Abstract
Apolipoprotein E3 (apoE3) is an anti-atherogenic apolipoprotein with the ability to exist in lipid-free and lipoprotein-associated states. During atherosclerosis, its function in promoting cholesterol efflux from macrophages via the ATP-binding cassette transporter A1 (ABCA1) takes a prominent role, leading to generation of nascent high density lipoprotein (nHDL) particles. The objective of this study is to understand the conformation adopted by apoE3 in macrophage-generated nHDL using a fluorescence spectroscopic approach involving pyrene. Pyrene-labeled recombinant human apoE3 displayed a robust ability to stimulate ABCA1-mediated cholesterol efflux from cholesterol-loaded J774 macrophages (which do not express apoE), comparable to that elicited by unlabeled apoE3. The nHDL recovered from the conditioned medium revealed the presence of apoE3 by immunoblot analysis. A heterogeneous population of nHDL bearing exogenously added apoE3 was generated with particle size varying from ~ 12 to ~19 nm in diameter, corresponding to molecular mass of ~ 450 to ~700 kDa. The lipid: apoE3 ratio varied from ~60:1 to 10:1. A significant extent of pyrene excimer emission was noted in nHDL, indicative of spatial proximity between Cys112 on neighboring apoE3 molecules similar to that noted in reconstituted HDL. Cross-linking analysis using Cys-specific cross-linkers revealed the predominant presence of dimers. Taken together the data indicate a double belt arrangement of apoE molecules on nHDL. A similar organization of the C-terminal tail of apoE on nHDL was noted when pyrene-apoEA277C(201-299) was used as the cholesterol acceptor. These studies open up the possibility of using exogenously labeled apoE3 to generate nHDL for structural and conformational analysis.
Keywords: Macrophage, apolipoproteins, nascent HDL, apoE3, pyrene fluorescence, cross-linking, reverse cholesterol transport
1. Introduction
The role of apoE3 in regulating plasma cholesterol homeostasis and as an anti-atherogenic agent is mediated by its ability to serve as a ligand for the LDL receptor family of proteins [1–3]. This facilitates cellular internalization of lipoproteins [4], thereby lowering plasma cholesterol levels. During atherosclerosis, apoE also functions in reverse cholesterol transport, which involves transport of cholesterol from peripheral tissues to liver via high density lipoproteins (HDL); under normal physiological conditions, this role is attributed to apolipoprotein AI (apoAI), a major exchangeable component of HDL [5]. However, in atherosclerosis apoE3 plays a dominant role with macrophages secreting large amounts of (lipid-free or lipid-poor) apoE3, which in turn presumably promotes cholesterol efflux via ATP-binding cassette transporter A1 (ABCA1) as a protective mechanism, with a resultant formation of nascent HDL (nHDL) containing apoE3 [6–8].
ApoE3 is composed of several amphipathic α-helices that are folded into an N-terminal domain (1-191) and a C-terminal domain (201-299) bearing high affinity binding sites for lipids [9]. In the lipid-free state, the former is folded as a 4-helix bundle wherein the hydrophobic face of each helix is sequestered towards the protein interior and the hydrophilic face is oriented towards the aqueous environment [10]. The amphipathic helices in the C-terminal domain mediate inter-molecular helix-helix interactions and protein tetramerization [11,12].
As exchangeable apolipoproteins, both apoAI and apoE3 exhibit the capability to exist in lipid-free and lipoprotein-bound states, with a conformational change accompanying the transition between the two states. Several biophysical approaches indicate that reconstitution of HDL using apoAI and synthetic phospholipids (typically DMPC or POPC) and cholesterol results in the protein adopting a double belt-like organization of α-helices circumscribing a bilayer of phospholipids [5,13,14]. Much less is known about the organization of lipid-associated apoE3 although the available data suggest a similar organization in reconstituted HDL (rHDL) [15–18]. While this “bottom-up” approach has yielded significant information regarding the functional organization of apolipoproteins, knowledge regarding their conformation in nHDL generated by macrophages is lacking. In this study, we adopt a “top-down” approach to investigate the conformation of exogenously added human apoE3 in nHDL by allowing J774 macrophages (which do not synthesize apoE) to assemble the lipoprotein particle in an ABCA1-dependent process. Other groups employing this approach used radiolabeled-apoAI or -lipids (due to the small amounts of HDL generated by macrophages) to study the functional composition of nHDL [19]. In the present study, we use single Cys containing apoE3 or its isolated C-terminal domain to study their organization in macrophage-assembled nHDL using a combination of spectroscopic and cross-linking approach.
