SUMMARY
The important human pathogen Streptococcus pyogenes (the group A streptococcus or GAS) produces many virulence factors that are regulated by the two-component signal transduction system CovRS (CsrRS). Dissemination of GAS infection originating at the skin has been shown to require production of streptokinase, whose transcription is repressed by CovR. In this work we have studied the interaction of CovR and phosphorylated CovR (CovR-P) with the promoter for streptokinase, Pska. We found that, in contrast to the other CovR-repressed promoters, Pska regulation by CovR occurs through binding at a single ATTARA consensus binding sequence (CB) that overlaps the −10 region of the promoter. Binding of CovR to other nearby consensus sequences occurs upon phosphorylation of the protein, but these other CBs do not contribute to the regulation of Pska by CovR. Thus, binding at a specific site does not necessarily indicate that site is involved in regulation by CovR. In addition, at Pska, CovR binding to the different sites does not appear to involve cooperative interactions, which simplifies the analysis of CovR binding and gives us insight into the modes of interaction that occur between CovR and its specific DNA binding sites. Finally, the observation that regulation of transcription from Pska occurs at a very low concentration of phosphorylated CovR may have important implications for the regulation of virulence gene expression during group A streptococcal infection.
INTRODUCTION
The group A streptococcus (GAS) or Streptococcus pyogenes is a strictly human pathogen that can cause many different illnesses ranging from mild localized infections to severe disseminated disease (for reviews, (Cunningham, 2000; Kwinn & Nizet, 2007; Musser & DeLeo, 2005; Tart et al., 2007). The variety of possible disease outcomes of a GAS infection is believed to be due in part to the regulated expression of streptococcal virulence factors that are either present on the surface of the bacterium or secreted by the organism. One of the secreted factors shown to be important for streptococcal virulence is streptokinase, encoded by the ska gene. GAS strains with specific ska alleles have been associated with cases of acute poststreptococcal glomerulonephritis and have been implicated in pathogenesis of this disease (Nordstrand et al., 2000). Streptokinase converts plasminogen into plasmin to activate it for fibrinolysis. The streptokinase produced by GAS is specific for the human form of plasmin, showing little or no activity on plasmin from mice or other animals. The importance of GAS streptokinase in pathogenesis was demonstrated using transgenic mice that express human plasminogen (Cole et al., 2006; Sun et al., 2004).
In GAS, as well as in Streptococcus equisimilis, streptokinase is expressed as a monocistronic operon (Malke et al., 2000). The promoter for the streptokinase gene, Pska, is activated by the Fas system (Kreikemeyer et al., 2001) and repressed by CovR (Federle et al., 1999; Heath et al., 1999). Both of these transcriptional regulators, which are conserved in all 12 GAS genome sequences available, are part of two-component signal transduction systems (TCSs; (Laub & Goulian, 2007; Stock et al., 2000). The CovRS two-component regulatory system is a global regulator of GAS gene expression that appears to have central importance in pathogenesis.
CovRS regulates expression of approximately 15% of the GAS genome (Dalton & Scott, 2004; Dalton et al., 2006; Graham et al., 2002) and mediates the bacterial response to several stress conditions (Dalton & Scott, 2004). In infection in primates, alterations in CovRS expression have been correlated with stages of infection, and maximal covRS transcription occurs at the transition from localized to disseminated systemic infection (Virtaneva et al., 2005). Although most two-component systems activate the promoters they regulate, CovR represses most of the genes it regulates, including ska (Federle et al., 1999; Miller et al., 2001). In addition to ska, other important virulence genes are repressed by CovR (Federle et al., 1999; Heath et al., 1999; Levin & Wessels, 1998), including genes required for capsule synthesis (has) (Ashbaugh et al., 2000; Husmann et al., 1997; Wessels & Bronze, 1994); the cytolysin streptolysin S (sag) (Salim et al., 2007); the Dnase streptodornase (sda) (Brinkmann et al., 2004; Buchanan et al., 2006; Sumby et al., 2005; Walker et al., 2007); and a cysteine protease (speB) (Collin & Olsen, 2001). Recently, the downstream transcriptional regulator RivR (encoded by rivR), which (indirectly) activates expression of the first recognized GAS virulence factor, the M protein, was also shown to be repressed by CovR (Roberts & Scott, 2007). CovR also represses its own expression as well as expression of the cognate CovS kinase, which is transcribed from the same promoter (Gusa & Scott, 2005).
Regulation of transcription of these genes is direct, through binding of CovR at specific sites near their promoters. Binding of CovR to the promoters that have been studied (Phas, Psag, Priv and Pcov) shows several common characteristics (Fig. 1; (Churchward, 2007): 1) multiple CovR binding sites are required at each promoter, 2) binding sites are located both upstream and downstream of the transcriptional start site, and 3) bound regions typically include the consensus sequence ATTARA (Federle & Scott, 2002; Gao et al., 2005; Gusa & Scott, 2005). Alteration of the consensus sequence by mutation of the TT pair to GG abolishes binding to at least part of the region containing the consensus sequence and uracil interference experiments show that these paired T residues are essential for CovR binding to this region (Federle & Scott, 2002; Gao et al., 2005; Gusa & Scott, 2005).
Figure 1. CovR binding sites at different promoters.
