Abstract
Nuclear-encoded proteins targeted to the chloroplast are typically synthesized with N-terminal transit peptides which are proteolytically removed upon import. Structurally related proteins of 145 and 143 kDa copurify with a soluble chloroplast processing enzyme (CPE) that cleaves the precursor for the major light-harvesting chlorophyll a/b binding protein and have been implicated in the maturation of the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase and acyl carrier protein. The 145- and 143-kDa proteins have not been found as a heterodimer and thus may represent functionally independent isoforms encoded by separate genes. Here we describe the primary structure of a 140-kDa polypeptide encoded by cDNAs isolated by using antibodies raised against the 145/143-kDa doublet. The 140-kDa polypeptide contains a transit peptide, and strikingly, a His-Xaa-Xaa-Glu-His zinc-binding motif that is conserved in a recently recognized family of metalloendopeptidases, which includes Escherichia coli protease III, insulin-degrading enzyme, and subunit beta of the mitochondrial processing peptidase. Identity of 25-30%, concentrated near the N terminus of the 140-kDa polypeptide, is found with these proteases. Expression of CPE in leaves is not light dependent. Indeed, transcripts are present in dark-grown plants, and the 145/143-kDa doublet and proteolytic activity are both found in etioplasts, as well as in root plastids. Thus, CPE appears to be a necessary component of the import machinery in photosynthetic and nonphotosynthetic tissues, and it may function as a general stromal processing peptidase in plastids.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abad M. S., Clark S. E., Lamppa G. K. Properties of a Chloroplast Enzyme that Cleaves the Chlorophyll a/b Binding Protein Precursor : Optimization of an Organelle-Free Reaction. Plant Physiol. 1989 May;90(1):117–124. doi: 10.1104/pp.90.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abad M. S., Oblong J. E., Lamppa G. K. Soluble Chloroplast Enzyme Cleaves preLHCP Made in Escherichia coli to a Mature Form Lacking a Basic N-Terminal Domain. Plant Physiol. 1991 Aug;96(4):1220–1227. doi: 10.1104/pp.96.4.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affholter J. A., Fried V. A., Roth R. A. Human insulin-degrading enzyme shares structural and functional homologies with E. coli protease III. Science. 1988 Dec 9;242(4884):1415–1418. doi: 10.1126/science.3059494. [DOI] [PubMed] [Google Scholar]
- Bassham D. C., Bartling D., Mould R. M., Dunbar B., Weisbeek P., Herrmann R. G., Robinson C. Transport of proteins into chloroplasts. Delineation of envelope "transit" and thylakoid "transfer" signals within the pre-sequences of three imported thylakoid lumen proteins. J Biol Chem. 1991 Dec 15;266(35):23606–23610. [PubMed] [Google Scholar]
