Abstract
Transcriptional enhancers are genomic sequences bound by transcription factors that act together with basal transcriptional machinery to regulate gene transcription. Several high-throughput methods have generated large datasets of tissue-specific enhancer sequences with putative roles in developmental processes. However, few enhancers have been deleted from the genome to determine their roles in development. To understand the roles of two enhancers active in the mouse embryonic limb bud we deleted them from the genome. Although the genes regulated by these enhancers are unknown, they were selected because they were identified in a screen for putative limb bud-specific enhancers associated with p300, an acetyltransferase that participates in protein complexes that promote active transcription, and because the orthologous human enhancers (H1442 and H280) drive distinct lacZ expression patterns in limb buds of embryonic day (E) 11.5 transgenic mice. We show that the orthologous mouse sequences, M280 and M1442, regulate dynamic expression in the developing limb. Although significant transcriptional differences in enhancer-proximal genes in embryonic limb buds accompany the deletion of M280 and M1442 no gross limb malformations during embryonic development were observed, demonstrating that M280 and M1442 are not required for mouse limb development. However, M280 is required for the development and/or maintenance of body size; M280 mice are significantly smaller than controls. M280 also harbors an “ultraconserved” sequence that is identical between human, rat, and mouse. This is the first report of a phenotype resulting from the deletion of an ultraconserved element. These studies highlight the importance of determining enhancer regulatory function by experiments that manipulate them in situ and suggest that some of an enhancer's regulatory capacities may be developmentally tolerated rather than developmentally required.
INTRODUCTION
The study of the proximate, molecular genetic mechanisms responsible for appendage development in numerous organisms has demonstrated that vertebrate limb development depends on the interplay of key conserved genes and signaling pathways, such as the homeobox (HOX) transcription factors, sonic hedgehog (SHH), bone morphogenetic proteins (BMP) and fibroblast growth factors (FGF) (Rabinowitz and Vokes 2012, Shubin N. et al. 1997, Zeller et al. 2009). Although a consensus picture of the genetic circuits underlying limb development has emerged from studies in humans, mice, and chick, less is known about the genetic components responsible for the execution and regulation of these circuits, and how the species-specific deployment of these circuits contributes to species-specific limb morphologies.
One important form of transcriptional regulation that may simultaneously contribute to the diversity of the deployment of limb-related genetic circuits and final species-specific limb morphologies is encoded in transcriptional enhancers, primary sequences of genomic DNA that contain clustered transcription factor (TF) binding sites (Levine 2010). When particular TFs or combinations of TFs bind to enhancer sequences they can act in concert with basal transcriptional machinery at a promoter to regulate the timing, location and quantity of gene expression (Banerji et al. 1981, Ohler and Wassarman 2010, Visel et al. 2009a). Enhancers may be located 5', 3', or within the introns of the genes they regulate (Chandler et al. 2007, Feng et al. 2008). Research aimed at uncovering the molecular genetic sources of divergent limb morphology, such as the role of enhancers, serves as a bridge between questions centered on proximate, or developmental and ultimate, or evolutionary explanations of vertebrate limb diversity (Cotney et al. 2013, Cretekos et al. 2008, Shubin N. et al. 2009, Shubin N. H. et al. 2006, Woltering and Duboule 2010).
Functional studies carried out in mice using gene targeting and chromosomal rearrangement methods have begun to establish the role of limb-specific enhancers or enhancer regions at several loci known to be integral to limb development, including Fgf8 (Marinic et al. 2013), the Hoxd cluster (Herault et al. 1998, Montavon et al. 2011, Spitz et al. 2005, Tarchini and Duboule 2006), Gremlin (Zuniga et al. 2004), Prrx1 (Cretekos et al. 2008), Shh (Sagai et al. 2005), Tcfap2a (Feng et al. 2008), and Tbx4 (Menke et al. 2008). Tempering our increased understanding of regulatory biology at these seven loci is the expanding use of high throughput methods to identify thousands of enhancer sequences with putative roles in tissue-specific development, including the limb (Ahituv et al. 2005, Cotney et al. 2012, Nobrega et al. 2003, Ruf et al. 2011, Taher et al. 2011, Visel et al. 2007a, Vokes et al. 2008). In one such study, chromatin immunoprecipitation with DNA sequencing (ChIP-seq) targeted at p300, a general transcription activator (Chan and La Thangue 2001, Kalkhoven 2004), was used to locate approximately 2,100 enhancer sequences that potentially regulated gene transcription specifically in embryonic day (E) 11.5 mouse limb buds (Visel et al. 2009b). Validation of their ChIP-seq approach to obtaining tissue-specific enhancer enrichment derives from the fact that human orthologs of the p300-associated enhancer sequences can drive reporter gene expression in E11.5 transgenic mouse embryos. Enhancer sequences and images of transgenic mice derived from the p300-ChIP-seq screen and other assays designed to identify enhancers (via extreme sequence conservation, for example) are available on the Vista Enhancer Browser (VEB, enhancer.lbl. gov) (Visel et al. 2007b).
Though the capacity for p300-associated enhancers to drive tissue-specific expression patterns in transgenic mouse assays confirms their regulatory potential, such experiments do not provide information about their role at their endogenous loci. To assess the role of each enhancer during limb development, and to begin to explore the role that such enhancers might play in contributing to the species-specific vertebrate limb morphologies in extant and extinct species, it is necessary to manipulate them at their endogenous loci. In this study we selected two p300-associated enhancers from the VEB and deleted them from the mouse genome, using gene targeting in embryonic stem (ES) cells to determine their contribution to limb development.
MATERIALS AND METHODS
Generation of transgenic mice
M280- and M1142-Hsp68lacZpA (M280-lacZ and M1442-lacZ) gene constructs were generated. Briefly, M280 and M1442 were PCR-amplified from 129S6/SvEv genomic DNA and introduced into a Gateway insertion site upstream of Hsp68lacZpA (Pennacchio et al. 2006). See Supplemental Information for the details of transgene construction. Transgenic mice were generated by pronuclear microinjection into FVB zygotes (Nagy et al. 2003). M280-lacZ PCR genotyping primers are: 280GWtg-L (5'-CTCTTGGGCCTTGGGGATAGTAG-3'), 280/1442GWtg-R2 (5'-AGTGCTGCCTCTGACCTCATGG-3'). M1442-lacZ PCR genotyping primers are: 1442GWtg-L2 (5'-AATGTCCTCTGACCTCCACATGC-3'), 280/1442GWtg-R2 (5'-AGTGCTGCCTCTGACCTCATGG-3').
Whole mount β-galactosidase staining
X-gal was used to detect lacZ expression in transgenic mice as previously described (Nagy et al. 2003). Salmon-gal (LabScientific, Inc.) was used for the weakly expressing M280-lacZ lines (Sundararajan et al. 2012).
Generation of enhancer deletion mice
Gene targeting vectors were generated to delete the enhancers in mouse ES cells. Briefly, a loxP-flanked neomycin resistance expression cassette (neo) replaces the enhancer sequence upon homologous recombination. Neo was subsequently removed by germline Cre expression (see below). See Supplemental Information for the details of gene targeting vector construction. Twenty-five μg of linearized gene targeting vector plasmid was electroporated into PC3 ES cells (O'Gorman et al. 1997), followed by selection in medium containing G418 and FIAU (Behringer et al. 1994). G418, FIAU-resistant ES cell colonies were picked and homologous recombinants were identified by Southern blot (Ramirez-Solis et al. 1992) (Supplementary Information Table SI.1).
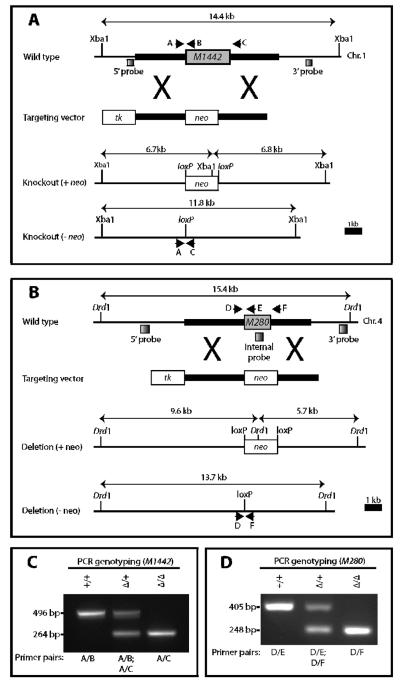
M1442 ES cell clone DNA was digested with Xba1 and hybridized with a 5'-external probe. Correctly targeted ES cell clones were identified by a 14.4 kb wild-type band and a 6.7 kb mutant band (Fig. SI.1A). Hybridization with a 3' external probe yielded the 14.4 kb wild-type band and a 6.8 kb mutant band (Fig. SI.1A). Two correctly targeted ES cell lines were injected into C57BL/6J blastocysts and transferred into pseudopregnant Swiss foster mothers to generate chimeras. PC3 ES cells are homozygous for a protamine-Cre transgene. Cre recombinase expression in the male chimera germline results in excision of the neo cassette (O'Gorman et al. 1997). Deletion of the M1442 enhancer was verified by Southern analysis of M1442 −/− genomic DNA digested with Xba1 and hybridization with either the 5'-external probe or 3'-external probe. The absence of the neo cassette at the targeted allele is shown by an 11.8 kb band, using either the 5'- or 3'-external probes (Fig. SI.1B). Confirmed transmitting male chimeras were bred to 129S6/SvEvTac females to maintain the deletion on a 129S6/SvEv inbred background. Offspring from all subsequent crosses were genotyped by PCR. The PCR reaction consisted of three primers: 1442-PreEnh-L2 (5'-AAAGCCCAAAGGTTTTTCTCAAGG-3'); 1442-wtEnh-R1 (5'-CAAACATCTCCTACAAGCACTCACG-3'); 1442-PostEnh-R2 (5'-GTGAACTCGGCAGGTAACAGACAG-3'). Primers 1442-PreEnh-L2 + 1442-wtEnh-R1 result in a 496 bp amplicon for the wild-type allele. Primers 1442-PreEnh-L2 + 1442-PostEnh-R2 result in a 264 bp amplicon for the deletion allele. Because the PC3 ES cells are homozygous for protamine-Cre, all F1 progeny from the male chimeras were heterozygous for protamine-Cre; thus F1 progeny were crossed to 129S6/SvEv mice to identify M1442 +/− mice lacking protamine-Cre.