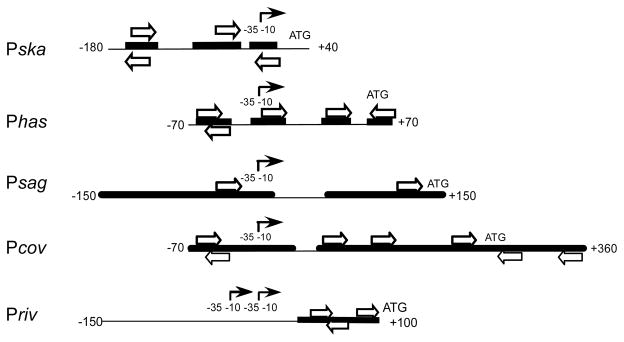
The interaction between CovR and different promoters is represented by regions protected by CovR from nuclease digestion (heavy black lines) and consensus or near-consensus CovR binding sequences (open arrows). (For clarity, not all near-consensus sequences are shown.) The promoters (indicated by the solid arrows) are aligned by their start points of transcription. Only one of the two closely overlapping promoters of Pska is shown.
Phosphorylation of CovR causes the protein to dimerize (Gusa et al., 2006) and increases its binding affinity to specific DNA regions (for review see (Churchward, 2007). In addition, phosphorylation of CovR often causes multimerization of the protein along the DNA to which it binds. Phosphorylation also affects the degree of repression of transcription from each promoter. Using an in vitro system that employs purified GAS RNA polymerase (Gusa et al., 2006), we have found that at some promoters there is good concordance between the amount of protein required to cause nuclease protection and to reduce transcription by 90% or more (Gao et al., 2005; Gusa & Scott, 2005). At other promoters, such as Phas (Gusa et al., 2006) and Priv (Roberts et al., 2007), phosphorylation has a larger effect on repression than on DNA binding, indicating that phosphorylation of CovR allows regulatory mechanisms in addition to simple DNA binding to come into play.
Transcription of streptokinase (ska) has been investigated extensively in Streptococcus equisimilis strain H46A (group C streptococcus or GCS, where it is called skc (Malke et al., 2000). In GAS, as well as in GCS, streptokinase is transcribed divergently from the adjacent gene, lrp, and the 274bp DNA sequence between these two genes is highly conserved (Fig. 2;(Frank et al., 1995). In GCS, two transcription initiation sites that are 8 bases apart were identified (Gase et al., 1995), and one of these corresponds to that identified for ska in GAS (Miller et al., 2001). In the work presented here, we have examined the interaction of CovR with the streptokinase promoter of GAS (Pska) and determined the effects of binding of both unphosphorylated and phosphorylated CovR on transcription from Pska.
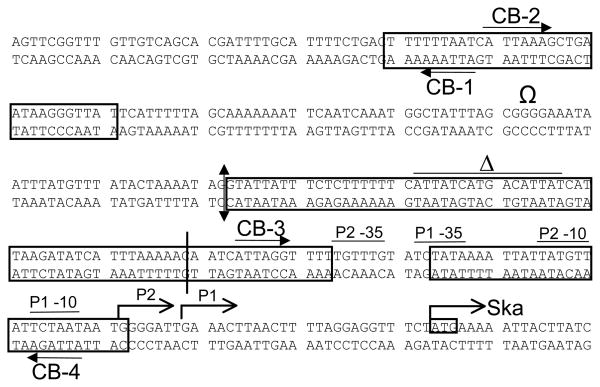
Figure 2. The ska promoter/operator region.
JRS4 sequence of the region upstream of the translation initiation site of ska which is indicated by the arrow labeled Ska. Possible transcription initiation sites are indicated by arrows labeled P1 and P2. The putative −35 and −10 regions associated with P1 and P2 are indicated. Consensus ATTARA CovR binding sites are indicated (CB-1, CB-2 and CB-4), as is a related non consensus sequence, ATTAR (CB-3). A 13 base pair sequence that is not present in group C streptococci is indicated by Δ. The vertical double-headed arrow indicates the center of bending of this region. The site of a 21 base pair insertion present in M1 and M3 strains is indicated by Ω. Regions protected from nuclease digestion by CovR are indicated by boxes. The vertical line upstream of the arrow labeled CB-3 indicates the 5′ end of the truncated promoter fragment used in the invitro transcription experiment described in figure 5B.
METHODS
Media
Escherichia coli DH5α and BL21 were grown in LB broth(Scott, 1972). GAS strains were grown at 37°C without agitation in Todd-Hewitt broth supplemented with 0.2% yeast extract (THY). Antibiotics were used at the following concentrations: ampicillin at 100 μg/ml for E. coli, kanamycin at 50 μg/ml for E. coli and 200 μg/ml for GAS, and spectinomycin at 100 μg/ml for both E. coli and GAS.
Strains
E. coli K-12 strain DH5α was used for cloning all plasmids. CovR was purified from BL21(DE3)(pLysS)(pEU7561) (Gusa & Scott, 2005) and the “housekeeping” sigma factor, RpoD, was purified from BL21(DE3)(pLysS)(pEU7534) (Gusa & Scott, 2005). GAS RNA polymerase was purified from strain JRSPolHis (Opdyke et al., 2001).
All GAS strains are derivatives of the M6 serotype strain JRS4 (Scott et al., 1986).
Oligonucleotide primers
All primers are listed in Supplementary Table 1.
Mutation of consensus binding sequences (CBs)
CB-1, CB-2, and CB-3 at Pska were mutated by replacing the thymine pairs of the ATTARA sequence with guanine residues by site-directed mutagenesis using complementary primer pairs. Each sense-strand primer was used with Pska A1 XhoI and each antisense primer was used with Pska S1 BamHI to amplify the ska promoter region from the JRS4 chromosome. These PCR products subsequently served as templates in an overlapping PCR with the outside primers Pska S1 BamHI and Pska A1 XhoI to produce the mutated Pska fragments with BamHI and XhoI sites for cloning into pJRS462 (Gusa & Scott, 2005). The primers Pska S1 BamHI and Pska A1 XhoI were used to amplify a 407 bp segment of Pska from the JRS4 chromosome. This segment was cloned between the BamHI and XhoI sites in pJRS462 (Gusa & Scott, 2005).
