Abstract
The aspartic protease cathepsin D (Clan AA, Family A1) is expressed in the schistosome gut where it plays an apical role in the digestion of hemoglobin released from ingested erythrocytes. In this report, RNA interference approaches were employed to investigate the effects of knockdown of schistosome cathepsin D. Cultured schistosomules of Schistosoma mansoni were exposed by square wave electroporation to double stranded RNA (dsRNA) specific for cDNA encoding S. mansoni cathepsin D. RNAi mediated reductions in transcript levels led to phenotypic changes including significant growth retardation in vitro and suppression of aspartic protease enzyme activity. In addition, black-pigmented heme, the end point by-product of normal hemoglobin proteolysis that accumulates in the schistosome gut, was not apparent within the guts of the treated schistosomules. Rather, their guts appeared to be red in color, rather than black, apparently indicating the presence of intact rather than digested host hemoglobin. These phenotypic effects were apparent when either of two forms of dsRNA, a long form spanning the entire target transcript or a short form specific for the 3’-region were employed. Off-target effects were not apparent in transcript levels of the gut-localized cysteine protease cathepsin B1. Finally, cathepsin D may be an essential enzyme in the mammal-parasitic stages of schistosomes because schistosomules treated ex vivo with dsRNA did not survive to maturity after transfer into Balb/c mice. These and earlier findings suggest that, given its essential function in parasite nutrition, schistosome cathepsin D could be developed as a target for novel anti-schistosomal interventions.
Keywords: schistosomes, cathepsin D, RNAi, schistosomules, hemoglobin proteolysis, RT-PCR
1. Introduction
Schistosomes feed on ingested host blood. Hemoglobin released from the ingested erythrocytes is essential for parasite development, growth and reproduction[1]. Identifying the key enzymes that facilitate host nutrient degradation is important not only to understand the biology and pathogenesis of schistosome infection but also to identify those enzymes that might be viable targets for development of new interventions. Cysteine and aspartic proteases have been described in secretions and/or worm extracts of schistosomules and adult worms [2], and previous studies have shown that inhibitors of either cysteine proteases [3] or aspartic proteases [4] block hemoglobin degradation and arrest schistosome development and egg production. A proteolytic cascade or network involving aspartic and cysteine proteases has been proposed as catalyzing hemoglobin degradation in schistosomes [5-7]. Aspartic proteases also play central roles in the degradation of ingested blood and hemoglobin in other blood-feeding parasitic worms, and sites of cleavage within hemoglobins by both hookworm and schistosome aspartic proteases have been mapped [1, 5, 8]. Moreover, from an evolutionary perspective, McKerrow and colleagues have hypothesized, based on findings with parasitic helminths, that early metazoans evolved a successful digestive network of proteases comprising cathepsins D, B, L, C and legumain that predated the evolution of the pancreas and the subsequent primacy of serine proteases as digestive enzymes in vertebrates [6].
A cathepsin D aspartic protease, able to degrade human and other mammalian hemoglobins, has been previously described in adults worm extracts from S. mansoni [9, 10]. Due to its physiological significance for schistosomes, deeper studies on its molecular and biochemical properties, gene structure and phylogeny have been undertaken [6, 11, 12]. Although cathepsin D-like protease (clan AA) has been involved in digestion of hemoglobin within the adult stages of S. mansoni, it remains unclear whether this enzyme is expressed in and functions in the schistosomula stage early in mammalian infection. Given the presence of an intact RNAi pathway in S. mansoni [13], it should be feasible to specifically investigate the role of S. mansoni cathepsin D (SmCD) by RNAi-mediated gene silencing. Skelly et al. [14] established the feasibility of experimental RNAi in S. mansoni by describing knockdown of the gut-associated cysteine protease cathepsin B1 after soaking cultured schistosomula in the dsRNA. Krautz-Peterson et al. [15] extended these studies by comparing and optimizing RNAi methodology for schistosomes and effectively silencing cathepsin B1 transcript levels. Also, they reported that square wave electroporation was exceedingly more efficient - 100 to1000 fold - than either soaking the parasites in an equivalent dsRNA dose or soaking them in an equivalent dose in the presence of lipofectamine reagents.