M280 ES cell clone DNA was digested with Drd1 and hybridized with a 5'-external probe. Correctly targeted ES cell clones were identified by a 15.4 kb wild-type band a 9.6 kb mutant band (Fig. SI.1C). Hybridization with a 3' external probe yielded the 15.4 kb wild-type band and a 5.7 kb mutant band (Fig. SI.1C). Two correctly targeted ES cell clones were injected into C57BL/6J blastocysts and transferred into pseudopregnant Swiss foster mothers to generate chimeras. Deletion of the M280 enhancer was verified by Southern analysis of M280 −/− genomic DNA digested with Ddr1 and hybridization with either the 5'-external probe or 3'-external probe (Fig. SI.1D). The absence of the neo cassette at the targeted allele is shown by a 13.7 kb band when using either the 5'-external or the 3'-external probe, and the absence of a band when using an M280-internal probe (Fig. SI.1E). Mice carrying the M280 deletion allele without protamine-Cre were maintained on a 129S6/SvEv background. The targeted allele was genotyping by PCR, using three primers: 280-PreEnh-Lb (5'-GGTTCAATCAGCCTTTCATTCCAG-3'); 280-wtEnh-R6 (5'-AGGCTTCCAGGGCTGATAACAAG-3'); 280-PostEnh-Rd (5'-AGGCAGCTCTGTCTCTGCAAAAG-3'). Primers 280-PreEnh-Lb + 280-wtEnh-R6 result in a 405 bp amplicon for the wild-type allele. Primers 280-PreEnh-Lb + 280-PostEnh-Rd result in a 248 bp amplicon for the deletion allele.
Limb-only bone and cartilage stain
We modified a standard alizarin red and alcian blue bone and cartilage staining protocol to examine newborn limbs (Nagy et al. 2003). See Supplemental Information for details of the protocol modifications.
Autopod measurements
Because the lacZ expression patterns driven by both M280 and M1442 did not intersect with the developing stylopod (radius and ulna) we expected the morphology of the radii to be unaffected by the enhancer deletions. Upon measuring the mineralized portion stained by alizarin red of E18.5 radii we found there to be no statistical difference in the length of radii between genotypes when corrected for body weight. Therefore, all subsequent measurements of autopod bone elements were normalized to an individual's radius length (as a proxy for embryo size/weight; Fig. SI.2). Bone lengths were measured in both right and left autopods in Photoshop (Adobe) using arbitrary units with the ruler tool. Measurements for left and right bone elements were averaged and used in the final calculations.
In accordance with the expression pattern in M280 transgenic mice, mineralized as well as non-mineralized bone elements of forearm digit III (third metacarpal, phalanges 1-3) were measured in E18.5 M280 −/− embryos and compared to wild-type controls. Concordant with the expression pattern in M1442 transgenic mice, the mineralized as well as non-mineralized portions of forearm digits I (phalanges 1–2) and V (fifth metacarpal and phalanges 1-3) were measured in E18.5 V442 −/− embryos and compared to wild-type controls. Bone elements for each digit were measured individually and then length measurements were combined for a total length that was normalized to radius length.
Histology
X-gal stained lacZ transgenic embryos were embedded in paraffin. 6–12 micron sections were counterstained with Nuclear Fast Red. E18.5 limbs were fixed in 10% formalin overnight, washed three times in phosphate buffered saline, and rinsed in 0.5M EDTA for 48 hours to remove calcium from bone. The limbs were embedded in paraffin and 6–10 micron sections were stained with hematoxylin and eosin (H&E).
Quantitative real-time PCR
M280 −/−, M1442 −/−, or 129S6/SvEv mice were intercrossed and E11.5 embryos were collected. Forelimb and hindlimb autopods were dissected from three embryos of the same genotype and pooled (1 sample = 12 autopods; 6 forelimbs and 6 hindlimbs). Pooled autopods were placed into 500 μl Ambion TRIzol Reagent (15596) and homogenized. Total RNA was obtained following the manufacture's guidelines (Ambion TRIzol Reagent) and RNA quality and concentration was assessed on a NanoDrop Spectrophotometer (260/280 nm ≥ 1.8; 260/230 nm ≥ 1.9). Five hundred ng of total RNA from each sample was used to synthesize cDNA with the Invitrogen SuperScript III First-Strand Synthesis System. Quantitative RT-PCR (qPCR) was carried out in duplicate or triplicate on oligo-dT-primed cDNA (n=8 biological replicates per genotype) on an ABI 7500 Real-Time System (Applied Biosystems). Retinoblastoma binding protein 4 (Rbbp4) was selected as an endogenous reference because it is a ubiquitously expressed, chromatin-associated protein (Ruthenburg et al. 2007) and because of its consistent transcriptional levels across numerous amniocyte stem cell lines (Maguire et al. 2013). The comparative CT method was employed to determine the relative expression of genes of interest (GOI) after normalization to Rbbp4 and calibration to wild-type GOI expression levels. Primer sequences for genes assessed are provided in Table SI.2. The primer set for Prx1 was obtained from Cretekos et al. (2008). All other primer sets were designed using Oligo Architect (http://www.sigmaaldrich.com/life-science/custom-oligos/dna-probes/product-lines/probe-design-services.html) or the Roche Universal ProbeLibrary Assay Design Center (https://www.rocheappliedscience.com/webapp/wcs/stores/servletCategoryDisplay?catalogId=10001&tab=Assay+Design+Center&identifier=Universal+Probe+Library&langId=−1).
Whole Mount In Situ Hybridization (WMISH)
WMISH was performed using standard protocols (Nagy et al. 2003). See Table SI.3 for details of the riboprobes used in the study.
Assessment of embryonic and postnatal growth
Images of whole E15.5 embryos were acquired using a Leica MZ0F microscope. The length of the desired structure was measured in Photoshop with the ruler tool using arbitrary units. Weights of males and females were recorded at noon at three and six weeks of age (Supplemental Information: SuppInfo_3and6wkWeights.xlsx). Linear mixed models were used to examine the differences in body weights at 3 week and 6 week among genotypes, adjusted for litter size. The linear mixed models included fixed effects of genotype and litter size, and a random effect of litter to account for the correlation between mice within the same litters. Pairwise comparisons between genotypes were carried out considering the differences in their least square means and adjusting for multiple comparisons within the model using Tukey's method. Quantile-quantile plots were used to assess the normality of the residuals in the linear models. All analyses were performed using SAS 9.3 (Cary, NC).
RESULTS
Selection of embryonic limb transcriptional enhancers for functional studies
Images of E11.5 transgenic mouse embryos on the VEB with limb-specific reporter gene expression guided us to select two enhancers, H1442 and H280, for further study (Visel et al. 2007b). We discuss the selection and characterization of H1442 first, and then do the same for H280.
H1442 and selection and characterization of M1442-lacZ transgenic mice
Images provided by the VEB showed that H1442 regulated a graded lacZ expression pattern in the limb bud, with strongest intensity in the posterior half of the limb bud and faded or absent expression in the anterior half (Fig.1A.a). This expression pattern is reminiscent of Shh, although Shh expression is even more restricted to the posterior edge of the limb bud (Masuya et al. 2007, Riddle et al. 1993). H1442 was originally identified in the p300-ChIP-seq screen described above (Visel et al. 2009b). Importantly, the VEB transgenic mice were generated using human sequences and were scored for lacZ expression at a single time point, E11.5. Therefore, it was necessary to assess whether the orthologous mouse enhancer, M1442, behaved similarly to its human ortholog. We selected a 2,723 bp sequence orthologous to H1442 from the mouse genome, M1442, for the production of transgenic mice and gene targeting (Table 1). A 1,440 core orthologous region of M1442 is 91.9% conserved with its human counterpart.
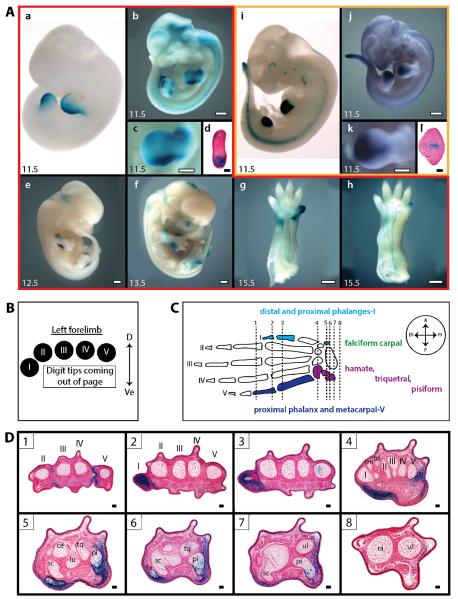
Figure 1. M1442 and M280 drive lacZ expression in the developing limb.