To construct the CB-4 mutation, pEU7227 containing the wild type ska promoter was used a template for site-directed mutagenesis using the SkaCB4mutF and SkaCB4mutR primers(Gusa & Scott, 2005). To construct the Pska-gusA fusions with TT to GG mutations at CB-1, CB-2 and CB-3, PCR products containing the mutated Pska regions with flanking BamHI and XhoI sites were cloned into plasmids which were then linearized and introduced into the chromosome of RTG229 (Geist et al., 1993).
Purification and phosphorylation of CovR
CovR was purified from E. coli and phosphorylated as previously described (Federle & Scott, 2002; Gusa et al., 2006). The protein concentration was determined using the BioRad protein assay reagent (BioRad) standardized with BSA.
DNaseI protection assays
Primers Pska S1 and Pska Xho A3 were used to amplify a 398 bp segment of the JRS4 chromosome including the ska promoter from −308 to +91 bp (with respect to the start of ska transcription). The primers in the PCR reaction were end-labeled (Munson & Scott, 1999) and nuclease protection assays were performed as described (Gusa & Scott, 2005).
In vitro transcription assays
Transcription reactions were performed and analyzed quantitatively as described (Gusa et al., 2006).
RESULTS
CovR binding to Pska
Examination of the ska promoter sequence in the serotype M6 GAS strain JRS4 shows four consensus CovR binding sequences (CBs) both upstream of and overlapping with the −10 RNA polymerase-binding regions of the promoter (Pska; Fig. 2). These sequences are conserved in all other GAS genomes whose sequence is publically available. To determine which sites bind CovR, we performed nuclease protection assays using a linear DNA fragment corresponding to bases −308 to +91 of Pska (Fig. 3A). We found that unphosphorylated CovR bound to site CB-4, the site that overlaps the −10 region of the promoter. Binding to this site required a concentration of unphosphorylated CovR greater than 1 μM. At 3 μM CovR, very weak protection of sites CB-1, CB-2 and CB-3 was also observed.
Figure 3. Effect of mutations on the binding of CovR and CovR-P to Pska.
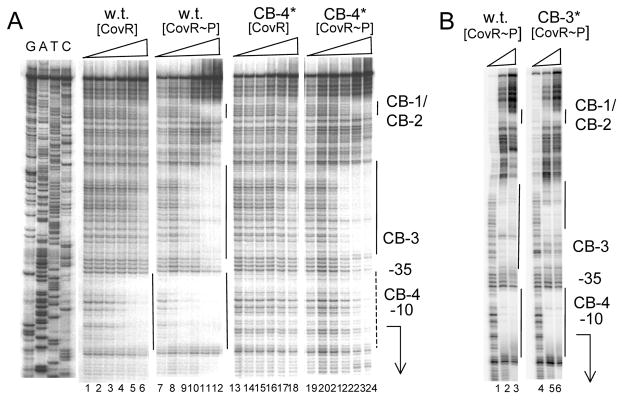
A: mutation in CB-4. Lanes 1–6 and 13–18: wild type DNA; lanes 7–12 and 19–24: mutant DNA with a TT to GG transversion in site CB-4 (denoted CB-4*). Lanes 1–6 and lanes 7–12: unphosphorylated CovR at 0, 0.25, 0.5, 1, 2 and 3 μM respectively. Lanes 13–18 and lanes 19–24: phosphorylated CovR at 0, 0.25, 0.5, 1, 2 and 3 μM respectively. The locations of the CovR binding sites CB-1 throgh CB-4 are indicated at the right of the figure. The solid lines indicate regions of DNaseI protection and the dashed line indicates partial DNaseI protection. The positions of the −10 and −35 regions of the promoter are indicated, and the arrow indicates the start point and direction of transcription. B: mutation in CB-3. Lanes 1–3: wild type DNA. Lanes 4–6: Mutant DNA with a TT to GG transversion in site CB-3 (denoted CB-3*). Lanes 1–3 and lanes 4–6; phosphorylated CovR at 0, 3, and 6 μM respectively. The location of the CovR binding sites CB1–CB4 are indicated at the right of the figure. The solid lines indicate regions of DNaseI protection. The positions of the −10 and −35 regions of the promoter are indicated, and the arrow indicates the start point and direction of transcription.
Effect of phosphorylation of CovR on binding to Pska
Previous work has shown that at other well-studied CovR-repressed promoters, with the exception of Psag, phosphorylation of the protein in vitro by incubation with acetyl phosphate led to a 2- to 3-fold increase in binding to the CBs within the promoter-containing DNA fragment (Churchward, 2007). In contrast to this, phosphorylated CovR (CovR-P) bound and protected CB-4 of Pska at a concentration of 0.25 μM, approximately 10-fold lower than that required for binding of unphosphorylated CovR (~3 μM) (Fig. 3A). This indicates that phosphorylation increases the binding affinity of CovR by 10-fold at this site.
At the other binding sites of Pska, significantly higher CovR-P protein concentrations were required for binding than for binding to site CB-4; protection at the CB-1/CB-2 sites was undetectable below 2 μM CovR-P. This indicates that CB-4 in the Pska promoter fragment has the strongest affinity both for CovR and for CovR-P.