2. Materials and methods
2.1 Purification of apoE3 and apoE(201-299)
Recombinant human apoE3(1-299) or apoE(201-299) with a hexa-His tag at the N-terminal end was purified as described previously [12]. ApoE(201-299) bears a single substituted cysteine at position 277 to enable labeling with an exogenous probe. Protein concentration was determined in a Nano-Drop spectrometer using molar extinction coefficient at 280 nm (44,460 M−1 cm−1 for apoE3(1-299) and 16,500 M−1 cm−1 for apoE(201-299).
2.2 Labeling with N-(1-pyrene)maleimide (NPM)
The naturally occurring single Cys112 in apoE3 and the substituted single cysteine in apoE(201-299) (A277C) were used to label with NPM [12,20]. The pyrene (pyr)-labeled sample was dialyzed against 10 mM sodium phosphate, pH 7.4, 150 mM NaCl (phosphate buffered saline, PBS). The stoichiometry of labeling was calculated to be ~ 1.0 using the molar extinction coefficient of pyrene at 345 nm in methanol (40,000 M−1 cm−1).
2.3 Biogenesis of nHDL containing apoE3
J774 mouse macrophages were grown in RPMI-1640 with 1% FBS for 48 h as described previously [21,22]. Cells from a confluent culture were plated with a 1:10 dilution onto twelve T175 flasks and exposed to 0.3 mM cAMP analogue (cpt-cAMP) for 18 h to up-regulate ABCA1 expression. They were rinsed extensively, and then exposed to 10 μg/ml of unlabeled or pyrene-labeled apoE3 or apoEA277C(201-299) in serum-free medium for 18 h. The conditioned medium (250 ml) was filtered through 0.45 μm PVDF membrane filter, and concentrated in a 10-kDa cut-off Amicon filter. Lipid-bound protein was separated from unbound protein by density gradient ultracentrifugation (435,000 × g, 5.5 h) using a KBr gradient (density range between 1.063–1.21 g/ml) as described previously [23] (0.5 g KBr/ml). About 10 fractions were collected and those containing both protein and lipid were pooled and concentrated. Under the conditions employed, fractions corresponding to the density range 1.10–1.13 g/ml are referred to as ‘Top’ (typically fractions 1 to 4) and those in 1.14–1.17 g/ml range as ‘Mid’ fractions (typically fractions 5 to 7) (designated based on identical conditions using PBS). The protein content of the lipoprotein complexes was measured using the DCTM protein assay kit (BioRad Laboratories, Hercules, CA).
2.4 Preparation of rHDL containing apoE3
rHDL containing unlabeled or pyrene-labeled apoE3 or apoEA277C(201-299), and, POPC or POPC/cholesterol was prepared by the cholate dialysis method with slight modifications [17]. Briefly, ~10 mg sodium deoxycholate was added to a thin film of POPC (10 mg) or POPC/cholesterol (between 10:1 and 1:1 ratio phospholipid: cholesterol, total lipid ~10 mg) in 1 ml PBS, vortexed until clear, incubated for 2 h with 1 ml of apoE (4 mg/ml) at 37 °C for 1 h and dialyzed extensively at 4 °C. Lipid-bound protein was isolated by density gradient ultracentrifugation and, characterized in terms of particle size and diameter. The protein and phospholipid content of rHDL was measured as described above; cholesterol content (both free cholesterol and cholesteryl ester, CE) was measured using the Amplex® Red kit (Life Technologies, Grand Island, NY).
2.5 Western blot
The nHDL and rHDL samples were electrophoresced on a SDS-PAGE 4–20% acrylamide gradient Tris-glycine gel and visualized by Western blot using goat anti-apoE-HRP antibody. The intact complexes were observed by electrophoresis for 20 h at 45 V under non-denaturing conditions using ~ 5 μg sample, followed by Western blot; the molecular mass and average particle size were determined using Amersham High Molecular Weight Standards.
2.6 Fluorescence measurements
Steady state fluorescence analyses were performed on a Perkin-Elmer LS55B fluorometer at 24 °C. Fluorescence emission spectra of pyrene-labeled samples (~10 μg) were recorded between 350 and 550 nm (excitation at 345 nm, excitation and emission slit-widths at 5 nm, and the scan speed at 50 nm/min).