(A) X-gal and Salmon-gal stained transgenic embryos. Numbers in the lower-left of panels indicate embryonic day. Panels bordered in red (A.a–h) correspond to H1442/M1442 data; panels bordered in gold (A.i–l) correspond to H280/M280 data. (A.a) E11.5 H1442 lacZ transgenic embryo (image from VEB, enhancer.lbl.gov) (Visel et al. 2007b). (A.b) E11.5 M1442-lacZ L2 embryo. (A.c) Closeup of left forelimb bud in (b). (A.d) Cross-section through E11.5 M1442-lacZ L2 limb bud showing lacZ expression in mesenchymal cells in the anterior and posterior limb bud margins. M1442-driven expression is strongest in the posterior limb, corresponding to the future location of digit V; scale bar is 50 μm. (A.e) E12.5 M1442-lacZ L2 embryo. (A.f) E13.5 M1442-lacZ L1 embryo. (A.g, h) Ventral view of E15.5 M1442-lacZ L1 forelimb and hindlimb, respectively.. (A.i) E11.5 H280 lacZ transgenic embryo (image from VEB, enhancer.lbl.gov) (Visel et al. 2007b). (A.j) E11.5 M280-lacZ L2 embryo. (A.k) Close-up of left forelimb bud in (j). (A.l) Cross-section through G0 E11.5 M280-lacZ limb bud showing lacZ expression in mesenchymal cells. M280-driven expression is strongest in the center of the forelimb bud, corresponding to the approximate future location of digit III; scale bar is 100 μm. Salmon-gal staining in A.j, k. Scale bars in (A.b–h, j, k) are 0.5 mm. (A.d, l) tissue from E11.5 embryos. (B) Orientation guide for cross-sections in D. D = dorsal; Ve = ventral. (C) Summary diagram of autopod elements that express lacZ in M1442-lacZ lines. Arabic numerals and associated vertical lines correspond to approximate plane of sections shown in D. Boxed Arabic numerals in D correspond to vertical lines in C. Color-coded autopod elements correspond to regions of positive lacZ signal in cross-sections shown in (D): light blue = anterior digit region; green = anterior carpal region; dark blue = posterior digit region; purple = anterior carpal region. Axes key: A = anterior; P = posterior; Di = distal; Pr = proximal. (D) Cross-sections through E15.5 M1442-lacZ L1 forelimb. Roman numerals indicate digit rays. Abbreviations: ce = centrate, fa = falciform carpal, lu = lunate, pi = pisiform, ra = radius, sc = scaphoid, td = trapezoid, tm = trapezium tq = triquetral, ul = ulna. Scale bars = 50 μm.
Table 1.
Summary of enhancer attributes
| M1442 |
M280 |
|
|---|---|---|
| Human ortholog (see enhancer.lbl.gov; Visel et al. 2007b) | H1442 | H280 |
| Method of Discovery | p300 association* | Ultraconservation^, p300 association* |
| Position in mouse genome (mm9) | Chr. 1: 165,827,290–165,830,012 | Chr. 4: 117,063,735–117,065,404 |
| Total length | 2,723 bp | 1,676 bp |
| Host gene | Kifap3 (19th intron) | Rnf220 (2nd intron) |
| Percent identity; mouse-on-human BLAT search (UCSC Genome Browser, mm9-on-hgl9) | 91.9 % (across 1,440 bp of total length) | 94.9 % |
The limb-specific transcriptional activity of M1442 was assessed in five generation zero (G0) transgenic mouse embryos and two stable transgenic lines, M1442-lacZ Lines 1 and 2 (M1442-lacZ L1 and L2). G0 embryos are derived directly from zygotes injected with gene constructs. Four of five E11.5 G0 transgenic mice displayed limb-specific lacZ expression patterns that overlapped with those driven by the orthologous H1442 sequence; in addition to driving posterior limb bud expression of lacZ, M1442 was capable of directing expression along the anterior margins of the limb bud (Fig. 1A; and data not shown). To characterize the transcriptional activity of M1442 beyond the E11.5 time point stable lines were made. Analysis of stable lines M1442-lacZ L1 and L2, both of which regulated lacZ similarly, showed that M1442 regulated dynamic limb-specific lacZ expression patterns throughout embryonic development (Fig. 1A.b–h). At E9–E9.5, lacZ expression is detected throughout the whole embryo, but it is particularly intense in the flanks in what may be the lateral plate mesoderm that gives rise to the mesenchymal portion of the limb bud (data not shown). By E10.5, reporter gene expression is weakly expressed in the limb bud in a diffuse manner, with a slightly more intense signal in the posterior half. By E11.5 intense lacZ expression was detectable in fore- and hindlimb buds similar to that seen in H1442 and M1442 G0 transgenic embryos (Fig. 1A.b, c). Histological sections of E11.5 M1442-lacZ limb buds showed intense lacZ expression in the mesenchymal cells of the posterior forelimb bud and weaker expression anteriorly (Fig. 1A.d). Also visible at this time point, is strong lacZ expression in the proximal-anterior margins of the forelimb bud, where the bud connects to the body wall – though this signal is less intense than that of the posterior limb bud. This anterior M1442-regulated expression, which persists into later stages, is not seen in any of the embryo images on the VEB that express lacZ under control of H1442. This indicates that the mouse enhancer has evolved species-specific regulatory capacities; alternatively, flanking sequence unique to M1442 may direct the anterior limb bud expression. In E12.5–13.5 embryos the posterior autopod expression is confined to the regions just proximal to and including the mesenchymal condensation that will give rise to digit V – tissue that likely corresponds to posterior wrist elements (Fig. 1A.e, f). Importantly, at E13.5, the anterior lacZ expression is now clearly detectable more distally, being present in developing digit I (Fig. 1A.f). Also at E13.5, anterior hindlimb expression is distinguishable in addition to the posterior expression pattern. By E15.5, expression is intense throughout forelimb digit I and in the wrist region just below the base of digit V (Fig. 1A.g). Hindlimbs at this time point are characterized by relatively intense lacZ expression in the tip of digit I (Fig. 1A.h). Expression is diminished greatly in digit I of forelimbs by E16.5, and is all but absent in the hindlimb footplate. Interestingly, expression is present in the skin throughout the entire limb and embryo in general at E16.5. Indeed, if the skin is removed one can see that there is very little expression in the underlying tissue. Histological sections of M1442-lacZ animals also show that M1442 can direct expression in the dermis of E15.5 embryos (data not shown). At E17.5 lacZ expression is absent in the limb (data not shown). Consistent lacZ expression was also detected in the genital tubercles (data not shown).
Thus, M1442-lacZ L1 and L2 embryos demonstrate that M1442's regulatory capacity is dynamic, active in specific, but changing locations of the developing limb, with a strong activity in the distal anterior and posterior margins of the limb bud. Therefore, phenotypes associated with deleted M1442 would likely result in aberrations in digits I and V, and in the wrist/ankle elements at the base of these digits.
Serial histological sections through E15.5 M1442-lacZ forelimbs revealed a somewhat complex lacZ expression pattern, one that interfaced with developing bone elements, likely dermal cell components and soft tissue surrounding both digits and wrist bones (Fig. 1B–D). Along the length of digits II and III, and digits IV and V, lacZ expression appears to be relatively constant in dermal layer of the skin (Fig. 1D). The distal and proximal phalanges of digit I are positive for lacZ expression, but the first metacarpal and carpal elements just proximal to the first metacarpal – the trapezium in particular – do not express lacZ. The falciform carpal, ventral to the trapezium, is lacZ positive. This indicates that on the anterior side of the limb, strong M1442-driven expression is restricted to digit I and the falciform carpal (Fig. 1D). The proximal phalanx and metacarpal of digit ray V are lacZ positive, as are the triquetral and pisiform proximal to digit V – lacZ expression in the posterior (ventral) margins of the pisiform are especially strong and consistent in the interior cells of the trabecular bone and in the thin layer of compact cells surrounding the spongy interior (Fig. 1D). On the posterior side of the limb, lacZ expression persisted in the soft tissues from at least the level of the fifth metacarpal all the way to the distal tip of the ulna; though lacZ-expressing cells do not appear in the distal ulnar tip itself, there is expression in the flanking dermal and mesenchymal cells (Fig. 1D).
H280 and selection and characterization of M280-lacZ transgenic mice
H280 drove a wedge-shaped lacZ expression pattern in the middle of the fore- and hindlimb buds (Fig. 1A.i). This distinct expression pattern corresponds approximately to the future location of digits II–III and potentially to elements of the central wrist and ankle. Understanding the regulatory inputs to this region of the limb bud interested us because of the diverse array of species-specific modifications to this region throughout evolutionary time; for example, in the bat, digits II–IV are highly elongated relative to digits I and V (Cretekos et al. 2005, Sears et al. 2006). Furthermore, numerous published embryonic and genetic manipulations in the chick and mouse focused on the posterior and anterior limb bud (Zeller et al. 2009); therefore it could be argued that less experimental attention has been directed at central autopod development. H280 was originally identified in a screen for highly conserved intergenic sequences (Visel et al. 2007b, Visel et al. 2008); in particular H280 encompasses a 256 base pair (bp) ultraconserved element (Bejerano et al. 2004, Visel et al. 2007b, Visel et al. 2008). Interestingly, a putative limb-specific enhancer sequence obtained in the p300-ChIP-seq screen carried out by Visel et al. (2009b) overlapped with 48 bp of the H280 ultraconserved element. We selected a 1,676 bp sequence orthologous to H280 and the overlapping p300 sequence from the mouse genome, M280, for the production of transgenic mice and gene targeting (Table 1). Along its entire length, M280 is 94.9% conserved with its human counterpart.