Binding of CovR and CovR-P to CB-4 is independent of binding at other sites of Pska
To determine if cooperative interactions occur between CovR molecules binding to different binding sites at Pska, we investigated binding of CovR and CovR-P to Pska DNA and to Pska DNA carrying mutations in different CovR-binding sites. To make the mutations, the pair of T residues in each consensus binding sequence was changed to a pair of G residues by site-directed mutagenesis. Figure 3A shows that a mutation in site CB-4 (designated CB-4*), which overlaps the −10 region of the promoter, abolished binding of CovR and greatly reduced binding of CovR-P to this site. However, when CB-4 was mutant, binding of CovR-P at sites CB-1, CB-2 and CB-3 was essentially unchanged. These results indicate that binding of CovR and CovR-P at other sites is independent of binding to site CB-4.
To show that binding to site CB-4 is independent of binding to other sites, we examined binding to the Pska DNA fragment containing a TT to GG transversion in CB-3 (CB-3*). CB-3 differs from the consensus binding sequence by a single base, but it appears to serve as a CovR-P binding site (Fig. 3B). The CB-3* mutation prevented binding of CovR-P to the CB-3 site, but did not affect binding to CB-4 (Fig. 3B). Thus binding to CB-4, which overlaps the −10 region of the promoter, is independent of binding to other sites in Pska. Furthermore, the CB-3* mutation did not affect binding of CovR-P to CB-1/CB-2 either, indicating that binding at these sites is also independent.
From Fig. 3B, it is apparent that CovR-P binds to a region upstream of CB-3, even when CB-3 is mutated. This suggests the existence of an additional CovR-P binding site in this region that does not include either a consensus ATTARA sequence or a sequence that differs from this by only a single base. However, binding to this region requires a higher CovR-P concentration than binding to CB-4 (Fig. 3A)
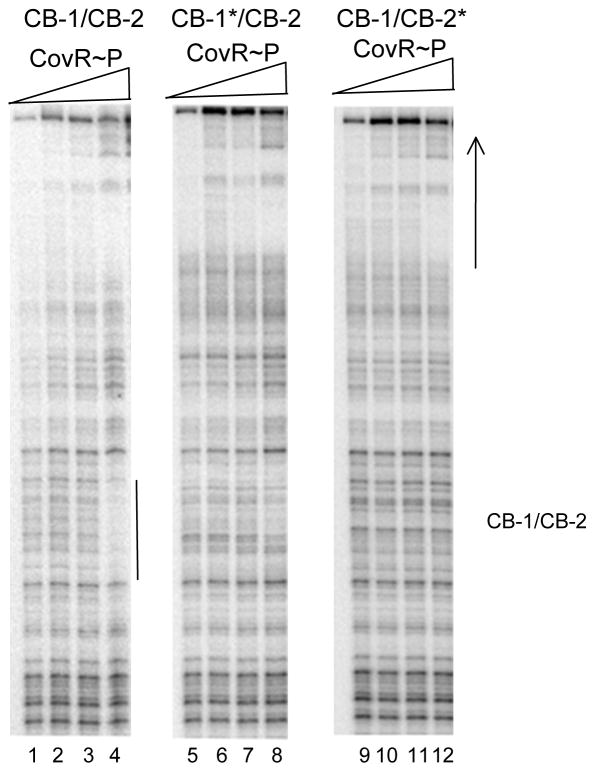
Since the palindromic arrangement of CB-1 and CB-2 suggests that binding might occur cooperatively to these two sites, we also examined the effect of mutations in these sites on CovR-P binding (Fig. 4). We found that a TT to GG mutation in either site abolished binding to this region. (Note that to better visualize binding to this region, in Fig. 4 we labeled the other strand of the DNA fragment compared to the experiments shown in Fig. 3.)
Figure 4. Effect of mutations on the binding of CovR and CovR-P to Pska: mutations in CB-1 and CB2.
Lanes 1–4:wild type DNA. Lanes 5–8: Mutant DNA with a TT to GG transversion in site CB-1 (CB-1*). Lanes 9–12: Mutant DNA with a TT to GG transversion in site CB-2 (CB-2*). Lanes 1–4, lanes 5–8 and lanes 9–12; phosphorylated CovR at 0, 0.25, 0.5, 1 and 3 μM respectively. The location of the CovR binding sites CB1 and CB2 are indicated at the right of the figure. The solid line indicates a region of DNaseI protection. The arrow indicates the direction of transcription.
In summary, site CB-4, which overlaps the −10 region of Pska, appears to be the primary binding site for both CovR and CovR-P. Furthermore, binding to this site appears to be independent of binding of CovR-P at sites further upstream of the promoter.