2.7 Cross-linking
The nHDL and rHDL samples (1–10 μg) were incubated with 5-fold molar excess of tris(2-carboxyethyl)phosphine at 24 °C for 30 min, followed by treatment with increasing concentrations of 1,6-bismaleimidohexane (BMH) (0-, 25-, 50-, 100-fold molar excess over protein) at 24 °C for 1 h. The cross-linked species were electrophoresced under denaturing, reducing conditions and visualized by Western blot.
3. Results and discussion
Pyr-apoE3 stimulated a robust ABCA1-dependent [3H]-cholesterol efflux in J774 macrophages, comparable to that of unlabeled protein, Fig 1A; the stimulation was dose- dependent at the concentrations used (5, 10 and 20 μg labeled protein/ml), Fig 1B. This indicates that the presence of the covalently attached exogenous probe, added for subsequent spectroscopic measurements, does not alter the functional ability of the protein. Previous studies have shown that the C-terminal domain of apoE3 encompassing residues 222-299 was necessary and sufficient to promote efficient ABCA1-mediated cholesterol efflux [21]. In the current study, we confirmed that apoE(201-299) served as an efficient acceptor of cholesterol efflux, Fig 1C, with the stimulation being dose dependent as well, Fig 1D.
Fig. 1.
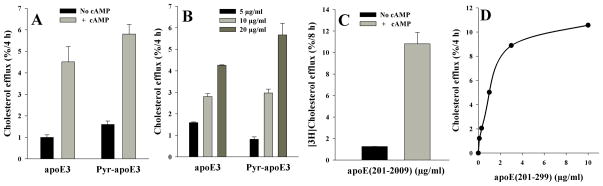
[3H]Cholesterol efflux mediated by unlabeled, pyr-apoE3 or apoE(201-299). A. J774 macrophages were labeled with [3H]cholesterol (1 μCi/mL) in RPMI-1640 with 1% FBS for 48 h. ABCA1 expression was up-regulated for 18 h with cpt-cAMP, followed by exposure to 10 μg/ml unlabeled or pyr-apoE3 in serum-free RPMI-1640 medium. The amount of [3]cholesterol appearing in the medium after 4 h was expressed as % of the radioactivity present in cells at time zero. A parallel experiment with no cpt-cAMP was conducted to assess efflux in the absence of ABCA1 up-regulation. Background release of [3H]cholesterol to serum-free medium was subtracted from values obtained with added proteins. Values are mean ± SD, n=3. B. Dose-dependent (5, 10 and 20 μg/ml) efflux for unlabeled apoE3 or pyr-apoE3. C. ABCA1-mediated cholesterol efflux promoted by apoE(201-299). All conditions employed were as described above except that the amount of [3]cholesterol appearing in the medium was followed 8 h after addition of 3 μg/ml apoE(201-299) as acceptor in the absence or presence of cpt-cAMP. Values are mean ± SD, n=3. D. Dose-dependent cholesterol efflux (0.1 – 10 μg/ml) promoted by apoE(201-299). Data shown are representative of 2 independent experiments.
These initial experiments laid the groundwork for subsequent studies using larger culture volumes bearing non-radioactive cholesterol for biophysical analysis. Following efflux, lipid-associated unlabeled or pyr-apoE3 was separated from lipid-free protein in the conditioned medium. The presence of apoE3 (in Top and Mid fractions) was confirmed by Western blot, Fig 2A. Unlabeled apoE3 in rHDL reconstituted with POPC is shown for comparison. Fig 2B shows nHDL bearing unlabeled apoE(201-299) or pyr-apoEA277C(201-299) (only the Top fraction) and the corresponding rHDL that were isolated as described above.
Fig. 2.