Here we investigated the expression and developmental importance of SmCD in the schistosomulum stage of the blood fluke using RNAi manipulations. With the aim of establishing a more effective gene silencing, we employed electroporation procedures (as recommended [14]) to introduce dsRNA into cultured schistosomules. This developmental stage is readily maintained in vitro [16] and the life cycle can be subsequently re-established in vivo by needle passage into mice of schistosomules that have been genetically transformed in vitro [17]. This system is attractive and tractable for RNAi based investigation of gene function in S. mansoni [17, 18]. Changes in SmCD gene expression were detected using reverse transcription PCR without apparent effect on unrelated transcripts. Suppression of SmCD transcripts was accompanied by profound reduction in cathepsin D enzyme activity. In vitro assays demonstrated that schistosomes treated with SmCD-dsRNA did not develop intestinal heme pigmentation, indicative of dysregulation of hemoglobin proteolysis, and the growth of the treated schistosomules was inhibited. Moreover, investigation of the fate in mice of SmCD-dsRNA treated schistosomules appeared to confirm that this aspartic protease is an essential gene for normal development of blood stage parasites and that silencing of SmCD resulted in a lethal outcome for the schistosome in vivo.
2. Materials and Methods
2.1. Parasites
Biomphalaria glabrata snails infected with the NMRI (Puerto Rican) strain of Schistosoma mansoni were supplied by Dr. Fred Lewis, Biomedical Research Institute, Rockville, MD. Schistosomules were mechanically transformed from cercariae released from infected B. glabrata snails. Briefly, cercariae were concentrated by centrifugation (2000 x rpm/ 10 min) and washed once with somule wash medium (RPMI 1640 supplemented with 200 U/ml Penicillin G sulfate, 200 μg/ml streptomycin sulfate, 500 ng/ml amphotericin B, 10 mM HEPES). Cercarial tails were sheared off by 20 passes through 22G emulsifying needles after which schistosomule bodies were isolated free from tails by Percoll gradient centrifugation [19]. Schistosomula were washed 3 times in wash medium and cultured at 37° C under 5% CO2 in air in modified Basch’s medium [16] supplemented with small quantities of washed human erythrocytes. In some experiments, parasites were incubated in wash medium at 37° C under 5% CO2 for 3 hours and recovered directly for electroporation.
2.2. Synthesis of dsRNAs
Full length (1.2 kb, residues 37 to 1,285 of the SmCD transcript) and 0.5 kb (residues 655 to 1154 of the transcript) dsRNAs of SmCD were synthesized from cDNA encoding S. mansoni cathepsin D (GenBank accession U60995) using gene-targeted primers containing T7 promoter sequences ( F: 5’- TAA TAC GAC TCA CTA TAG GGT GAA GTG GTT AGG ATC CCT CT-3’; R:5’- TAA TAC GAC TCA CTA TAG GGT CAA ACT TCA TCT GAA AGA AG-3’ and F: 5’- TAA TAC GAC TCA CTA TAG GGT GAC CTG ATG ATT GGT GG-3’; R:5’- TAA TAC GAC TCA CTA TAG GGT CTG GAT GAA GAG CTT TCG C-3’) (Figure 1). Control luciferase