Repression of transcription from Pska by CovR in vitro
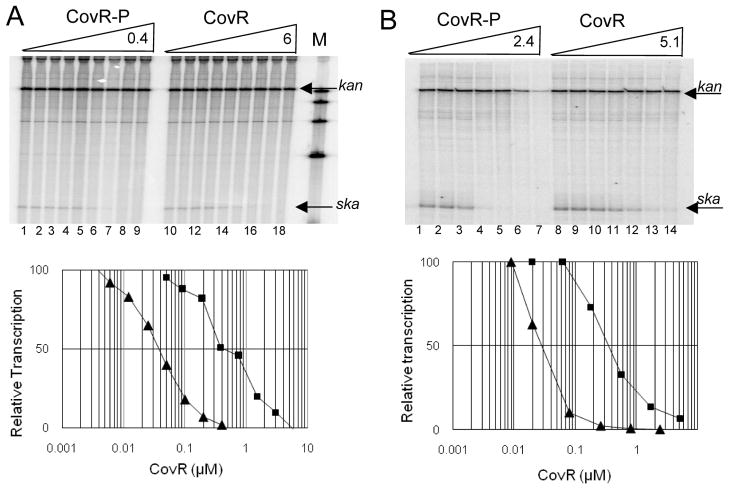
The results described in the two sections above are consistent with the notion that at Pska, in contrast to other CovR-regulated promoters, regulation occurs primarily by binding of CovR-P to a single site that overlaps the −10 region of the promoter. To test this directly, we used an in vitro transcription system containing purified GAS RNA polymerase (Gusa & Scott, 2005) to compare repression of a wild-type promoter sequence with a truncated sequence carrying a minimal promoter with a single CovR binding site. Experiments used a linearized template and a control promoter (Pkan) present on a separate DNA fragment was present in each reaction mix. All results are expressed relative to that of the control kan transcript. As shown in Fig. 5A, both CovR-P and CovR repressed transcription from Pska. The amounts of CovR-P and CovR required to reduce transcription by 50% were 0.04 μM and 0.4 μM respectively. Maximal repression (>90%) occurred at protein concentrations similar to those required for nuclease protection in the DNA binding experiments shown in Fig 3. Thus, in vitro, CovR-P repressed transcription from Pska 10-fold more effectively than CovR. This is in agreement with the 10-fold increase in binding affinity produced by phosphorylation of CovR.
Figure 5. Repression of Pska by CovR and CovR-P in vitro.
A. Run-off transcriptions using a wild-type template. Upper panel: Lanes 1–9 ; CovR-P at concentrations of 0, 0.003, 0.006, 0.012, 0.025, 0.050, 0.1, 0.2 and 0.4 μM respectively. Lanes 10–18: CovR at concentrations of 0, 0.05, 0.09, 0.19, 0.38, 0.75, 1.5, 3.0 and 6.0 μM respectively. Lane M: molecular weight markers. The transcripts originating from Pska and Pkan are indicated by the arrows. Lower panel. Densitometric analysis of the median transcript volume. Data is plotted as the ratio of ska/kanR transcript in the presence of CovR-P (triangles) or CovR (squares). The amount of transcript generated in the absence of CovR was defined as 100% and relative transcription was determined by the amount of transcript generated at a given CovR or CovR~P concentration divided by the amount of transcript with no CovR or CovR~P present. B. Run-off transcriptions using a truncated template. Upper panel: Lanes 1–7; CovR-P at concentrations of 0, 0.009, 0.2, 0.008, 0.026, 0.8, and 2.4 μM respectively. Lanes 8–14: CovR at concentrations of 0, 0.02, 0.06, 0.18, 0.56, 1.7 and 5.1 μM respectively. The transcripts originating from Pska and Pkan are indicated by the arrows. Lower panel. Densitometric analysis of the median transcript volume as described in A above.
To determine if this repression was due primarily to the interaction of CovR and CovR-P with the CB-4 site of the promoter region, we repeated the transcription experiments using a truncated template carrying a minimal promoter sequence. This minimal promoter sequence lacked all CovR-binding sites upstream of Cb-3 (Fig. 2). Because CB-3 is close to the −35 region of the promoter, the minimal promoter fragment retained most of the CB-3 binding site. Therefore, to study CB-4 alone, we inactivated CB-3 in the truncated template by mutating its TT residues to GG, as we did for the binding studies described above. Using this truncated template carrying CB-3*, we found that the amount of CovR-P and CovR required to reduce transcription by 50% were 0.03 μM and 0.3 μM respectively (Fig. 5B). This is almost identical to the amounts required to repress transcription from the intact promoter carrying all five CovR binding sites (Fig. 5A). Thus, efficient repression of transcription in vitro from Pska by either CovR-P or CovR requires only a minimal promoter fragment with a single CovR binding site. We conclude that the contributions to CovR-mediated repression of Pska by CovR binding sites upstream of CB-4 are negligible.
Repression of transcription from Pska by CovR in vivo
To determine whether CB-1, CB-2 and CB-3 affect repression of Pska, we tested the effect of the CB-1*, CB-2* and CB-3* mutations in a covR+ strain using both a transcriptional reporter fusion to β-glucuronidase (Federle et al., 1999) and quantitative RT-PCR to detect ska mRNA. We did not test the effect of a mutation in CB-4 on transcription in vivo since this mutation would be predicted to destroy the −10 region and thus to inactivate the promoter. In wild type bacteria expressing CovR, at the stage of growth at which Pska is maximally transcribed, we found no significant difference in the amount of transcript between that produced from wild type Pska and from each of the three CB* mutant promoters (data not shown). This is consistent with our model that CovR-mediated repression of Pska occurs primarily through binding to site CB-4.
DISCUSSION
An understanding of the mechanism of binding and regulation by CovR is central to an understanding of GAS virulence, since the CovRS TCS has been found to be a pivotal global regulator for this pathogen (Graham et al., 2005; Graham et al., 2006; Sumby et al., 2006; Virtaneva et al., 2005). As a contribution to unraveling the complex regulatory network controlling virulence in GAS, in this work we have studied binding and repression of CovR and CovR-P at the Pska promoter, which controls transcription of the critical GAS virulence factor, streptokinase. We have found several significant differences between the mechanism of CovR-mediated repression of transcription at Pska from the mechanism of repression at other promoters whose response to CovR we have characterized previously (Churchward, 2007).
Unique aspects of CovR binding at Pska
Although the pattern of nuclease protection by CovR at Pska is superficially similar to that at other promoters, at Pska the only CB to which unphosphorylated CovR binds is CB-4, which overlaps the −10 sequence of the promoter. In addition, phosphorylated CovR has a much higher affinity for CB-4 than for the other CBs in the Pska region of the DNA. We have not observed such a site preference in CovR and CovR-P binding at other promoters studied. Furthermore, the affinity of CovR-P for CB-4 of Pska is significantly greater than that for sites at the other promoters.