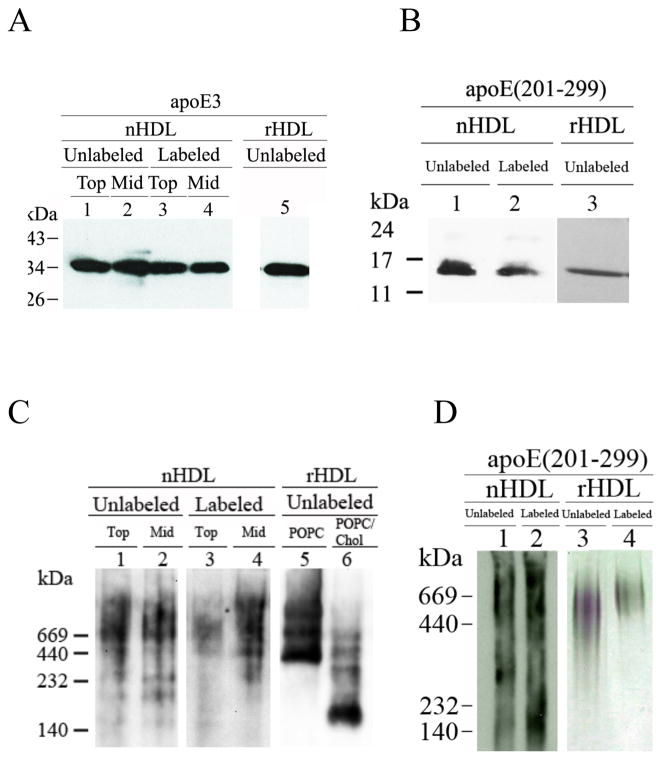
Western blot analysis of nHDL and rHDL containing apoE3 or apoE(201-299). A. SDS-PAGE of apoE3 containing nHDL and rHDL. The nHDL samples containing unlabeled (lanes 1 & 2) or pyr-apoE3 (lanes 3 & 4) (1 μg protein) were electrophoresced on a SDS-PAGE gel under reducing conditions. Lanes 1 & 3 represent Top, while lanes 2 & 4 represent the Mid fractions. Unlabeled apoE3 in rHDL (POPC/apoE3) is shown for comparison (lane 5). ApoE was detected using apoE-HRP antibody. B. SDS-PAGE of apoE(201-299) containing nHDL and rHDL. nHDL containing apoE(201-299) (lane 1) or pyr-apoEA277C(201-299) (lane 2) were electrophoresced as described under A and detected with 3H1 as the primary antibody. Unlabeled apoEA277C(201-299) in rHDL reconstituted with POPC is shown for comparison (lane 3). C. Non-denaturing PAGE of apoE3 containing nHDL and rHDL. The nHDL samples containing unlabeled (lanes 1 & 2) or pyr-apoE3 (lanes 3 & 4) (5 μg protein) were electrophoresced under non-denaturing conditions. Lanes 1 & 3 represent Top, while lanes 2 & 4 represent the Mid fractions. Unlabeled apoE in rHDL reconstituted with POPC (lane 5) or POPC/cholesterol (lane 6) are shown for comparison. D. Non-denaturing PAGE of apoE(201-299) containing nHDL and rHDL. nHDL containing apoE(201-299) (lane 1) or pyr-apoEA277C(201-299) (lane 2) were electrophoresced as described under C and visualized as in B. Unlabeled and pyr-apoEA277C(201-299) in rHDL reconstituted with POPC are shown for comparison (lanes 3 & 4, respectively). The Stokes’ diameter of the standards shown in C and D (from high to low) are 17, 12.2, 9.2 and 8.1 nm.
The intact nHDL bearing unlabeled or labeled apoE3 was observed by Western blot under non-denaturing conditions (Fig 2C, lanes 1-4). In both cases, a heterogeneous pool of nHDL particles was observed in the Top and Mid fractions; the mass varied from 450 to 700 kDa (12–17 nm diameter); in addition, a minor population with molecular mass ~ 250 kDa (9 nm diameter) was observed in the Mid fraction containing apoE3. The lipid: protein ratio in the Top and Mid fractions was found to be 60:1 and 10:1, respectively; the presence of the probe did not affect this ratio. Similarly, rHDL samples (Fig 2C) containing POPC and apoE3 also contained large diameter particles (lane 5), while those containing POPC, cholesterol and apoE3 (lane 6) revealed the additional presence of smaller diameter (8–9 nm) particles. The molecular mass and average diameter of the nHDL particles bearing unlabeled or pyrene-labeled apoE(201-299), (Fig 2D, lanes 1 and 2, respectively) were found to be 140 – 700 kDa range corresponding to 8–17 nm particle diameter. For comparison, rHDL bearing POPC, cholesterol and unlabeled or pyrene-labeled apoE(201-299) is shown, Fig 2D, lanes 3 and 4, respectively. Their mass ranged from 450 to 700 kDa corresponding to 12–17 nm diameter. We did not detect any significant CE in these nHDL, regardless of the presence or absence of C-terminal domain or pyrene label.
