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seaver LC, Imlay JA. Alkyl Hydroperoxide Reductase Is the Primary Scavenger of Endogenous Hydrogen Peroxide in Escherichia coli . J Bacteriol. 2001;183:7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Imlay JA. Pathways of Oxidative Damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 71.Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: countering the phagocyte oxidative burst. Mol Microbiol. 2004;52:1291–1302. doi: 10.1111/j.1365-2958.2004.04078.x. [DOI] [PubMed] [Google Scholar]
- 72.Sund CJ, Rocha ER, Tzianabos AO, Wells WG, Gee JM, Reott MA, O’Rourke DP, Smith CJ. The Bacteroides fragilis transcriptome response to oxygen and H2O2: the role of OxyR and its effect on survival and virulence. Mol Microbiol. 2008;67:129–142. doi: 10.1111/j.1365-2958.2007.06031.x. [DOI] [PubMed] [Google Scholar]
- 73.Stohl EA, Criss AK, Seifert HS. The transcriptome response of Neisseria gonorrhoeae to hydrogen peroxide reveals genes with previously uncharacterized roles in oxidative damage protection. Mol Microbiol. 2005;58:520–532. doi: 10.1111/j.1365-2958.2005.04839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palma M, DeLuca D, Worgall S, Quadri LEN. Transcriptome Analysis of the Response of Pseudomonas aeruginosa to Hydrogen Peroxide. J Bacteriol. 2004;186:248–252. doi: 10.1128/JB.186.1.248-252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Adimora NJ, Jones DP, Kemp ML. A Model of Redox Kinetics Implicates the Thiol Proteome in Cellular Hydrogen Peroxide Responses. Antioxid Redox Signal. 2010;13:731–743. doi: 10.1089/ars.2009.2968. A recent kinetic model of the intracellular H2O2 chemical network in T cells. It includes several important redox maintenance enzymes, and is a good initial step toward a comprehensive model of the intracellular H2O2 network.
- 76.Makino N, Sasaki K, Hashida K, Sakakura Y. A metabolic model describing the H2O2 elimination by mammalian cells including H2O2 permeation through cytoplasmic and peroxisomal membranes: comparison with experimental data. Biochim Biophys Acta. 2004;1673:149–159. doi: 10.1016/j.bbagen.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 77.Johnson RM, Goyette G, Ravindranath Y, Ho YS. Hemoglobin autoxidation and regulation of endogenous H2O2 levels in erythrocytes. Free Radic Biol Med. 2005;39:1407–1417. doi: 10.1016/j.freeradbiomed.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 78.Svensson L, Poljakovic M, Save S, Gilberthorpec N, Schon T, Strid S, Corker H, Poole RK, Persson K. Role of flavohemoglobin in combating nitrosative stress in uropathogenic Escherichia coli - Implications for urinary tract infection. Microb Pathog. 2010;49:59–66. doi: 10.1016/j.micpath.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 79.Richardson AR, Dunman PM, Fang FC. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol Microbiol. 2006;61:927–939. doi: 10.1111/j.1365-2958.2006.05290.x. [DOI] [PubMed] [Google Scholar]