At several promoters, CovR binding to one site increases its affinity for a neighboring site, indicating a cooperative binding interaction. This is not seen at Pska. Mutation of CB-4 has no visible effect on binding to the other sites and mutation of the nearest site, CB-3, does not affect binding to CB-4. Thus, the high affinity of CovR-P binding at CB-4 is not caused by cooperativity.
Binding at sites other than CB-4 does not affect transcription from Pska
The most striking difference between CovR regulation at Pska compared to regulation at other promoters is that, in vitro, a unique CB, CB-4, is the only one required for complete repression. In vivo as well, in a covR+ strain, mutation of CB-1, CB-2 or CB-3 has no effect on repression at Pska. Since the other CBs in the Pska DNA fragment are bound and protected in vitro by CovR-P, this surprising result indicates that binding alone is not sufficient to cause repression. Given the high affinity of CovR-P for CB-4 and the fact that this binding site overlaps the −10 region of the promoter, binding to this site would be expected to reduce transcription. However, at Phas, although one of the CovR binding sites also overlaps the −10 region of the promoter, both upstream and downstream sites have major effects on repression in vivo, as demonstrated by mutation of these sites (Federle & Scott, 2002). Similarly, at Pcov, mutation of the upstream binding sites reduces repression by CovR-P both in vivo and in vitro (Gusa & Scott, 2005). An important conclusion from the lack of effect of mutations in CB-1, CB-2 and CB-3 is that binding of CovR to a specific site in the DNA near a promoter does not automatically imply that binding to this site plays a regulatory role. Instead it appears that the orientation of the bound CovR-P dimers relative to the promoter might be critical in determining the efficiency of repression.
Effect of phosphorylation on CovR binding at Pska compared to other promoters
At the CovR-regulated promoters Phas, Pcov and Priv, phosphorylation extends the region protected from Dnase, indicative of some cooperativity in binding, and increases the affinity of the protein for the DNA only about 2–4-fold (Federle & Scott, 2002; Gusa & Scott, 2005; Roberts et al., 2007). In contrast, we find that phosphorylation of CovR results in about a 10-fold increase in binding to CB-4 of Pska. The only other CovR-regulated promoter at which a large increase in binding affinity occurs upon phosphorylation of the protein is Psag (Gao et al., 2005), at which binding is highly cooperative (Gao et al., 2005). This cooperativity enables CovR and CovR-P to bind long stretches of Psag DNA (~100 bp) that contain only a single consensus or near-consensus binding site. Since binding to site CB-4 of Pska is notable for its independence from protein binding at adjacent sites, the strong effect of phosphorylation on binding affinity at CB-4 cannot be due to cooperativity, as it is at Psag.
Interactions of CovR with DNA: a model for binding at Pska
To explain the high affinity of CovR-P for CB-4 of Pska, we examined the sequence of this binding site. We have shown that nearly all regions of DNA protected from nuclease cleavage by CovR contain the consensus sequence ATTARA or a near-consensus sequence containing 5/6 of these bases (Federle & Scott, 2002; Gao et al., 2005; Gusa & Scott, 2005; Roberts et al., 2007). We have also shown by uracil interference assays that the T residues of this consensus sequence and the central T residue of its complement (TYTAAT) are frequently necessary (though not sufficient) for binding (Federle & Scott, 2002; Gao et al., 2005). CB-4 is composed of a consensus and a related sequence in inverted orientation separated by five base pairs: ATTATgttatTCTAAT. This places the T bases crucial for CovR binding 11 base-pairs, or one turn of the DNA helix, apart. Of all the CovR binding sites we have examined at different promoters where binding is not affected by interactions with other binding sites, this is the only site with potential binding sites in inverted orientation that also has this spacing. Since phosphorylation of CovR causes it to form a dimer (Gusa et al., 2006), we therefore propose that, upon phosphorylation, the two DNA binding domains of the CovR-P dimer bind CB-4 in an inverted orientation (“head-to-head”). CovR is structurally related to OmpR and its relatives, and the DNA binding domains of these proteins are characterized by a single winged helix structure that interacts with a single DNA binding site (Martinez-Hackert & Stock, 1997). We propose that the winged helix of each CovR DNA binding domain in the CovR-P dimer interacts with each consensus or near-consensus sequence of CB-4. The 10-fold stimulation by phosphorylation suggests that an individual monomer can only bind CB-4 with low affinity.
At other CovR binding sequences we have examined that contain consensus or near-consensus sequences and that show no cooperative interactions with other nearby sites, the consensus and near-consensus sequences are in direct rather than inverted orientation, as at site CB-3 of Pska. At CB-3, there are two near-consensus sequences separated by 7 bases: TTTAAAaacaatcATTAGG and the crucial T residues are 12 bases apart. At the other sites the spacing varies between 9 and 12 base pairs. We propose that at these sites the DNA binding domains of adjacent bound CovR or CovR-P molecules are in direct rather than in the inverted orientation we suggest for site CB-4 of Pska.
Thus, similarly to what has been suggested fo OmpR binding to DNA (Rhee et al., 2008), our analysis of CovR binding to independent sequences at Pska supports our previous proposal (Churchward, 2007) that, depending upon the DNA sequence, CovR and CovR-P can bind either with their DNA binding domains in head-to-head or head-to-tail orientation. We suggest that sites that allow CovR binding in inverted orientation will be bound with higher affinity, as seen at CB-4 of Pska. CovR and CovR-P also bind with high affinity to arrays of sites in direct orientation, such as those at Psag, provided that the sites are appropriately spaced along the DNA to permit a high degree of binding cooperativity.