Having established the lipid-associated state of pyr-apoE3 in nHDL, its organization was evaluated by comparing the fluorescence emission spectra with that of lipid-free protein. NPM was the fluorophore of choice because: (i) it has a high molar extinction coefficient, which enables us to employ physiologically relevant amounts of labeled protein, which is often not the case in spectroscopic measurements; (ii) it offers information regarding spatial proximity, exhibiting featureless excimer emission at ~460 nm when two pyrene moieties are ~ 10 Å from each other; this is in addition to the monomeric fluorescence emission (an ensemble of 5 major vibronic bands), corresponding to peaks at ~375, 379, 385, 395 and 405 nm, respectively, and, (iii) the peak at 385 nm is exquisitely sensitive to the polarity of the probe’s microenvironment, which offers definitive information about the lipid-associated state of the protein. The appearance of a higher 385 nm emission peak (compared to 375 nm) is indicative of a hydrophobic microenvironment in the probe vicinity [24,25].
Lipid-free pyr-apoE3 shows a strong excimer emission (a) indicative of ~ 10 Å proximity between C112 in neighboring monomeric units in a tetramer, Fig 3A. Significant excimer emission was noted in both Top (b) and Mid (c) fractions of nHDL, suggesting ~ 10 Å proximity between C112 in neighboring apoE3 molecules in nHDL. Further, nHDL samples revealed a prominent peak at 385 nm (compared to 375 nm); this suggests that the pyrene moiety is located in a hydrophobic microenvironment, confirming lipid-association of apoE3. Fluorescence emission spectra of rHDL reconstituted with pyr-apoE3 and POPC (d) or POPC/cholesterol (e) are shown for comparison; the extent of excimer emission and 385 nm peak in these cases are comparable to that observed in nHDL samples.
Fig. 3.
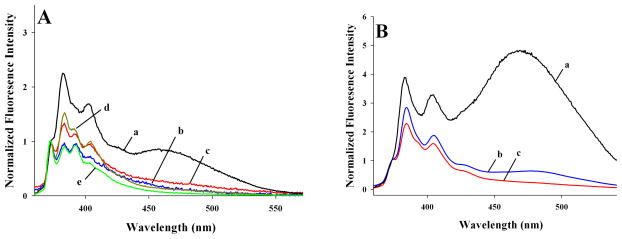
Fluorescence emission spectra of nHDL- and rHDL-associated pyrene-labeled apoE. A. Fluorescence emission spectra of pyr-apoE3 (~ 5 μg/ml protein) were recorded in lipid-free state (a), nHDL Top (b) and Mid (c) fractions and rHDL-associated states with POPC (d) or POPC/cholesterol (e). B. Fluorescence emission spectra of pyr-apoEA277C(201-299) were recorded in lipid-free state (a), nHDL (b) and rHDL-associated states with POPC/cholesterol (c). The spectra shown are average of 4 scans (excitation at 345 nm) and normalized with respect to the peak at 375 nm.
Lipid-free pyr-apoEA277C(201-299) displays a dramatic excimer emission, Fig 3B (a), as reported previously [12]; this is due to tetramerization mediated by the CT domain, which would align position 277 from four monomeric units [11,12] and bring 4 pyrene rings into close proximity. In nHDL bearing pyr-apoEA277C(201-299), a significant excimer emission peak was also noted (b), similar to that seen in the corresponding rHDL prepared with POPC/cholesterol (10:1 weight ratio) (c), Fig 3B. This is indicative of juxtaposition of position 277 from neighboring molecules in lipid-associated apoEA277C(201-299).