We assessed the ability of M280 to regulate lacZ expression in the limb and elsewhere throughout embryonic development in 10 G0 transgenic embryos and two stable transgenic lines. The lacZ expression pattern in the E11.5 M280-lacZ G0 transgenic embryos suggested that M280 regulates limb-specific gene expression similar to H280 (data not shown), therefore we proceeded to generate stable transgenic lines, M280-lacZ Line 1 and M280-lacZ Line 2 (M280-lacZ L1 and L2). Though the expression pattern of lacZ in the M280-lacZ L1 and L2 embryos was consistent with that seen in the G0 transgenic embryos, the expression in the stable lines was relatively weak and necessitated the use of Salmon-gal for lacZ staining. This resulted in greater overall background in stained M280-lacZ L1 and L2 but also sharpened the limb-specific expression pattern.
Embryos assayed for lacZ expression from both lines indicated that the limb-specific expression pattern regulated by M280 spans a narrow window of embryonic development, from E10.5 to E11.5. In both lines, limb bud expression was first detected at E10.5 and only persisted through to E11.5 (Fig. 1A.j, k). In all E12.5 M280-lacZ embryos collected lacZ expression was not detected in the developing limb (data not shown). The expression pattern in E10.5 limb buds presaged the wedge-shaped pattern seen in E11.5 embryos. Moreover, lacZ expression was just as intense at E10.5 as it was at E11.5 (data not shown).
Expression of lacZ was also detected in the posterior neural tube and tail bud as early as E9.5 (Fig. 1A.j). Expression in the tail bud is maintained until E12.5, and is occasionally detected at the very tip of the tail in E13.5 embryos (data not shown). lacZ expression is also present in the pharyngeal arches at E10.5, particularly in the oral groove of the first arch. The expression in the arches is weaker at E11.5 (Fig. 1A.j), and concomitant with the development of more advanced facial structures at E12.5 the expression in the craniofacial region is absent or weak in embryos of this age.
To gain a better understanding of the spatial extent of the expression pattern regulated by M280, we sectioned forelimb buds from E11.5 M280-lacZ embryos. Mesenchymal cells positive for lacZ were observed in the central mesenchymal cells of the E11.5 limb buds (Fig. 1A.l). Expression was especially intense in a central core group of expressing cells and weakened towards the anterior and posterior edges of the limb bud; cells positive for lacZ were not observed in the ectoderm.
Generation of M1442 and M280 deletion mice
To functionally test the role of M1442 and M280 during limb development we deleted either M1442 (M1442tm1) or M280 (M280tm1) from the mouse genome using gene targeting in ES cells (Fig. 2A, B). Mice homozygous for either enhancer deletion are referred to as −/− or null. Correct targeting and deletion of the enhancers was verified by Southern blot (Fig. S1). PCR assays were developed to genotype heterozygotes, nulls, and wild-type mice (Fig. 2C, D). All mice used in this study for phenotype analysis were on a 129/SvEv inbred background.
Figure 2. Gene targeting strategies for the generation of M1442tm1 and M280tm1 knockout mice.
(A) M1442 and (B) M280 deletion strategies. Thick lines correspond to 5' and 3' arms of homology. PCR genotyping primers, black arrows; Southern blot probes, shaded boxes. Sizes of DNA fragments recognized by Southern probes for each allele are shown. (C, D) PCR genotyping to distinguish between heterozygous and homozygous deletion and wild-type mice. Primers A–F, black arrows shown in A. neo, PGKneobpA-lox; tk, MC1tkpA.
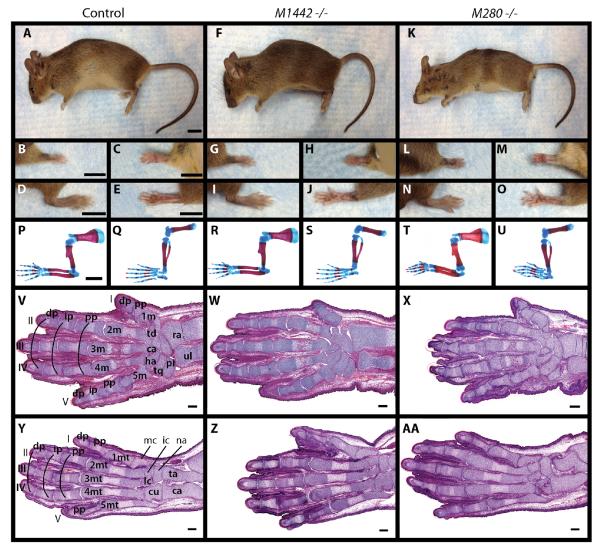
M1442tm1/+ were intercrossed to generate M1442tm1 null mice. Heterozygous and homozygous M1442tm1 mice are born at expected Mendelian ratios, are viable, fertile, and exhibit no obvious external limb phenotypes (Fig. 3A–J). To assess potential limb skeletal phenotypes that were not observed upon gross evaluation we stained newborn M1442tm1 null pups (postnatal day 0 or P0) with alizarin red and alcian blue for bone and the cartilage, respectively. M1442 null limbs appeared similar to control littermates (Fig. 3P–S). Although observations from M1442-lacZ embryos suggested that phenotypes would most likely arise in digit rays I and V and their proximal carpal elements we noted no aberrations in the cartilage and bone elements of these digit rays. Moreover, histological sections through E18.5 fore- and hindlimbs showed that digit and carpal elements in null mice were comparable to those in controls, including the presence of ossification centers in the elements of the digit rays (Fig. 3V, Y, W, Z). Therefore, initial external and internal observations of limb morphology suggested that M1442 is not required for limb development.
Figure 3. Comparison of newborn and adult M1442 and M280 null mice with control littermates.
(A–O) Adult female mice. (A–E) Controls. (F–J) M1442 −/−. (K–O) M280 −/−. (B, G, L) Dorsal and (C, H, M) ventral forelimbs. (D, I, N) Dorsal and (E, J, O) ventral hindlimbs. Scale bars in (A–E) = 9 mm. (P–U) Alizarin red and alcian blue stain for bone and cartilage in newborn pups. (P, Q) controls, (R, S) M1442 −/−. (T–U) M280 −/−. (P, R, T) forelimbs; (Q, S, U) hindlimbs. Scale bar in (P–U) = 1 mm. (V, Y) H&E stained planar sections through control E15.5 forelimb and hindlimb autopods, respectively. (W, Z) H&E stained planar sections through E15.5 1442tm1 −/− forelimb and hindlimb autopods, respectively. (X, AA) H&E stained planar sections through E15.5 280tm1 −/− forelimb and hindlimb autopods, respectively. ca, capitate; cl, calcaneus; cu, cuboid; dp, distal phalanx; ha, hamate; ic, intermediate cuneiform; ip, intermediate phalanx; lc, lateral cuneiform; mc, medial cuneiform; m, metacarpal; mt, metatarsal; na, navicular; pi, pisiform; pp, proximal phalanx; ra, radius; ta, talus; td, trapezoid; tq, triquetral; ul, ulna. Roman numerals correspond to digit rays. Images in (V-AA) are composites of serial sections through a single individual. Scale bars in (V-AA) = 100 μm.
Similar results were obtained when M280 null mice were generated from heterozygous M280tm1 intercrosses. Expression patterns in our M280-lacZ mice suggested that M280 null mice could manifest phenotypes associated with digits II–IV. However, adult mouse fore- and hindlimbs looked unaffected in null mice, and central digit and carpal elements were present and unaltered in alcian blue and alizarin red stained P0 pups (Fig. 3A–E, K–O, P, Q, T, U). Concordantly, histological sections of E18.5 null autopods demonstrated that ossification centers in all digit rays were unaffected by the absence of M280 (Fig. 3V, Y, X, AA). Therefore, initial external and internal observations of limb morphology suggested that M280 is not required for proper limb development.
Quantitative analysis of transcription in enhancer-null mice
Though no gross anatomical aberrations accompanied the deletion of either M1442 or M280 we predicted that gene transcription would be altered at the mutated loci. Both H/M1442 and H/M280 possess transcriptional enhancer activity, as demonstrated in transgenic mice. Moreover, M1442 is located in the 19th intron of Kifap3, whereas M280 lies within the second intron of Rnf220 (Fig. 4A, B). It is possible that both enhancers regulate the limb-specific transcription of their host gene (Donner and Williams 2006, Zhang and Williams 2003). On the other hand, it is possible the enhancers regulate one or several neighboring genes (Zuniga et al. 2004).
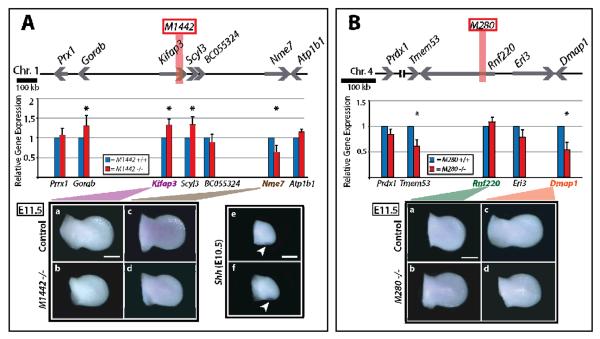
Figure 4. Enhancer-proximal transcript levels and whole mount in situ hybridization in M1442 −/−, M280 −/− and control E11.5 limb buds.
(A) M1442 locus. Assayed genes represented by gray arrows. The location of M1442 represented by vertical, orange bar. (B) M280 locus. Assayed genes represented by gray arrows. The location of M280 represented by vertical, orange rectangle. Significant transcript level changes at P = 0.05 represented by asterisks. WMISH of E11.5 limb buds for enhancer-proximal genes. Shh WMISH signal in E10.5 control limb bud (top) and M1442 −/− limb bud (bottom) indicated by white arrows. Scale bars = 0.5 mm.