- Becker A. B., Roth R. A. An unusual active site identified in a family of zinc metalloendopeptidases. Proc Natl Acad Sci U S A. 1992 May 1;89(9):3835–3839. doi: 10.1073/pnas.89.9.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J., Jenkins G. I., Hartley M. R. Differential regulation of the accumulation of the light-harvesting chlorophyll a/b complex and ribulose bisphosphate carboxylase/oxygenase in greening pea leaves. J Cell Biochem. 1984;25(1):1–13. doi: 10.1002/jcb.240250102. [DOI] [PubMed] [Google Scholar]
- Chen N. Y., Jiang S. Q., Klein D. A., Paulus H. Organization and nucleotide sequence of the Bacillus subtilis diaminopimelate operon, a cluster of genes encoding the first three enzymes of diaminopimelate synthesis and dipicolinate synthase. J Biol Chem. 1993 May 5;268(13):9448–9465. [PubMed] [Google Scholar]
- Clark S. E., Lamppa G. K. Determinants for cleavage of the chlorophyll a/b binding protein precursor: a requirement for a basic residue that is not universal for chloroplast imported proteins. J Cell Biol. 1991 Aug;114(4):681–688. doi: 10.1083/jcb.114.4.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler-Blum G., Meier M., Frank J., Müller G. A. Reduction of background problems in nonradioactive northern and Southern blot analyses enables higher sensitivity than 32P-based hybridizations. Anal Biochem. 1993 May 1;210(2):235–244. doi: 10.1006/abio.1993.1189. [DOI] [PubMed] [Google Scholar]
- Finch P. W., Wilson R. E., Brown K., Hickson I. D., Emmerson P. T. Complete nucleotide sequence of the Escherichia coli ptr gene encoding protease III. Nucleic Acids Res. 1986 Oct 10;14(19):7695–7703. doi: 10.1093/nar/14.19.7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavel Y., von Heijne G. A conserved cleavage-site motif in chloroplast transit peptides. FEBS Lett. 1990 Feb 26;261(2):455–458. doi: 10.1016/0014-5793(90)80614-o. [DOI] [PubMed] [Google Scholar]
- Gehm B. D., Kuo W. L., Perlman R. K., Rosner M. R. Mutations in a zinc-binding domain of human insulin-degrading enzyme eliminate catalytic activity but not insulin binding. J Biol Chem. 1993 Apr 15;268(11):7943–7948. [PubMed] [Google Scholar]
- Guan K. L., Dixon J. E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991 Feb 1;192(2):262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Hawlitschek G., Schneider H., Schmidt B., Tropschug M., Hartl F. U., Neupert W. Mitochondrial protein import: identification of processing peptidase and of PEP, a processing enhancing protein. Cell. 1988 Jun 3;53(5):795–806. doi: 10.1016/0092-8674(88)90096-7. [DOI] [PubMed] [Google Scholar]
- Hirsch S., Muckel E., Heemeyer F., von Heijne G., Soll J. A receptor component of the chloroplast protein translocation machinery. Science. 1994 Dec 23;266(5193):1989–1992. doi: 10.1126/science.7801125. [DOI] [PubMed] [Google Scholar]
- Kirwin P. M., Elderfield P. D., Williams R. S., Robinson C. Transport of proteins into chloroplasts. Organization, orientation, and lateral distribution of the plastocyanin processing peptidase in the thylakoid network. J Biol Chem. 1988 Dec 5;263(34):18128–18132. [PubMed] [Google Scholar]
- Kuo W. L., Gehm B. D., Rosner M. R. Cloning and expression of the cDNA for a Drosophila insulin-degrading enzyme. Mol Endocrinol. 1990 Oct;4(10):1580–1591. doi: 10.1210/mend-4-10-1580. [DOI] [PubMed] [Google Scholar]