dsRNA template encoding the full length 1,672 bp transcript was synthesized from plasmid pGL3-Basic (Promega, Madison, WI) which encodes Photinus pyralis (firefly) luciferase (F: 5’-TAA TAC GAC TCA CTA TAG GGT GCG CCC GCG AAC GAC ATT TA-3’; R: 5’-TAA TAC GAC TCA CTA TAG GGG CAA CCG CTT CCC CGA CTT CCT TA-3’). Leucine aminopeptidase 1 (GenBank U83906) (LAP1) and leucine aminopeptidase 2 (S. mansoni Gene DB Smp_083870.2) (LAP2) cDNAs were employed as templates for synthesis of LAP1-dsRNA and LAP2-dsRNA using gene-targeted primers containing T7 promoter sequences (F: 5’-TAA TAC GAC TCA CTA TAG GGA CGA ACA TTA GCA CGA GAT ATT-3’; R: 5’- TAA TAC GAC TCA CTA TAG GGC ATA ACC ATT CTA CCT TCA GCA-3’ spanning coding DNA positions 543-1077; and F: 5’- TAA TAC GAC TCA CTA TAG GGG CTGAAGTCC TGG GTT GGT T-3’; R: 5’- TAA TAC GAC TCA CTA TAG GGC CAT TCG ACC TTC AGC ATC A-3’ spanning coding DNA positions 502-1117, respectably). dsRNA was synthesized and purified using the Megascript RNAi kit (Ambion, Austin, TX) according to the manufacturer’s instructions. dsRNA was precipitated with 1 volume of 5 M ammonium acetate and 2.5 volumes of 95% ethanol after which the RNA pellet was dissolved in water. Integrity of the dsRNAs was verified by non-denaturing 1% agarose gel electrophoresis, and concentration and purity determined with a spectrophotometer (ND-1000, NanoDrop Technologies, Wilmington, DE).
Figure 1.
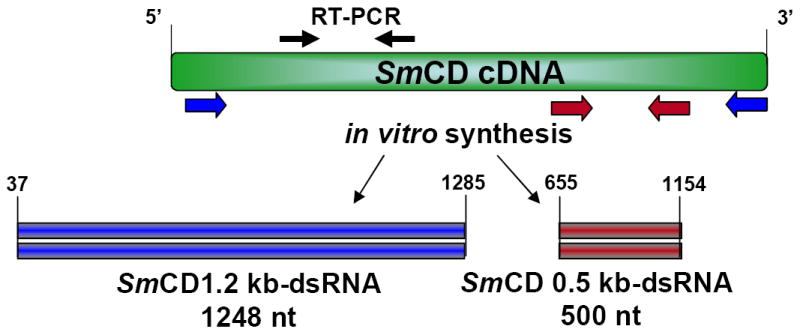
Schematic representation of the Schistosoma mansoni cathepsin D (SmCD) transcript and open reading frames (ORFs) illustrating the regions from which the double-stranded RNAs (red and blue) used in this study were synthesized. Solid black arrows indicate the relative positions of the primers used in the RT-PCRs.
2.3. Electroporation of schistosomules
For electroporation of dsRNA, the schistosomula were removed from culture at three hour and at seven, nine and 11 days after cercarial transformation. Electroporation was performed as described [20]. In brief, at the time of electroporation, dsRNA was added at a final concentration 0.1 μg/μl to 3 hour old schistosomules, and at a final concentration 0.3μg/μl for seven, nine and 11 day old somules. Electroporations using a single 20 ms square wave pulse at 125V were accomplished in 4 mm gap cuvettes (BTX, San Diego, CA) using a BTX ElectroSquarePorator™ ECM830 with ~2,000 schistosomules suspended in 100 μl of wash medium containing 10 μg or 30 μg of dsRNA. Immediately following electroporation, parasites were transferred to pre-warmed Basch’s medium and maintained in culture, as above.
In some experiments, dsRNA transformed schistosomules were used to infect female Balb/c mice. For this, ~500 schistosomules in ~ 200 μl Modified Eagle’s Medium were injected into the thigh muscle of mice using a 22G syringe, as described [18]. Sixty days later, the mice were euthanized, adult worms recovered by portal vein perfusion, gross pathology of livers examined, and schistosome eggs recovered from the livers by digestion with bacterial collagenase [21].