Because our selection of M1442 and M280 was not based on knowledge of the genes they regulate we created a framework to determine which genes to assay for transcriptional differences between null and control embryos. Criteria were developed to determine which enhancer-proximal genes to assay by qPCR: 1) The host gene and at least one gene immediately 5' and 3' of the host gene; 2) Additional genes beyond the immediate neighbors of the host gene if they appeared in a PubMed and/or Google Scholar search in association with the key words “limb” and “development;” and 3) genes that were found up to 1 Mb upstream and downstream of the enhancers that were both a) expressed at a statistically significant level in limb buds relative to other cell types as reported by Cotney et al. (2012), and b) associated with the key words “limb” and “development” in a PubMed and/or Google Scholar search. The latter criterion stems from the observation that enhancers contributing to the transcriptional regulation of Shh and the Hoxd gene cluster are located at distances approaching 1 Mb (Lettice et al. 2003, Montavon et al. 2011, Sagai et al. 2004). To investigate whether deletion of M1442 and M280 affected gene transcription we performed qPCR on null and control E11.5 limb buds.
Transcriptional changes in M1442 null mice
We quantitatively assessed transcript levels at seven loci Atp1b1, BC055324, Gorab, Kifap3, Nme7, Prrx1, and Scyl3 near M1442 (Table SI.4). The host gene for M1442, Kifap3, encodes a component of a kinesin motor; such motors participate in ciliary mediated Shh expression (Kolpakova-Hart et al. 2007, Wong et al. 2009). Interestingly, deletion of M1442 altered the transcription levels of numerous genes located approximately 500 kb 5' or 3' of M1442 (Fig. 4A). In M1442 null limb bud transcript levels of Kifap3, Gorab, and Scy13 increased by 30%. Gorab and Scyl3 are located roughly 500 kb 5' and 30 kb 3' from M1442, respectively. In contrast, the expression level of Nme7, which is located ~410 kb 3' of M1442, was decreased to 65% of control levels (Fig. 4A). Deletion of M1442 had no effect on Prrx1, BC055324, and Atp1b1 transcript levels (Fig. 4A). In addition to altering average transcript levels, the absence of an enhancer may affect the transcriptional variance of a particular locus. Deletion of M1442 significantly reduced the variation in transcript levels at two loci, BC055324 and Nme7. Interestingly, in both cases transcriptional variation, determined from the cycle threshold standard deviations, was reduced in the mutant limbs, suggesting that either M1442 harbors elements that promote variation in transcription at these two loci or that the removal of M1442 brought BC055324 and Nme7 nearer to regulatory elements that foster transcriptional robustness (F-test: BC055324, p=0.02; Nme7, p=0.02; Table SI.7).
We used whole mount in situ hybridization (WMISH) to determine if Kifap3 and Nme7 are expressed in a limb-specific manner that overlaps with that spatially regulated by M1442 and to visually detect transcriptional alternations in E11.5 M1442 null limb buds. Additionally, Shh expression was monitored in E10.5 null and control limbs. WMISH indicated that Kifap3 is not expressed in a restricted manner in E11.5 wild-type limb buds (Fig. 4A.a, b). Thus the M1442 regulation of Kifap3 indicated by qPCR is not likely to be a required input for limb development. Nme7 does not appear to be expressed in a spatially restricted manner in the E11.5 limb bud (Fig. 4A.c, d). Visual detection of Shh transcription in null limbs was indistinguishable from controls; therefore the absence of the M1442 does not directly impact posterior limb bud Shh expression (Fig. 4A.e, f).
Transcriptional changes in M280 null mice
We quantitatively assessed transcript levels at five loci near M280 including Prdx1, Tmem53, Rnf220, Eri3 and Dmap1 (Table SI.5). Upon deletion of M280 the transcripts levels of its host gene, Rnf220, was unaffected in E11.5 limb buds, as was the transcription of Prdx1 and Eri3 (Fig. 4B). Tmem53 and Dmap1 transcript levels were significantly reduced by 60% and 50%, respectively (Fig. 4B). Currently, there is no reported role for Tmem53 or Dmap1 in limb development. Additionally, deletion of M280 significantly reduced the variation in transcript levels at one locus, Prdx1; as was the case in the M1442 null animals, Prdx1 transcriptional variation was reduced in mutant M280 limbs (F-test: Prdx1, p=0.03; Table SI.7). Thus M280 can regulate multiple genes within a given region in a selective manner.
We also used WMISH to determine if M280's host gene, Rnf220, or Dmap1 are expressed in a limb-specific manner that overlaps with that directed by M280 exhibited in the M280-lacZ mice. Neither Rnf220 nor Dmap1 were expressed in a restricted pattern in wild-type limb buds (Fig. 4B.a–d). We concluded that neither Rnf220 nor Damp1 are the primary regulatory targets of M280.
Analysis of autopod bone elements in null mice
Initial gross observations of our null mice showed that limb development was unperturbed despite the transcriptional changes detected near each deleted enhancer. The relatively few limb enhancer deletion experiments that have been conducted suggest that phenotypes may be subtle, (Menke et al. 2008, Montavon et al. 2011). Presumably this is because numerous enhancers may regulate the expression of a gene necessary in a particular anatomical structure, simultaneously mitigating large phenotypic shifts and enabling a gradual change in form as enhancers acquire mutations (Guenther et al. 2008). Empirical findings and theoretical explanations assert that separable enhancers regulate gene expression within a single anatomical element, often in a very incremental fashion (Guenther et al. 2008, Marinic et al. 2013). We decided to conduct finer-scaled measurements of digit elements in M1442 and M280 null embryos to determine whether subtle, though significant alterations in bone elements accompanied the loss of the enhancers.
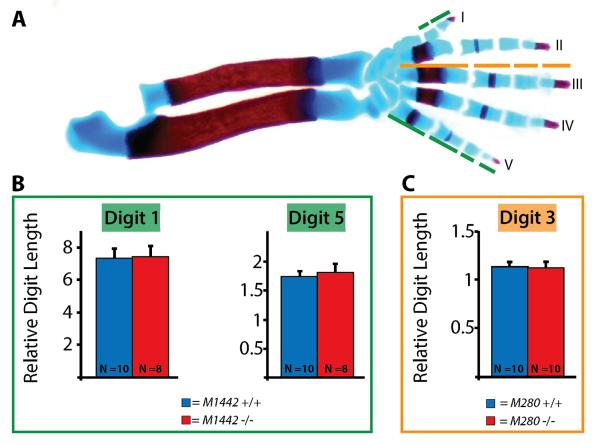
To assess whether M1442 null mice manifested minor, but significant limb phenotypes we collected eight null and ten control E18.5 embryos and dissected alcian blue and alizarin red stained forelimbs (Fig. 5A). Our M1442-lacZ lines indicated that digits I and V were likely to be affected by the targeted deletion of M1442. Lengths taken from each of the three individual elements of digit ray I (1st metacarpal, and the proximal and distal phalanges) and each of the four individual elements of digit ray V (4th metacarpal, and the proximal, intermediate and distal phalanges) were added together to obtain total digit I and V lengths (Fig. 5B). After digit I and V lengths were normalized to radii length (as a proxy for embryo size/weight; Fig. SI.2) we determined that the lengths of digits I and IV were not significantly different between M1442 null and control embryos (Fig. 5B).
Figure 5. Digit lengths in M1442 −/− and M280 −/− mice.
(A) Control E18.5 alizarin red and alcian blue stained forelimb. Green line segments indicate digit elements measured in control and M1442 −/− forelimbs. Orange line segments indicate digit elements measured in control and M280 −/− forelimbs. Digit lengths normalized to radius lengths. (B) Digit I and IV lengths in E18.5 M1442 −/− forelimbs are indistinguishable from those of control forelimbs. (C) Digit III lengths in E18.5 M280 −/− forelimbs are indistinguishable from those of control forelimbs.
We also collected ten M280 null E18.5 embryos and dissected alcian blue and alizarin red stained forelimbs to compare with controls. Our M280-lacZ lines indicated that digit III was likely to be affected by the targeted removal of M280 from the mouse genome. Our measurements and comparisons showed that normalized M280 −/− digit III lengths were no different than control digit III lengths (Fig. 5C).
Having not detected differences in gross embryonic bone morphology between either M1442 null or M280 null and control mice, we evaluated bone structure in adults. Forelimbs from three 5-month-old control, M1442 null, and M280 null female mice were subjected to a microCT scan and bone mass density (BMD) values for each forelimb autopod were collected. There was no statistical difference in BMD between either control and M1442 null mice or control and M280 null mice (Students t-Test: control and M1442 null, p=0.38; control and M280 null, p=0.68; Fig. SI.3).
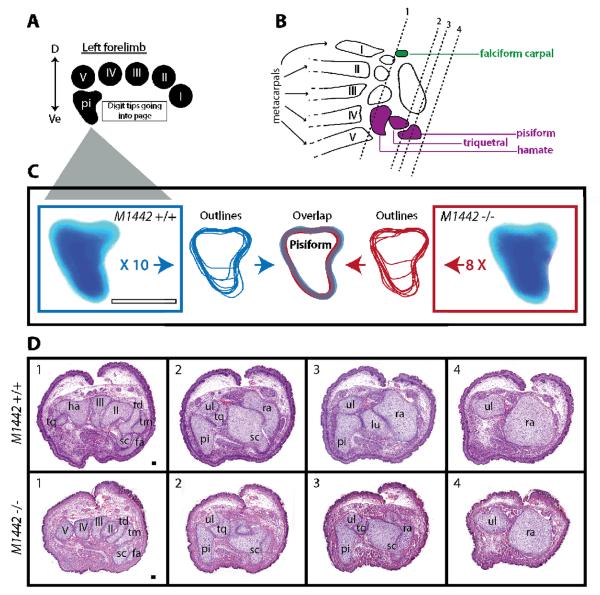
Assessment of carpal elements in M1442 null embryos
In addition to driving expression in the first and fifth digits, M1442 also regulates expression in anterior and posterior carpal elements, specifically in the falciform carpal and pisiform bones. Assessment of the pisiform bone was carried out by dissecting and imaging this carpal element from eight E18.5 M1442 null and ten control embryos (Fig. 6A, C). Each pisiform was analyzed closely under a dissecting microscope and images were taken of each bone. In Adobe Illustrator each pisiform was outlined and superimposed on outlines from other samples of like genotype. Finally, the grouped outlines from each genotype, M1442 null and control, were overlaid onto one another and we noted that the variation in size and shape from M1442 null individuals was minimal and comparable to that seen in pisiforms from controls (Fig. 6C). Consequently, we concluded that development of the pisiform is unaffected by the loss of M1442 gene regulation. We also assessed the shape, presence and relative size of the falciform carpal, hamate and triquetral elements in at least three E18.5 M1442 null individuals and noted no differences from controls.