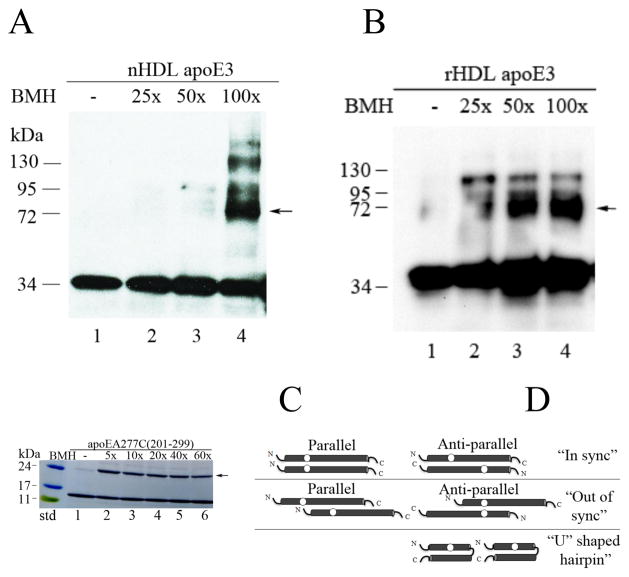
To independently confirm the juxtaposition of apoE3 molecules in the lipid-associated state, nHDL (Mid fractions) bearing apoE3 was subjected to cross-linking using increasing molar concentrations of BMH, Fig 4A. BMH is a homobifunctional agent that reacts predominantly with free sulfhydryls forming stable non-reducible thioether cross-links, and leading to dimeric proteins if the Cys are ~10 Å apart. The results obtained were compared with cross-linking carried out under similar conditions using rHDL, Fig 4B. In both cases, a major band corresponding to dimeric apoE3 was noted (arrow). In addition, a minor band corresponding to ~ 130 kDa was noted, the reason for which is unclear since apoE3 has a single Cys. It may be due to cross-linking between amino groups on tandem apoE molecules leading to appearance of tetrameric species: although maleimide groups are ~1000-fold more reactive with -SH than -NH3+ at pH 7.0, it is possible that BMH reacts with primary amines. A dimeric form was noted upon cross-linking in rHDL bearing POPC/cholesterol and apoEA277C(201-299) at all concentrations of BMH, Fig 4C.
Fig. 4.
Cross-linking of nHDL and rHDL. nHDL (A) or rHDL (B) bearing 1 μg unlabeled apoE3 was cross-linked as described under Methods by treatment with increasing concentrations of BMH as indicated. The samples were electrophoresced by SDS-PAGE under reducing conditions, and visualized by Western blot using apoE-HRP antibody. C. rHDL bearing 10 μg unlabeled apoEA277C(201-299) was cross-linked as above, electrophoresced and visualized with Amido Black. Arrows draw attention to bands corresponding to cross-linked dimers of apoE3 or apoEA277C/(201-299). D. Possible models of apoE organization of nHDL. The circles represent location of single Cys.
Taken together, our results show that J774 macrophages generate nHDL using exogenously added fluorescently-labeled apoE3 or apoEA277C(201-299) that can be examined by biophysical approaches. Where possible, we compared data obtained with nHDL with that from rHDL bearing the corresponding apoE. A direct comparison is not possible at this juncture since the apoE-containing nHDL generated herein are highly heterogeneous in terms of particle composition. This appears to be a characteristic feature of ABCA1-mediated nHDL biogenesis, as exemplified by studies carried out with apoAI with different cell types: baby hamster kidney or HEK293 cells [19] expressing ABCA1. In general, for reasons not clearly understood at present, apoE3 appears to generate predominantly larger discoidal nHDL particles, a trend also noted in rHDL bearing POPC and apoE3. In contrast, apoAI can give rise to both small and large particles with POPC [19]. Further, other labs have shown that in contrast to cells, which have the capability to incorporate large amounts of cholesterol in the phospholipid bilayer of discoidal HDL with apoAI, it is difficult to achieve rHDL particles containing > 30 mol % cholesterol [19]. We made a similar observation with apoE3; J774 cells were able to generate apoE3-containing nHDL with 1:1 phospholipid: cholesterol, while the reconstitution procedure yields rHDL with 10:1 ratio under the conditions employed herein, regardless of the initial ratio of phospholipid: cholesterol used in reconstitution protocol (2:1 to 1:10) (data not shown).
Our current understanding of rHDL containing apoE3 has evolved based on observations from different biophysical and spectroscopic data using apoE3 and DMPC. ApoE3 undergoes a conformational change upon lipid association, wherein the C-terminal domain unfurls away from the N-terminal domain, and the N-terminal 4-helix bundle undergoes opening with the helices moving apart from each other. Our current results with rHDL suggest a similar conformational re-organization of apoE3 upon interaction with POPC or POPC/cholesterol. This would allow the hydrophobic sides of the amphipathic helices to face the lipid environment to yield a stable lipoprotein complex. The helices of apoE3 appear to circumscribe the periphery like a belt around a bilayer of phospholipid (and cholesterol) molecules (~ 40 Å high) like a belt to form a discoidal lipoprotein complex.