Regulation of Pska and GAS pathogenesis
The properties of CovR-mediated regulation of Pska described here may have importance in the pathogenesis of GAS. Our results suggest that Pska can be regulated by lower concentrations of phosphorylated CovR than the other Cov-regulon promoters investigated. Further, it seems to require less unphosphorylated CovR to repress Pska than to repress other promoters. This leads to the prediction that ska would be repressed under some conditions in which other Cov-regulon genes were expressed. However, it should be remembered that other regulatory factors affect the amount of streptokinase. In addition to the amount of ska transcript, the amount of streptokinase produced will be affected by the amount of the Cov-regulated cysteine protease SpeB, and probably by other proteases secreted by GAS. The way in which these additional regulatory factors interact to allow production of streptokinase at the time and place at which its activity is important to the GAS will require further analysis of these complex regulatory networks.
Supplementary Material
Acknowledgments
This work was supported by grant AI020723 from NIH. C.B. was supported in part by IRACDA NIH/NIGMS grant K12 GM000680 to Emory University and the Atlanta University Center. A.A.G. was supported in part by NIH training grant T32 AI07470 and by a UNCF-Merck Graduate Science Research Dissertation Fellowship.
References
- Ashbaugh CD, Moser TJ, Shearer MH, White GL, Kennedy RC, Wessels MR. Bacterial determinants of persistent throat colonization and the associated immune response in a primate model of human group A streptococcal pharyngeal infection. Cellular Microbiology. 2000;2:283–292. doi: 10.1046/j.1462-5822.2000.00050.x. [DOI] [PubMed] [Google Scholar]
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, Kotb M, Feramisco J, Nizet V. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr Biol. 2006;16:396–400. doi: 10.1016/j.cub.2005.12.039. [DOI] [PubMed] [Google Scholar]
- Churchward G. The two faces of Janus: virulence gene regulation by CovR/S in group A streptococci. Mol Microbiol. 2007;64:34–41. doi: 10.1111/j.1365-2958.2007.05649.x. [DOI] [PubMed] [Google Scholar]
- Cole JN, McArthur JD, McKay FC, et al. Trigger for group A streptococcal M1T1 invasive disease. Faseb J. 2006;20:1745–1747. doi: 10.1096/fj.06-5804fje. [DOI] [PubMed] [Google Scholar]
- Collin M, Olsen A. Effect of SpeB and EndoS from Streptococcus pyogenes on human immunoglobulins. Infect Immun. 2001;69:7187–7189. doi: 10.1128/IAI.69.11.7187-7189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham MW. Pathogenesis of GroupA Streptococcal Infection. ClinMicrobiolRev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton TL, Scott JR. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J Bacteriol. 2004;186:3928–3937. doi: 10.1128/JB.186.12.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton TL, Collins JT, Barnett TC, Scott JR. RscA, a Member of the MDR1 Family of Transporters, Is Repressed by CovR and Required for Growth of Streptococcus pyogenes under Heat Stress. J Bacteriol. 2006;188:77–85. doi: 10.1128/JB.188.1.77-85.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federle MJ, McIver KS, Scott JR. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J Bacteriol. 1999;181:3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federle MJ, Scott JR. Identification of binding sites for the group A streptococcal global regulator CovR. Mol Microbiol. 2002;43:1161–1172. doi: 10.1046/j.1365-2958.2002.02810.x. [DOI] [PubMed] [Google Scholar]
- Frank C, Steiner K, Malke H. Conservation of the organization of the streptokinase gene region among pathogenic streptococci. Med Microbiol Immunol (Berl) 1995;184:139–146. doi: 10.1007/BF00224351. [DOI] [PubMed] [Google Scholar]
- Gao J, Gusa AA, Scott JR, Churchward G. Binding of the global response regulator protein CovR to the sag promoter of Streptococcus pyogenes reveals a new mode of CovR-DNA interaction. J Biol Chem. 2005;280:38948–38956. doi: 10.1074/jbc.M506121200. [DOI] [PubMed] [Google Scholar]
- Gase K, Ellinger T, Malke H. Complex transcriptional control of the streptokinase gene of Streptococcus equisimilis H46A. Mol Gen Genet. 1995;247:749–758. doi: 10.1007/BF00290407. [DOI] [PubMed] [Google Scholar]
- Geist RT, Okada N, Caparon MG. Analysis of Streptococcus pyogenes promoters by using novel Tn916-based shuttle vectors for the construction of transcriptional fusions to chloramphenicol acetyltransferase. J Bacteriol. 1993;175:7561–7570. doi: 10.1128/jb.175.23.7561-7570.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MR, Smoot LM, Migliaccio CA, et al. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci U S A. 2002;99:13855–13860. doi: 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MR, Virtaneva K, Porcella SF, Barry WT, Gowen BB, Johnson CR, Wright FA, Musser JM. Group A Streptococcus transcriptome dynamics during growth in human blood reveals bacterial adaptive and survival strategies. Am J Pathol. 2005;166:455–465. doi: 10.1016/S0002-9440(10)62268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MR, Virtaneva K, Porcella SF, et al. Analysis of the transcriptome of group A Streptococcus in mouse soft tissue infection. Am J Pathol. 2006;169:927–942. doi: 10.2353/ajpath.2006.060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusa AA, Scott JR. The CovR response regulator of group A streptococcus (GAS) acts directly to repress its own promoter. Mol Microbiol. 2005;56:1195–1207. doi: 10.1111/j.1365-2958.2005.04623.x. [DOI] [PubMed] [Google Scholar]