Figure 6. Analysis of E18.5 M1442 null and control forelimb carpal regions.
(A) Orientation guide for images in C and cross-sections in D. D, dorsal; Ve, ventral. (B) Summary diagram of carpal elements potentially affected by the deletion of M1442. green, anterior carpal region; purple, posterior carpal region. Arabic numerals and associated vertical lines correspond to approximate plane of sections shown in D. Boxed Arabic numerals in D correspond to vertical lines in B. Roman numerals indicate digit rays. (C) Cartilaginous pisiforms dissected from control E18.5 forelimbs outlined in blue; pisiforms from E18.5 M1442 null forelimbs outlined in red. Outlines that don't extend the entire dorsal-ventral axis of the schematized pisiform (center) indicate that a particular pisiform was severed during dissection; however, the severed pisiforms were still useful in determining the dorsal contours of the element in control and mutant animals. Scale bar = 0.25 mm. (D) H&E stained sections of forelimb carpal regions in control and M1442 −/− E18.5 embryos. Roman numerals refer to digit rays. fa, falciform carpal; lu, lunate; pi, pisiform; ra, radius; sc, scaphoid; td, trapezoid; tm, trapezium; tq, triquetral; ul, ulna. Scale bar = 50 μm.
Conclusions drawn from the above analysis were confirmed in histological sections of E18.5 carpal elements from M1442 null and control animals (Fig. 6A, B, D). In frontal cross sections of null carpal elements we noted the presence of each carpal element expected to be possibly affected by the targeted deletion of M1442, including the falciform carpal and scaphoid in the anterior carpal region, and the hamate, triquetral and pisiform in the posterior carpal region (Fig. 6D). These histological observations confirmed our earlier findings that autopod development is unaffected by the deletion of M1442.
Growth phenotype in postnatal M280 null mice
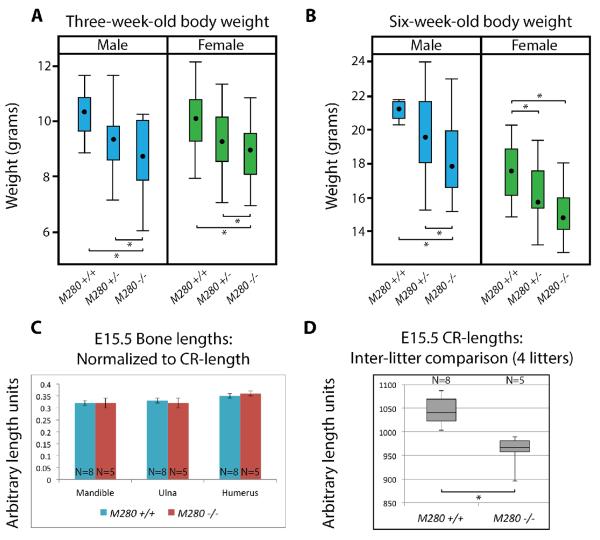
Early on in our experiments we noticed that M280 null individuals seemed to be consistently smaller than heterozygous and control littermates (Fig. 3A, K). To quantitatively confirm this observation we set up heterozygous M280tm1 intercrosses and recorded the weights of all male and female offspring at three weeks of age (weaning) and at six weeks of age (sexual maturity). We collected weights from 26 litters ranging in size from 3–10 pups (N=163 total mice). A statistical analysis confirmed our initial observations (SuppInfo_3and6wkWeights.xlsx). Female and male M280tm1 −/− mice weighed less than control mice at 3 weeks and 6 weeks (Fig. 7A, B; Females: at 3 weeks p=0.05; at 6 weeks p<0.001; Males: at 3 weeks p=0.003; at 6 weeks p=0.001; Tukey-Kramer adjustment for multiple comparisons used for all p-values). Moreover, the weight differences between female and male heterozygous and homozygous M280tm1 mice were also either significant or marginally significant, with null mice weighing less than heterozygous mice. Likewise, the weight differences between female and male heterozygous M280tm1 and control littermates were also significant or marginally significant, with heterozygous mice weighing less than controls. At 3 weeks M280tm1 −/− mice weigh, on average, about 1 gram (females) to 1.5 grams (males) less than control littermates (Fig. 7A). At six weeks male and female M280tm1 −/− mice weigh, on average, approximately 2.5 grams less than control littermates (Fig. 7B). Therefore, M280 null mice weigh approximately 10% less than control mice at six weeks of age. These results demonstrate that M280 is required for development and/or maintenance of postnatal body weight.
Figure 7. Adult and embryonic M280 null animals are smaller than controls.
(A) Box plot showing distribution of three-week-old body weight in control, heterozygous, and homozygous M280 mice. (B) Box plot showing distribution of six-week-old body weight in control, heterozygous, and homozygous M280 mice. (C) Graph shows average lengths of indicated cartilaginous elements normalized to CR-lengths at E15.5. Student's t-Test applied to normalized lengths indicated no significant differences; error bars indicate standard deviation. (D) Box plot showing distribution of E15.5 crown rump-lengths (CR-length) in M280 null and control embryos. Asterisks in (A), (B) and (D) indicate significant difference determined via Student's t-Test.
M280-lacZ LI and L2 embryos consistently expressed lacZ in the oral groove of the first pharyngeal arch at E11.5 (Fig. 1A.e, f). We hypothesized that development of the mandible and maxilla might be affected in null mice, perhaps leading to poor feeding abilities and ultimately to reduced weight. We observed no malocclusion of the teeth in adult mice. We also measured mandibular length in E15.5 null (N=5) and control (N=8) animals in 4 litters from M280tm1/+ intercrosses. When normalized to embryonic crown-rump length (CR-length) there is no significant difference in mandibular length between M280 nulls and controls (Fig. 7C).
Interestingly, while carrying out this analysis it was noticed that CR-lengths in mutants were always shorter relative to controls (Fig. 7D). Indeed, CR-lengths in E15.5 M280 mutants are significantly shorter than control CR-lengths (Fig. 7D; Student's t-Test, p<0.001; p-value still significant if inter-litter size variation is accounted for; moreover, the statistical analysis performed on adult body weights found no significant correlation between litter size and weight differences between genotypes; SuppInfo_ 3and6wkWeights.xlsx). Concordant with these findings is that raw, non-normalized mandibular and ulnar lengths are shorter in M280 mutants than in controls; and the lengths of mutant humeri are also generally shorter than controls (Fig. SI.4A, B). But, as with the mandible, if E15.5 ulnar and humeri lengths are normalized to CR-length the M280 mutant cartilaginous elements are indistinguishable from those of the controls (Fig. 7C). Therefore, we conclude from these observations that at E15.5 whole M280 embryos are smaller than control embryos, but that the proportional lengths of individual bone elements to overall body size, such as the mandible or ulna, are maintained in the mutants. Thus the weight differences observed between mutant and control postnatal mice are presaged by embryonic size differences by at least E15.5.
These findings corroborate results obtained while assaying for transient developmental defects in null limbs. We collected null and control embryos from at least three litters at E13.5, E14.5 and E15.5 and stained them for cartilage. We did not find any delay in the formation of cartilaginous precursors of autopod elements in M1442 or M280 null embryos at any time point but we noted that the majority of M280 mutant forelimbs were shorter than control forelimbs only at the E15.5 time point (data not shown). In conjunction with the CR-length data presented above, this observation bolsters the idea that size differences in M280 individuals begin around E15.5.
M280-flanking genes were queried for an association with the regulation of body size using CoCiter, a tool that identifies published research that co-cites a gene or members of a gene list with a term list (Qiao et al. 2013). When a gene list containing all genes ± 1 megabase pair upstream and downstream from M280 is searched against a term list containing body growth-related terms in CoCiter no publications are returned that link M280-flanking genes with body size (p=0.664; Supplemental Information).
DISCUSSION
The decision to assess the role of M280 and M1442 during mouse limb development stems from two initial observations: 1) in mouse limb buds both sequences were associated with p300, which is known to be associated with active enhancers and contributes to the acetylation of H3K27, a chromatin modification that is a signature of active transcription (Chan and La Thangue 2001, Kalkhoven 2004, Visel et al. 2009b); and 2) their orthologous human sequences, H1442 and H280, directed reporter gene expression in the limbs of E11.5 transgenic mice (Visel et al. 2007b). We demonstrated that M1442 and M280, like their human sequence counterparts, direct reporter gene expression in transgenic mouse limb buds. Importantly, we showed that transcription regulated by these enhancers begins before E11.5 (both M1442 and M280) and extends beyond this time as well (M1442). Thus, in further research aimed at characterizing enhancer sequences obtained in high throughput methods it will be important to detail the entire temporal and spatial range of an enhancer's regulatory capacity.