Further, we propose a similar conformational re-organization of apoE3 in nHDL as well. The presence of an excimer peak in nHDL suggests that Cys112 from two neighboring molecules are juxtaposed, with the apoE3 molecules wrapped around the periphery of the lipid bilayer. The Cys-specific cross-linking data that show the formation of a dimer support this conclusion. The isolated C-terminal domain of apoE also appears to adopt a similar organization in nHDL and in rHDL with POPC/cholesterol.
During HDL biogenesis, the nHDL undergoes maturation to spherical HDL comprising a monolayer of amphipathic lipid and protein encompassing a core of CE. The conversion is catalyzed by lecithin-cholesterol acyltransferase (LCAT), which transfers a fatty acyl chain from phospholipid to cholesterol to form CE that transitions to the core. Since we did not detect any significant levels of CE in nHDL, and since the culture medium lacks LCAT, we believe that the nHDL generated herein is discoidal in geometry.
Given the particle heterogeneity, the precise number of apoE molecules per discoidal complex is not known at present. Based on particle mass and compositional analysis, we estimate that there would be 4 to 6 apoE3 molecules per particle. Regardless of the number, they appear to be organized as a parallel “in sync” or anti-parallel “out of sync” double belt circumscribing the periphery of discoidal nHDL, Fig 4D. If apoE molecules are arranged in a perfectly anti-parallel manner (the N-terminal end of one molecule juxtaposed to the C-terminal end of the neighboring molecule), the distance between C112 in apoE3 (or A277C in apoE(201-299)) would be far ≫10Å and would not form an excimer or a cross-link; therefore the anti-parallel “in sync” and parallel “out of sync” (beyond 10 Å) orientation are excluded. Based on this data, we also exclude a helical “hairpin” conformation wherein a single apoE3 molecule folds on itself.
Taken together, the current data suggest that neighboring apoE3 molecules are arranged like a double belt in nHDL. Our study offers the opportunity to investigate the conformation of apolipoproteins in macrophage-assembled nHDL in fine detail using rational design since it allows introduction of a spectroscopic probe of choice at any desired location on the protein.
Pyrene-labeled human apoE3 mediates efficient macrophage ABCA1 cholesterol efflux
Nascent HDL generated by J774 macrophages bears exogenously added pyrene-apoE3
Pyrene excimer emission indicates spatial proximity between 2 apoE3 on nascent HDL
Cross-linking reveal apoE3 spatial proximity in nascent and reconstituted HDL
Powerful new approach to examine HDL biogenesis by biophysical and spectroscopic tools
Acknowledgments
This work was funded by NIH-GM105561, Maria Erlinda Co Sarno (SHK) and McAbee-Overstreet Graduate Research award (SK).
ABBREVIATIONS
- ABCA1
ATP-binding cassette transporter A1
- ApoE3
Apolipoprotein E3
- nHDL
Nascent high density lipoprotein
- POPC
Palmitoyloleoylphosphatidylcholine
- rHDL
reconstituted high density lipoprotein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhang SH, Reddick RL, Piedrahita JA, et al. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science. 1992;258:468–471. doi: 10.1126/science.1411543. [DOI] [PubMed] [Google Scholar]
- 2.Ghiselli G, Schaefer EJ, Gascon P, et al. Type III hyperlipoproteinemia associated with apolipoprotein E deficiency. Science. 1981;214:1239–1241. doi: 10.1126/science.6795720. [DOI] [PubMed] [Google Scholar]
- 3.Rudenko G, Deisenhofer J. The low-density lipoprotein receptor: ligands, debates and lore. Curr Opin Struct Biol. 2003;13:683–689. doi: 10.1016/j.sbi.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Mahley RW, Rall SC., Jr Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 5.Lund-Katz S, Phillips MC. High density lipoprotein structure-function and role in reverse cholesterol transport. Subcell Biochem. 2010;51:183–227. doi: 10.1007/978-90-481-8622-8_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Eckardstein A, Nofer JR, Assmann G. High density lipoproteins and arteriosclerosis. Role of cholesterol efflux and reverse cholesterol transport. Arterioscler Thromb Vasc Biol. 2001;21:13–27. doi: 10.1161/01.atv.21.1.13. [DOI] [PubMed] [Google Scholar]