2.4. Gene expression analysis
The endogenous expression of SmCD was determined in schistosomula cultured for 48 hours, seven days and 14 days. RNA was extracted from the worms using the RNAqueus-Micro Kit (Ambion) following the manufacturer’s instruction. Any residual DNA remaining in the RNA preparations was removed by DNase digestion using a TurboDNase kit (Ambion). cDNA was synthesized using 10-fold serial dilutions of schistosomula RNA starting with 50 ng RNA, using the iScript™ cDNA Synthesis Kit (BioRad, Hercules, CA). SmCD. cDNA was amplified using F: 5’- CGG TGA TAT AAC GAT TGG TAC GCC-3’; R: 5’- GAA TAC AGG AGT AAC GCC ATC CAC (spanning cathepsin D coding DNA positions 203-575). Cathepsin B1 (GenBank AJ506157) F: 5’-ATA TCA ATG AAC ATC CGA ATG CTG-3’; Rev: 5’-CCC AAG CAG GAC CAA GGA TAC CAC-3’ [18] (spanning cathepsin B1 coding region, residues 126 to 522) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (GenBank M92359) F:5’- GTG GTT TCT ACC GCA TCA TG-3’; R: 5’-CAG CCT TCG CAT CAA AGA TA-3’ (spanning GAPDH coding region, residues 773 to 1235). The PCR conditions included an initial denaturation at 94° C for one minute followed by 30 cycles of 30 sec at 94° C, 30 sec at 51° C, 30 sec at 72° C and a final extension at 72° C for 10 min. Images of PCR products in agarose gels were captured using Versadoc software (BioRad).
2.5 Cathepsin D enzyme activity assay
Soluble protein extracts from ~1,000 schistosomula were prepared by sonication-induced lysis (5 × 5 second bursts on ice, output control value 3, model W-220F Sonicator, Heat Systems – Ultrasonics, Inc, Plainview, New York) in 100 mM sodium formate, pH 3.5. After centrifugation of the lysate for 10 minutes at 4° C at 14,000 rpm, the supernatant was employed as soluble schistosome extract. The protein concentration in the soluble fraction was determined using the bicinchoninic acid assay (BCA kit, Pierce, Rockford, IL). A total of 1μg of soluble protein from each group was added in triplicate to a 100 μl reaction mixture of 100 mM sodium formate pH 3.5 and the fluorogenic peptide 7-methoxycoumarin-4-acetyl-Gly-Lys-Pro-Ile-Leu-Phe-Phe-Arg-Leu-Lys(DNP)-D-Arg-amide (Sigma-Aldrich, St. Louis, MO) at a final concentration of 2 μM and incubated at 37°C for 30 minutes. Fluorescence from substrate hydrolysis was measured in a fluorometer at 393 nm with excitation at 328 nm [22]. To confirm that cathepsin D activity was being measured, parasite extract was pre-incubated for 10 min with 20 μM concentration of the broad-spectrum aspartic protease inhibitor, pepstatin A (Sigma), prior to the addition of the substrate.
2.6. Parasite measurements
Schistosomules were fixed overnight at 4° C in 10% formalin in phosphate buffered saline after which they were photographed using an inverted microscope (Nikon Eclipse TS100) fitted with a digital camera (CoolPix 5700, Nikon). The area occupied by individual schistosomules was measured using Image J Version 1.37 software [23]. Student’s t-test was employed to assess the statistical significance of differences observed.