There was good reason to believe that both M280 and M1442 were required for limb development. In addition to being associated with p300, M280 harbors a 256 bp ultraconserved element (Bejerano et al. 2004, Visel et al. 2008) and its sequence identity along its entire length when compared to its homologous human sequence is 94.9%. Similarly, M1442 is associated with p300 and its sequence identity along a 1,440 bp stretch that aligns to the human ortholog is 91.9%. Moreover, both M280 and M1442 and their human counterparts drive lacZ expression in the developing limb. Despite these observations neither M280 nor M1442 are required for the development of a functional limb in the mouse. Much of our work focused on assessing the effect that M280/M1442 deletion might have on bone development. However, in histological sections, adult dissections, and observations of the mutant animals we did not see any indication that limb soft tissue was defective – mutant mice were active and mobile on par with control littermates.
The lack of a limb phenotype in the M280 and M1442 null mice represent only the second and third examples, respectively, of an undetected embryonic phenotype when a limb enhancer is deleted via gene targeting – mice lacking the Prrx1-limb-enhancer manifested no observed limb defects and limb-specific Prrx1 expression was unchanged in limb buds (Cretekos et al. 2008). Hence, although p300 is known to associate with sequences involved in active transcription this association does not in and of itself ensure that a p300-affiliated enhancer is required for the development of a specific tissue.
The absence of a limb phenotype in M280 is especially puzzling, given that in addition to its association with p300 it also contains an ultraconserved element (Bejerano et al. 2004, Visel et al. 2008). In addition to the ultraconserved element within M280, four other ultraconserved elements have been deleted from the mouse genome – none of them drive expression in the limb. These four other ultraconserved elements were selected because of their proximity to genes that have been shown to be required for particular developmental processes in knockout mouse experiments. However, none of the phenotypes resulting from the targeted deletion of the four genes is recapitulated in the ultraconserved element-knockout mice (Ahituv et al. 2007). To date, therefore, deletion of ultraconsered intergenic regions in the mouse genome has not led to predicted phenotypes. It will be interesting to see if this trend continues in future research; it might be possible that ulraconserved elements have additional roles in the genome beyond their regulatory abilities. It is intriguing that though our M280 mice possess no obvious defects in limb development they do manifest a general deficiency in body size/weight.
These results underscore several important concepts in enhancer biology. Firstly, just because an enhancer regulates gene expression in a particular embryonic tissue, such as the limb, does not mean that its main function is to direct gene expression in that tissue during embryonic development. Such an enhancer may actually contribute to development before the time at which it is assayed and/or contribute to tissue maintenance after birth regardless of its regulatory potential during development. Therefore, one could say that at specific time points and in specific tissues, enhancers, such as M1442 and M280, are developmentally tolerated. That is, they attract metabolically expensive pro-transcriptional machinery – such as p300, and likely a menagerie of TFs – and yet the result of this pro-transcriptional milieu, which is gene transcription in a particular tissue, such as the limb bud, is not necessary for the elaboration of that tissue and is perhaps secondary to a more important organismal role, such as growth regulation (Fig. SI.5). It will be interesting to determine if M280 directs gene transcription in extra-embryonic tissues or in adult tissues that might help in understanding the mechanism responsible for the reduced body size and weight seen in null animals. For example, previous studies have linked defects in the placenta with embryonic and adult body size (Li and Behringer 1998, Murthi et al. 2012, Rodriguez et al. 2004).
The results of the M280 knockout experiment are relevant for another reason. Recently, Cotney et al. (2013) demonstrated that H280, the homologous region in the human genome to M280, possessed increased H3K27ac marks relative to orthologous sequences in rhesus macaques and mice. Authors of this study suggested that the increased H3K27ac specifically in the human lineage, like those observed at H280, likely correspond to increased gene expression and thus to transcriptional differences that may account for the evolution of human-specific traits (Cotney et al. 2013). The deletion of M280 in the mouse indicates that predicting what roles an enhancer plays in evolution and development will be difficult without manipulating such an enhancer at its endogenous loci. So, for example, though the orthologous H280 sequence has a significant H3K27ac signature in human embryonic limbs perhaps an important role for this enhancer in evolutionary processes has been in the divergence of body size rather than limb morphology.
M280 −/− and M1442 −/− mice had molecular phenotypes. We demonstrated that significant transcriptional changes were detected in genes within the vicinity of each enhancer. As would be expected if enhancer activity were disrupted, particular genes near both M1442 and M280 were down-regulated (Nme7 near M1442; Tmem53 and Dmap1 near M280). Of interest at the M1442 locus is the observation that three genes that span a 600 kb region experienced an increase in transcripts levels, including Kifap3, which hosts the M1442 enhancer (the other genes are Gorab and Scyl3). This is a somewhat unexpected result in that we know from our M1442-lacZ mice that M1442 should activate transcription, not repress it. One interpretation of these results is that it is not the absence of a repressor element within M1442 per se that is causing the minor increase in transcription, but the topographical shift of the promoter regions of the affected genes (Spitz et al. 2005). Perhaps the removal of the approximately 3 kb M1442 sequence moved the promoter regions of these genes nearer to another enhancer element. Alternatively, it is possible that M1442 harbors repressor and enhancer sequences and in its endogenous context the repressor elements override the enhancer elements for at least Kifap3, Scyl3 and Gorab. Yet one more possibility is that additional enhancer sequences might act in concert with M1442 to properly regulate transcription in this genomic window and in the absence of M1442 this balanced regulatory interaction is thrown off and an up-regulation of nearby genes results. We also note that we collected the entire E11.5 limb bud for the qPCR assays and therefore the unexpected results may be a result of differential gene expression from changed proportions of specific limb bud cell subpopulations that were altered as a result of the M1442 deletion.
Transcriptional alterations in M280 and M1442 null mice ranged from 30% increases to 45% decreases. Similar magnitudes of transcriptional change have been noted with the deletion of other limb enhancer loci, and have been accompanied by observable phenotypes, including enhancers regulating the Hoxd cluster (Montavon et al. 2011) and Tbx4 (Menke et al. 2008). Therefore, whether the main role of M280 and M1442 is to regulate the genes that are affected by their absence or not, the more interesting observation is that there are transcriptional changes in the limb and yet there are no accompanying detectable phenotypes.
A common explanation for the lack of phenotypes, or the presence of only minor phenotypes, in enhancer knockout experiments is that additional enhancers compensate for the loss of those that were deleted (Cretekos et al. 2008). Certainly, distinct enhancers, often referred to as shadow enhancers, capable of driving similar and overlapping expression patterns have been well documented in multiple species, from fruit flies to mice (Hong et al. 2008, Marinic et al. 2013, Montavon et al. 2011). Also, the evidence for networks of enhancers that contribute to the expression of a single gene or gene groups has been well documented (Guillaume Andrey 2013, Montavon et al. 2011).
It is also possible, however, that the unaffected limb development in our mutant mice is a result of redundancy in genetic pathway activity rather than regulatory activity. For example, although M1442 may be required for the proper limb expression of a particular gene, the effects of reduced transcription of that gene in mutant mice may be compensated for by another redundantly expressed gene that feeds into the same genetic circuit. Redundant gene function has been amply demonstrated for numerous gene families during limb development, including Hoxa and Hoxd cluster members, Bmps and Fgfs (Bandyopadhyay et al. 2006, Davis et al. 1995, Fromental-Ramain et al. 1996, Niswander et al. 1993). But even this possibility does not satisfactorily explain why M280 and M1442 possess so many independent signatures of enhancer activity (Bejerano et al. 2004, Visel et al. 2007b, Visel et al. 2008, Visel et al. 2009b) and yet are not required in limb development.
For this reason we to return to the concept of developmental tolerance and the possibility that embryonic enhancer activity does not always predict well an enhancer's role throughout the life of an organism. Our results and those of others at least suggest that increasing our understanding of the role of enhancer regulation in both development and evolution will require more experiments aimed at disrupting enhancers at their endogenous loci. New experimental approaches may also be required to subject enhancer-deleted and enhancer-manipulated mouse lines to proxy ecological conditions to enhance detection of phenotypes dependent on gene regulation that is called upon in the daily and strenuous exertions of animals in natural conditions.
Supplementary Material
Acknowledgements
We thank Jan Parker-Thornburg and Chad Smith (MD Anderson Cancer Center) for helpful guidance with mice; Kendra Alton, Lindsey Minter, and Sabrina Stratton (MD Anderson Cancer Center) and Colin Maguire (University of Utah) for guidance with qPCR; and Nadav Ahituv (UCSF) for providing pGS-HSP-LZ. Supported by the Ben F. Love Endowment to R.R.B. M.J.N. was supported by NIH T32 grant HD07325 and a Hearst Foundation Fellowship. Veterinary services, microCT scans, and DNA sequencing were supported by NIH grant CA16672.