- 7.Mahley RW, Huang Y, Weisgraber KH. Putting cholesterol in its place: apoE and reverse cholesterol transport. J Clin Invest. 2006;116:1226–1229. doi: 10.1172/JCI28632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yokoyama S. ABCA1 and biogenesis of HDL. J Atheroscler Thromb. 2006;13:1–15. doi: 10.5551/jat.13.1. [DOI] [PubMed] [Google Scholar]
- 9.Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Wilson C, Wardell MR, Weisgraber KH, et al. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 11.Westerlund JA, Weisgraber KH. Discrete carboxyl-terminal segments of apolipoprotein E mediate lipoprotein association and protein oligomerization. J Biol Chem. 1993;268:15745–15750. [PubMed] [Google Scholar]
- 12.Patel AB, Khumsupan P, Narayanaswami V. Pyrene fluorescence analysis offers new insights into the conformation of the lipoprotein-binding domain of human apolipoprotein E. Biochemistry. 2010;49:1766–1775. doi: 10.1021/bi901902e. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Lyles DS, Thomas MJ, et al. Structural determination of lipid-bound ApoA-I using fluorescence resonance energy transfer. J Biol Chem. 2000;275:37048–37054. doi: 10.1074/jbc.M005336200. [DOI] [PubMed] [Google Scholar]
- 14.Davidson WS, Hilliard GM. The spatial organization of apolipoprotein A-I on the edge of discoidal high density lipoprotein particles: a mass specrometry study. J Biol Chem. 2003;278:27199–27207. doi: 10.1074/jbc.M302764200. [DOI] [PubMed] [Google Scholar]
- 15.Narayanaswami V, Kiss RS, Weers PM. The helix bundle: a reversible lipid binding motif. Comp Biochem Physiol Part A. 2010;155:123–133. doi: 10.1016/j.cbpa.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narayanaswami V, Szeto SS, Ryan RO. Lipid association-induced N- and C-terminal domain reorganization in human apolipoprotein E3. J Biol Chem. 2001;276:37853–37860. doi: 10.1074/jbc.M102953200. [DOI] [PubMed] [Google Scholar]
- 17.Narayanaswami V, Maiorano JN, Dhanasekaran P, et al. Helix orientation of the functional domains in apolipoprotein e in discoidal high density lipoprotein particles. J Biol Chem. 2004;279:14273–14279. doi: 10.1074/jbc.M313318200. [DOI] [PubMed] [Google Scholar]
- 18.Raussens V, Fisher CA, Goormaghtigh E, et al. The low density lipoprotein receptor active conformation of apolipoprotein E. Helix organization in N-terminal domain-phospholipid disc particles. J Biol Chem. 1998;273:25825–25830. doi: 10.1074/jbc.273.40.25825. [DOI] [PubMed] [Google Scholar]
- 19.Lund-Katz S, Lyssenko NN, Nickel M, et al. Mechanisms responsible for the compositional heterogeneity of nascent high density lipoprotein. J Biol Chem. 2013;288:23150–23160. doi: 10.1074/jbc.M113.495523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bains GK, Kim SH, Sorin EJ, et al. The extent of pyrene excimer fluorescence emission is a reflector of distance and flexibility: analysis of the segment linking the LDL receptor-binding and tetramerization domains of apolipoprotein E3. Biochemistry. 2012;51:6207–6219. doi: 10.1021/bi3005285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vedhachalam C, Narayanaswami V, Neto N, et al. The C-terminal lipid-binding domain of apolipoprotein E is a highly efficient mediator of ABCA1-dependent cholesterol efflux that promotes the assembly of high-density lipoproteins. Biochemistry. 2007;46:2583–2593. doi: 10.1021/bi602407r. [DOI] [PubMed] [Google Scholar]
- 22.Bielicki JK, Zhang H, Cortez Y, et al. A new HDL mimetic peptide that stimulates cellular cholesterol efflux with high efficiency greatly reduces atherosclerosis in mice. J Lipid Res. 2010;51:1496–1503. doi: 10.1194/jlr.M003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamamizu-Kato S, Cohen JK, Drake CB, et al. Interaction with amyloid beta peptide compromises the lipid binding function of apolipoprotein E. Biochemistry. 2008;47:5225–5234. doi: 10.1021/bi702097s. [DOI] [PubMed] [Google Scholar]
- 24.Dong DC, Winnik MA. The Py scale of solvent polarities. Can J Chem. 1984;62:2560–2565. [Google Scholar]
- 25.Karpovich DS, Blanchard GJ. Relating the polarity-dependent fluorescence response of pyrene to vibronic coupling. Achieving a fundamental understanding of the py polarity scale. J Phys Chem. 1995;99:3951–3958. [Google Scholar]