- Gusa AA, Gao J, Stringer V, Churchward G, Scott JR. Phosphorylation of the Group A Streptococcal CovR Response Regulator Causes Dimerization and Promoter-Specific Recruitment by RNA Polymerase. J Bacteriol. 2006;188:4620–4626. doi: 10.1128/JB.00198-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath A, DiRita VJ, Barg NL, Engleberg NC. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infection & Immunity. 1999;67:5298–5305. doi: 10.1128/iai.67.10.5298-5305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husmann LK, Yung DL, Hollingshead SK, Scott JR. Role of putative virulence factors of Streptococcus pyogenes in mouse models of long-term throat colonization and pneumonia. Infect Immun. 1997;65:1422–1430. doi: 10.1128/iai.65.4.1422-1430.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreikemeyer B, Boyle MD, Buttaro BA, Heinemann M, Podbielski A. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol Microbiol. 2001;39:392–406. doi: 10.1046/j.1365-2958.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- Kwinn LA, Nizet V. How group A Streptococcus circumvents host phagocyte defenses. Future Microbiol. 2007;2:75–84. doi: 10.2217/17460913.2.1.75. [DOI] [PubMed] [Google Scholar]
- Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- Levin JC, Wessels MR. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- Malke H, Steiner K, Gase K, Frank C. Expression and regulation of the streptokinase gene. Methods. 2000;21:111–124. doi: 10.1006/meth.2000.0982. [DOI] [PubMed] [Google Scholar]
- Martinez-Hackert E, Stock AM. The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure. 1997;5:109–124. doi: 10.1016/s0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- Miller AA, Engleberg NC, DiRita VJ. Repression of virulence genes by phosphorylation-dependent oligomerization of CsrR at target promoters in S. pyogenes. Mol Microbiol. 2001;40:976–990. doi: 10.1046/j.1365-2958.2001.02441.x. [DOI] [PubMed] [Google Scholar]
- Munson GP, Scott JR. Binding site recognition by Rns, a virulence regulator in the AraC family. JBacteriol. 1999;181:2110–2117. doi: 10.1128/jb.181.7.2110-2117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser JM, DeLeo FR. Toward a genome-wide systems biology analysis of host-pathogen interactions in group A Streptococcus. Am J Pathol. 2005;167:1461–1472. doi: 10.1016/S0002-9440(10)61232-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrand A, McShan WM, Ferretti JJ, Holm SE, Norgren M. Allele substitution of the streptokinase gene reduces the nephritogenic capacity of group A streptococcal strain NZ131. Infect Immun. 2000;68:1019–1025. doi: 10.1128/iai.68.3.1019-1025.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdyke JA, Scott JR, Moran CP., Jr A secondary RNA polymerase sigma factor from Streptococcus pyogenes. Mol Microbiol. 2001;42:495–502. doi: 10.1046/j.1365-2958.2001.02657.x. [DOI] [PubMed] [Google Scholar]
- Rhee JE, Sheng W, Morgan LK, Nolet R, Liao X, Kenney LJ. Amino acids important for DNA recognition by the response regulator OmpR. J Biol Chem. 2008;283:8664–8677. doi: 10.1074/jbc.M705550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Churchward GG, Scott JR. Unraveling the Regulatory Network in Streptococcus pyogenes: The Global Response Regulator CovR Represses rivR Directly. J Bacteriol. 2007;189:1459–1463. doi: 10.1128/JB.01026-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Scott JR. RivR and the small RNA RivX: the missing links between the CovR regulatory cascade and the Mga regulon. Mol Microbiol. 2007;66:1506–1522. doi: 10.1111/j.1365-2958.2007.06015.x. [DOI] [PubMed] [Google Scholar]
- Salim KY, de Azavedo JC, Bast DJ, Cvitkovitch DG. Role for sagA and siaA in quorum sensing and iron regulation in Streptococcus pyogenes. Infect Immun. 2007;75:5011–5017. doi: 10.1128/IAI.01824-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JR. A new gene controlling lysogeny in phage P1. Virology. 1972;48:282–283. doi: 10.1016/0042-6822(72)90139-0. [DOI] [PubMed] [Google Scholar]
- Scott JR, Guenthner PC, Malone LM, Fischetti VA. Conversion of an M-group A streptococcus to M+ by transfer of a plasmid containing an M6 gene. J Exp Med. 1986;164:1641–1651. doi: 10.1084/jem.164.5.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Sumby P, Barbian KD, Gardner DJ, et al. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc Natl Acad Sci U S A. 2005 doi: 10.1073/pnas.0406641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of Group A Streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathogens. 2006;2:41–49. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Ringdahl U, Homeister JW, et al. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science. 2004;305:1283–1286. doi: 10.1126/science.1101245. [DOI] [PubMed] [Google Scholar]
- Tart AH, Walker MJ, Musser JM. New understanding of the group A Streptococcus pathogenesis cycle. Trends Microbiol. 2007;15:318–325. doi: 10.1016/j.tim.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Virtaneva K, Porcella SF, Graham MR, et al. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc Natl Acad Sci U S A. 2005;102:9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MJ, Hollands A, Sanderson-Smith ML, et al. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nature medicine. 2007;13:981–985. doi: 10.1038/nm1612. [DOI] [PubMed] [Google Scholar]
- Wessels MR, Bronze MS. Critical role of the group A streptococcal capsule in pharyngeal colonization and infection in mice. Proc Natl Acad Sci U S A. 1994;91:12238–12242. doi: 10.1073/pnas.91.25.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.