3. Results
3.1. Schistosomules treated with cathepsin D dsRNA show retarded growth after 14 days post electroporation
At the outset, we investigated whether we could suppress expression of SmCD via the RNAi pathway and if so, second, would a visible phenotype manifest in the SmCD gene silenced schistosomula. As illustrated in Figure 1, two different length dsRNAs were employed in our studies. First, SmCD 1.2 kb-dsRNA was employed for treatment of 3-hour-old schistosomules that were cultured for the next 14 days. dsRNA of this length was employed because earlier studies with cathepsin B1 of S. mansoni had shown that dsRNA of at least one kb in length induced strong knockdown and phenotypic effects against a proteolytic enzyme, which like cathepsin D, is gut-localized and participates in hemoglobin proteolysis [6, 18]. Second, a SmCD dsRNA of 0.5 kb in length was prepared for use with seven, nine and 11 day old somules that were cultured for seven days after dsRNA treatment. We tested the shorter dsRNA probe because it had become apparent that shorter dsRNAs of 300 to 700 bp, and indeed very short 21 bp short interfering RNAs (siRNAs), triggered profound gene suppression of gut-associated proteases of S. mansoni [6, 15], and because there exists the possibility that residual dsRNAs might interfere with downstream analysis of gene-silenced cells, specifically by serving as spurious templates in RT-PCRs employed to investigate levels of target gene transcripts [24].
Three hours after cercarial transformation, schistosomules were electroporated with SmCD 1.2 kb-dsRNA or Luc-dsRNA (1,673 bp) and the worms were maintained thereafter in Basch’s medium supplemented with washed human erythrocytes. A visible morphological phenotype was clearly apparent with SmCD1.2 kb-dsRNA after culture for 14 days. The parasites electroporated with SmCD 1.2 kb-dsRNA exhibited stunted growth, measured as body surface area, compared with the Luc-dsRNA controls (Figure 2A). In particular, when the surface area of each of up to 200 randomly selected schistosomules was measured, the mean surface area values were 6,150 ± 1,756 and 4,183 ± 1,106 μm2 (mean ± standard deviation; n = 133 and 201) for the control Luc-dsRNA and experimental SmCD dsRNA treatment groups, respectively. This analysis revealed that the size of the majority of SmCD 1.2 kb-dsRNA treated schistosomules was significantly smaller than the controls (p ≤ 0.0001) (Figure 2). By contrast, motility and movement of the SmCD dsRNA treated worms did not appear different from that of the control schistosomules (not shown).
Figure 2.
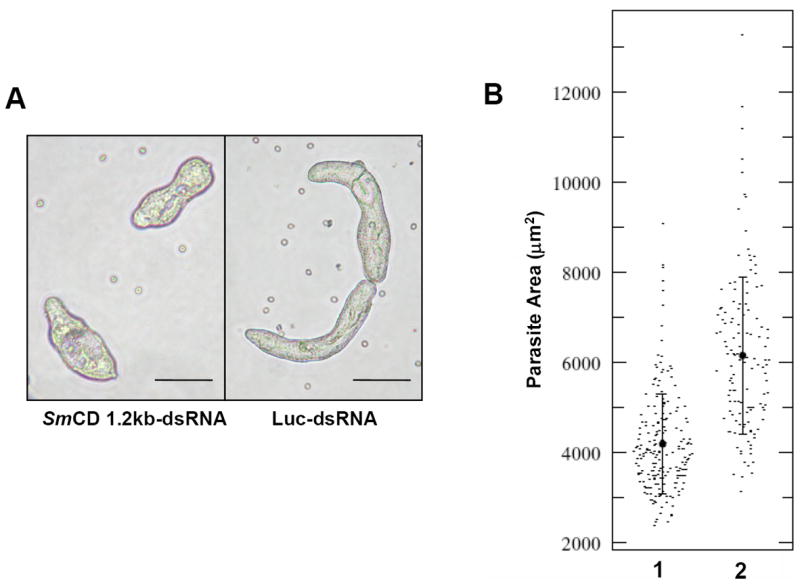
SmCD is essential for normal worm growth. (A) Examples of schistosomes at day 14 following electroporation with either 10 μg of SmCD 1.2 kb-dsRNA (left) or Luc-dsRNA (right). (Scale bars, 100 μm). (B) Population-wide quantitation of parasite size (as measured by the area occupied by parasites on a 2D image) on day 14 following dsRNA treatment; (1) SmCD 1.2 kb-dsRNA, (2) Luc-dsRNA; (p ≤ 0.0001).