- Lamppa G. K., Abad M. S. Processing of a wheat light-harvesting chlorophyll a/b protein precursor by a soluble enzyme from higher plant chloroplasts. J Cell Biol. 1987 Dec;105(6 Pt 1):2641–2648. doi: 10.1083/jcb.105.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamppa G. K., Morelli G., Chua N. H. Structure and developmental regulation of a wheat gene encoding the major chlorophyll a/b-binding polypeptide. Mol Cell Biol. 1985 Jun;5(6):1370–1378. doi: 10.1128/mcb.5.6.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
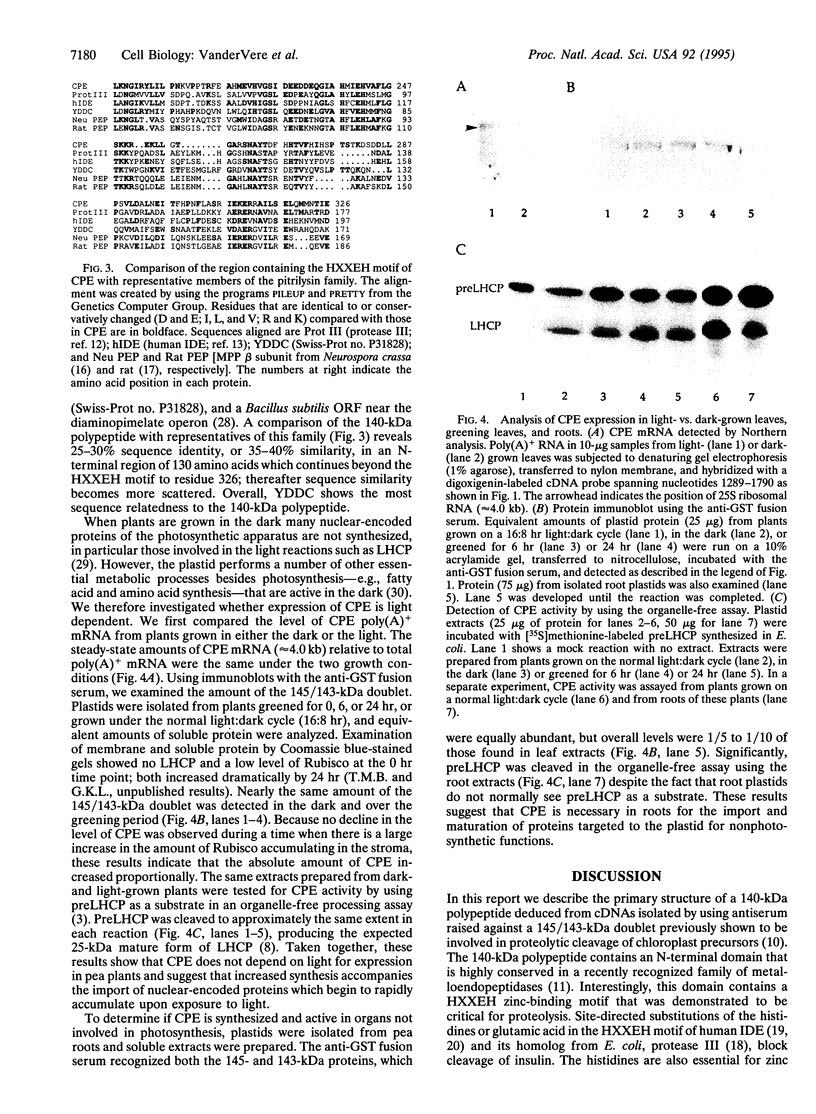
- Oblong J. E., Lamppa G. K. Identification of two structurally related proteins involved in proteolytic processing of precursors targeted to the chloroplast. EMBO J. 1992 Dec;11(12):4401–4409. doi: 10.1002/j.1460-2075.1992.tb05540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paces V., Rosenberg L. E., Fenton W. A., Kalousek F. The beta subunit of the mitochondrial processing peptidase from rat liver: cloning and sequencing of a cDNA and comparison with a proposed family of metallopeptidases. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):5355–5358. doi: 10.1073/pnas.90.11.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman R. K., Gehm B. D., Kuo W. L., Rosner M. R. Functional analysis of conserved residues in the active site of insulin-degrading enzyme. J Biol Chem. 1993 Oct 15;268(29):21538–21544. [PubMed] [Google Scholar]
- Pierotti A. R., Prat A., Chesneau V., Gaudoux F., Leseney A. M., Foulon T., Cohen P. N-arginine dibasic convertase, a metalloendopeptidase as a prototype of a class of processing enzymes. Proc Natl Acad Sci U S A. 1994 Jun 21;91(13):6078–6082. doi: 10.1073/pnas.91.13.6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N. D., Barrett A. J. Evolutionary families of peptidases. Biochem J. 1993 Feb 15;290(Pt 1):205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C., Ellis R. J. Transport of proteins into chloroplasts. Partial purification of a chloroplast protease involved in the processing of important precursor polypeptides. Eur J Biochem. 1984 Jul 16;142(2):337–342. doi: 10.1111/j.1432-1033.1984.tb08291.x. [DOI] [PubMed] [Google Scholar]
- Schnell D. J., Kessler F., Blobel G. Isolation of components of the chloroplast protein import machinery. Science. 1994 Nov 11;266(5187):1007–1012. doi: 10.1126/science.7973649. [DOI] [PubMed] [Google Scholar]
- Witte C., Jensen R. E., Yaffe M. P., Schatz G. MAS1, a gene essential for yeast mitochondrial assembly, encodes a subunit of the mitochondrial processing protease. EMBO J. 1988 May;7(5):1439–1447. doi: 10.1002/j.1460-2075.1988.tb02961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M. J., Geli V., Oppliger W., Suda K., James P., Schatz G. The MAS-encoded processing protease of yeast mitochondria. Interaction of the purified enzyme with signal peptides and a purified precursor protein. J Biol Chem. 1991 Apr 5;266(10):6416–6423. [PubMed] [Google Scholar]