REFERENCES
- Ahituv N, Prabhakar S, Poulin F, Rubin EM, Couronne O. Mapping cis-regulatory domains in the human genome using multi-species conservation of synteny. Hum Mol Genet. 2005;14:3057–3063. doi: 10.1093/hmg/ddi338. [DOI] [PubMed] [Google Scholar]
- Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A, Tsuji K, Cox K, Harfe BD, Rosen V, Tabin CJ. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2:e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Behringer RR, Finegold MJ, Cate RL. Mullerian-inhibiting substance function during mammalian sexual development. Cell. 1994;79:415–425. doi: 10.1016/0092-8674(94)90251-8. [DOI] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- Chandler RL, Chandler KJ, McFarland KA, Mortlock DP. Bmp2 transcription in osteoblast progenitors is regulated by a distant 3' enhancer located 156.3 kilobases from the promoter. Mol Cell Biol. 2007;27:2934–2951. doi: 10.1128/MCB.01609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Leng J, Oh S, DeMare LE, Reilly SK, Gerstein MB, Noonan JP. Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome Res. 2012;22:1069–1080. doi: 10.1101/gr.129817.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Leng J, Yin J, Reilly SK, Demare LE, Emera D, Ayoub AE, Rakic P, Noonan JP. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell. 2013;154:185–196. doi: 10.1016/j.cell.2013.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretekos CJ, Wang Y, Green ED, Martin JF, Rasweiler JJt, Behringer RR. Regulatory divergence modifies limb length between mammals. Genes Dev. 2008;22:141–151. doi: 10.1101/gad.1620408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretekos CJ, Weatherbee SD, Chen CH, Badwaik NK, Niswander L, Behringer RR, Rasweiler JJt. Embryonic staging system for the short-tailed fruit bat, Carollia perspicillata, a model organism for the mammalian order Chiroptera, based upon timed pregnancies in captive-bred animals. Dev Dyn. 2005;233:721–738. doi: 10.1002/dvdy.20400. [DOI] [PubMed] [Google Scholar]
- Davis AP, Witte DP, Hsieh-Li HM, Potter SS, Capecchi MR. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature. 1995;375:791–795. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- Donner AL, Williams T. Frontal nasal prominence expression driven by Tcfap2a relies on a conserved binding site for STAT proteins. Dev Dyn. 2006;235:1358–1370. doi: 10.1002/dvdy.20722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Huang J, Zhang J, Williams T. Identification and analysis of a conserved Tcfap2a intronic enhancer element required for expression in facial and limb bud mesenchyme. Mol Cell Biol. 2008;28:315–325. doi: 10.1128/MCB.01168-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromental-Ramain C, Warot X, Messadecq N, LeMeur M, Dolle P, Chambon P. Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development. 1996;122:2997–3011. doi: 10.1242/dev.122.10.2997. [DOI] [PubMed] [Google Scholar]
- Guenther C, Pantalena-Filho L, Kingsley DM. Shaping skeletal growth by modular regulatory elements in the Bmp5 gene. PLoS Genet. 2008;4:e1000308. doi: 10.1371/journal.pgen.1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume Andrey TM, Bénédicte Mascrez, Federico Gonzalez, Daan Noordermeer, Marion Leleu, Didier Trono, François Spitz, Denis Duboule. A Switch Between Topological Domains Underlies HoxD Genes Collinearity in Mouse Limbs. Science. 2013;340 doi: 10.1126/science.1234167. [DOI] [PubMed] [Google Scholar]
- Herault Y, Rassoulzadegan M, Cuzin F, Duboule D. Engineering chromosomes in mice through targeted meiotic recombination (TAMERE) Nat Genet. 1998;20:381–384. doi: 10.1038/3861. [DOI] [PubMed] [Google Scholar]
- Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkhoven E. CBP and p300: HATs for different occasions. Biochem Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- Kolpakova-Hart E, Jinnin M, Hou B, Fukai N, Olsen BR. Kinesin-2 controls development and patterning of the vertebrate skeleton by Hedgehog- and Gli3-dependent mechanisms. Dev Biol. 2007;309:273–284. doi: 10.1016/j.ydbio.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20:R754–763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Behringer RR. Esx1 is an X-chromosome-imprinted regulator of placental development and fetal growth. Nat Genet. 1998;20:309–311. doi: 10.1038/3129. [DOI] [PubMed] [Google Scholar]
- Maguire CT, Demarest BL, Hill JT, Palmer JD, Brothman AR, Yost HJ, Condic ML. Genome-wide analysis reveals the unique stem cell identity of human amniocytes. PLoS One. 2013;8:e53372. doi: 10.1371/journal.pone.0053372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinic M, Aktas T, Ruf S, Spitz F. An integrated holo-enhancer unit defines tissue and gene specificity of the Fgf8 regulatory landscape. Dev Cell. 2013;24:530–542. doi: 10.1016/j.devcel.2013.01.025. [DOI] [PubMed] [Google Scholar]
- Masuya H, et al. A series of ENU-induced single-base substitutions in a long-range cis-element altering Sonic hedgehog expression in the developing mouse limb bud. Genomics. 2007;89:207–214. doi: 10.1016/j.ygeno.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Menke DB, Guenther C, Kingsley DM. Dual hindlimb control elements in the Tbx4 gene and region-specific control of bone size in vertebrate limbs. Development. 2008;135:2543–2553. doi: 10.1242/dev.017384. [DOI] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Murthi P, Kalionis B, Rajaraman G, Keogh RJ, Da Silva Costa F. The role of homeobox genes in the development of placental insufficiency. Fetal Diagn Ther. 2012;32:225–230. doi: 10.1159/000339657. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R, editors. Manipulating the Mouse Embryo: A Laboratory Manual. 2003. [Google Scholar]
- Niswander L, Tickle C, Vogel A, Booth I, Martin GR. FGF-4 replaces the apical ectodermal ridge and directs outgrowth and patterning of the limb. Cell. 1993;75:579–587. doi: 10.1016/0092-8674(93)90391-3. [DOI] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci U S A. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U, Wassarman DA. Promoting developmental transcription. Development. 2010;137:15–26. doi: 10.1242/dev.035493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Qiao N, Huang Y, Naveed H, Green CD, Han JJ. CoCiter: an efficient tool to infer gene function by assessing the significance of literature co-citation. PLoS One. 2013;8:e74074. doi: 10.1371/journal.pone.0074074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz AH, Vokes SA. Integration of the transcriptional networks regulating limb morphogenesis. Dev Biol. 2012;368:165–180. doi: 10.1016/j.ydbio.2012.05.035. [DOI] [PubMed] [Google Scholar]
- Ramirez-Solis R, Rivera-Perez J, Wallace JD, Wims M, Zheng H, Bradley A. Genomic DNA microextraction: a method to screen numerous samples. Anal Biochem. 1992;201:331–335. doi: 10.1016/0003-2697(92)90347-a. [DOI] [PubMed] [Google Scholar]
- Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- Rodriguez TA, et al. Cited1 is required in trophoblasts for placental development and for embryo growth and survival. Mol Cell Biol. 2004;24:228–244. doi: 10.1128/MCB.24.1.228-244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S, Symmons O, Uslu VV, Dolle D, Hot C, Ettwiller L, Spitz F. Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat Genet. 2011;43:379–386. doi: 10.1038/ng.790. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- Sagai T, Masuya H, Tamura M, Shimizu K, Yada Y, Wakana S, Gondo Y, Noda T, Shiroishi T. Phylogenetic conservation of a limb-specific, cis-acting regulator of Sonic hedgehog (Shh) Mamm Genome. 2004;15:23–34. doi: 10.1007/s00335-033-2317-5. [DOI] [PubMed] [Google Scholar]
- Sears KE, Behringer RR, Rasweiler JJt, Niswander LA. Development of bat flight: morphologic and molecular evolution of bat wing digits. Proc Natl Acad Sci U S A. 2006;103:6581–6586. doi: 10.1073/pnas.0509716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubin N, Tabin C, Carroll S. Fossils, genes and the evolution of animal limbs. Nature. 1997;388:639–648. doi: 10.1038/41710. [DOI] [PubMed] [Google Scholar]
- Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- Shubin NH, Daeschler EB, Jenkins FA., Jr The pectoral fin of Tiktaalik roseae and the origin of the tetrapod limb. Nature. 2006;440:764–771. doi: 10.1038/nature04637. [DOI] [PubMed] [Google Scholar]
- Spitz F, Herkenne C, Morris MA, Duboule D. Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat Genet. 2005;37:889–893. doi: 10.1038/ng1597. [DOI] [PubMed] [Google Scholar]
- Sundararajan S, Wakamiya M, Behringer RR, Rivera-Perez JA. A fast and sensitive alternative for beta-galactosidase detection in mouse embryos. Development. 2012;139:4484–4490. doi: 10.1242/dev.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taher L, McGaughey DM, Maragh S, Aneas I, Bessling SL, Miller W, Nobrega MA, McCallion AS, Ovcharenko I. Genome-wide identification of conserved regulatory function in diverged sequences. Genome Res. 2011;21:1139–1149. doi: 10.1101/gr.119016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarchini B, Duboule D. Control of Hoxd genes' collinearity during early limb development. Dev Cell. 2006;10:93–103. doi: 10.1016/j.devcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Visel A, Bristow J, Pennacchio LA. Enhancer identification through comparative genomics. Semin Cell Dev Biol. 2007a;18:140–152. doi: 10.1016/j.semcdb.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009a;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007b;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009b;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes SA, Ji H, Wong WH, McMahon AP. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 2008;22:2651–2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltering JM, Duboule D. The origin of digits: expression patterns versus regulatory mechanisms. Dev Cell. 2010;18:526–532. doi: 10.1016/j.devcel.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Wong SY, Seol AD, So PL, Ermilov AN, Bichakjian CK, Epstein EH, Jr., Dlugosz AA, Reiter JF. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med. 2009;15:1055–1061. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller R, Lopez-Rios J, Zuniga A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat Rev Genet. 2009;10:845–858. doi: 10.1038/nrg2681. [DOI] [PubMed] [Google Scholar]
- Zhang J, Williams T. Identification and regulation of tissue-specific cis-acting elements associated with the human AP-2alpha gene. Dev Dyn. 2003;228:194–207. doi: 10.1002/dvdy.10365. [DOI] [PubMed] [Google Scholar]
- Zuniga A, et al. Mouse limb deformity mutations disrupt a global control region within the large regulatory landscape required for Gremlin expression. Genes Dev. 2004;18:1553–1564. doi: 10.1101/gad.299904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.