3.2. RNAi-mediated knockdown of SmCD protease activity
In addition to the phenotype of retarded growth, we investigated cathepsin D protease activities in the schistosomules harvested 14 days later after electroporation of 3-hour-old schistosomules with dsRNAs. To assess enzyme activity in soluble extracts of the schistosomules, we employed the cathepsin D-diagnostic peptide MCA-Gly-Lys-Pro-Ile-Leu-Phe-Phe-Arg-Leu-Lys(DNP)-Arg as the substrate. Proteolytic activity was markedly reduced in SmCD-dsRNA treated worms, with Relative Fluorescence Units (RLUs)/ μg schistosome protein of 1,138 ± 4.98 and 415 ± 18.4 (mean ± SD; three replicates) RLUs/ μg for the control (Luc-dsRNA) and experimental (SmCD1.2 kb-dsRNA) groups, respectively (p ≤ 0.05). This represented a reduction of ~64 % of cathepsin D protease activity in the SmCD dsRNA-treated schistosomules.
The reduced levels of cathepsin D enzyme activity in schistosomula paralleled reduction in mRNA levels. Total RNA extracted from representatives of the SmCD 1.2 kb-dsRNA and Luc-dsRNA treated groups was 10-fold diluted serially and these dilutions were employed as the template for RT-PCR. As illustrated in Figure 3B, the inhibitory effect mediated by the dsRNA was specific for SmCD, since SmCB1 transcript levels appeared to be unaffected by SmCD-dsRNA treatment. Other workers have also reported that RNAi gene silencing targeting gut-associated proteases of schistosomes delivers specific effects on the target gene without off-target effects on other schistosome genes including genes encoding other gut localized enzymes [6, 15].
Figure 3.
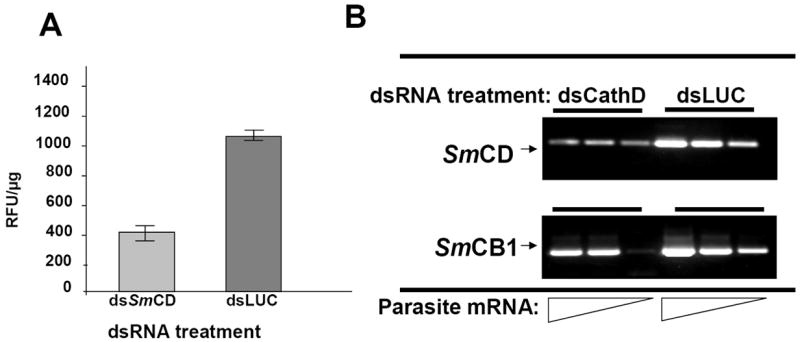
Induction of specific RNAi following treatment with cathepsin D dsRNA. (A) SmCD enzyme activity in extracts of schistosomula. Three hour old schistosomula were electroporated with 10 μg of SmCD 1.2 kb-dsRNA or Luc-dsRNA, and cultured for 14 days post-electroporation in the presence of human erythrocytes. (B) Shows relative cathepsin D gene expression of the same treatment group. RNA was 10 fold serially diluted, and assayed by RT-PCR for SmCD and SmCB1. The dilution series reflects input of 50.0, 5.0, 0.05 ng of total RNA into the RT-PCRs. In all cases, statistically significant differences between experimental and control groups were observed (p<0.05).
3.3 Absence of accumulation of gut localized hemozoin indicative of disruption of hemoglobin proteolysis
Under normal conditions in vivo or in vitro, from about one week after cercarial transformation, the gut of schistosomules becomes patent and fills with an obvious black pigment, hemozoin (= hematin) which is the end product of hemoglobin proteolysis [18]. In order to test the role of SmCD at the early stages of hemoglobin digestion, seven, nine and 11 day old somules were treated with SmCD 1.2 kb-dsRNA or Luc-dsRNA. For the next seven days the appearance of these schistosomules was monitored microscopically. We observed that normal looking hemozoin pigment accumulated in the guts of the schistosomules treated with Luc-dsRNA (Figure 4, panels B and D). By contrast, schistosomula treated with SmCD 1.2 kb-dsRNA exhibited a different phenotype; their guts contained red colored contents rather than the expected black-colored hemozoin (Figure 4 panels A and C). We anticipate that the red-colored material in their guts was non-digested hemoglobin. S. mansoni cathepsin D makes the initial cleavage of the hemoglobin monomer, after which other proteases including cathepsins B1, L1, L2 and C and leucine aminopeptidase digest the hemoglobin to short peptides or free amino acids [6, 11, 25]. In view of the markedly reduced protease activity in extracts of SmCD 1.2 kb-dsRNA treated schistosomules (Figure 3) and the red-colored gut contents, these findings indicated that the RNAi treatment results in inhibition of hemoglobin proteolysis in cultured schistosomules. Moreover, they represent an additional visible phenotype.
Figure 4.
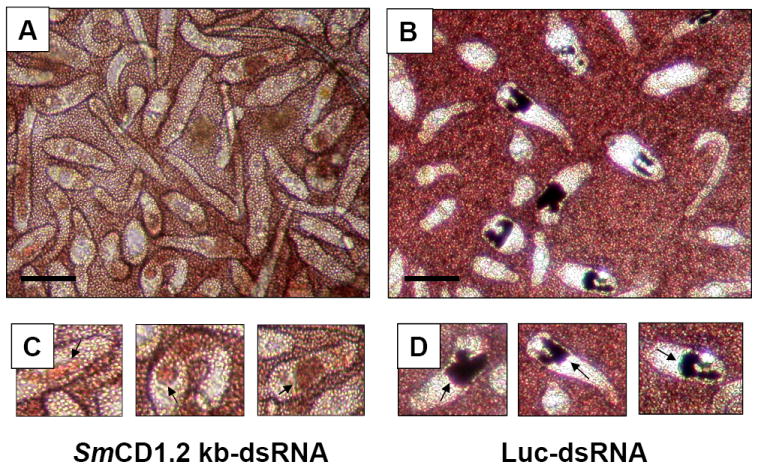
Cathepsin D may be required for normal hemoglobin proteolysis. Seven, nine and 11 day old schistosomules were transformed by electroporation with 30 μg of SmCD 1.2kb-dsRNA or Luc-dsRNA, after which they were cultured for seven days in Basch’s medium supplemented with human erythrocytes. Examples of the 11 day old schistosomes at seven days after electroporation are presented: panel A, SmCD-dsRNA; panel B, Luc-dsRNA; panel C, higher magnifications of three worms from the SmCD-dsRNA-treated population, with arrows pointing to the worm’s gut; panel D, higher magnifications of three worms from the Luc-dsRNA-treated population. Scale bars = 100 μm. Similar findings were apparent in the seven- and nine-day-old schistosomules (not shown).
3.4.Robust gene suppression also induced by shorter SmCD 0.5 kb-dsRNA
In some experiments we employed a different form of SmCD dsRNA, a 0.5 kb dsRNA (Figure 1). We introduced this probe because we were concerned that residual SmCD 1.2 kb-dsRNA may have confounded analysis of RNAi treated schistosomes by serving as a spurious template for reverse transcription. Indeed, we had observed a weak signal in RT-PCR (Fig. 3B) and were unsure the signal represented the presence of target gene transcripts rather than reflecting residual SmCD 1.2 kb-dsRNA. Other investigators have noted similar artifacts [18, 24]. To obviate this potential problem, RNAi investigations can be designed so that the dsRNA probe and the transcript region targeted in RT-PCRs span discontinuous regions of the target transcript [6, 15, 26, 27]. Thus the SmCD 0.5 kb-dsRNA spans residues 655 to 1154 while the RT-PCR targeted residues 203-575 of the cDNA. SmCD 1.2 kb-dsRNA spans residues 37 to 1285, effectively the entire transcript (Figure 1). Schistosomules of seven, nine and 11days old were maintained in culture for two or seven days after exposure to SmCD 0.5kb-dsRNA or SmCD 1.2 kb-dsRNA. Luc-dsRNA treated controls were included in all experiments. Although no visual phenotype was yet apparent, enzymatic analysis of parasites at 48 hours post-electroporation showed ≥50% declines in cathepsin D activity levels in the SmCD 0.5 kb-dsRNA or SmCD 1.2 kb-dsRNA treated worms compared with the Luc-dsRNA controls (not shown). RT-PCR analysis revealed inconsistent effects of SmCD 1.2 kb-dsRNA on SmCD transcripts, however with SmCD 0.5kb-dsRNA markedly reduced levels of SmCD mRNA were observed (not shown). By seven days after treatment, SmCD 0.5 kb dsRNA treated parasites showed a consistent and highly significant decrease in SmCD mRNA levels of >95% when compared to levels of SmCD mRNA in the Luc-dsRNA treated controls (Fig. 5A). The silencing of transcription of SmCD by SmCD 0.5-kb-dsRNA did not influence transcription of cathepsin B1 or GAPDH (Fig. 5A), which confirmed both the quality of RNA template and specificity of SmCD RNAi knockdown. The gene suppression was accompanied by reduced levels of cathepsin D enzyme activity (Fig. 5B). The day seven treated parasites showed ~ 72% relative decline in cathepsin D activity. In addition, an apparent age-associated refractoriness to gene silencing was evident in that the silencing was not as effective in day nine somules (~50% reduction in protease activity) and even less effective in 11 day old somules (~33% reduction). Alternatively, or additionally, increasing levels of cathepsin D activity at seven, nine and 11 days of age may reflect the development of the nascent gastrodermis and gut. Nonetheless, in all three age treatment groups, the knockdown of cathepsin D protease activity was statistically significant when compared to the matched Luc-dsRNA controls (p ≤ 0.05).
Figure 5.
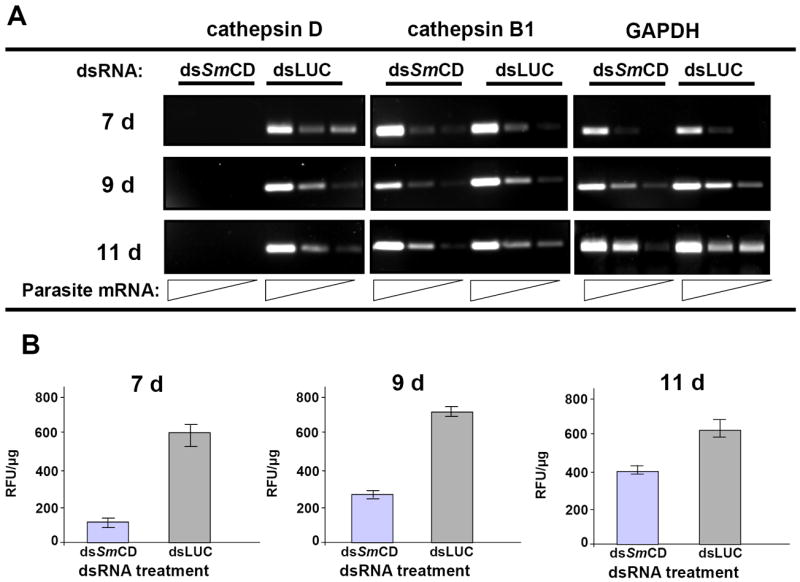
Gene suppression of S. mansoni cathepsin D (SmCD) in schistosomules. Seven, nine and 11day old schistosomes were transformed by electroporation with 30 μg of SmCD 0.5 kb-dsRNA or Luc-dsRNA, and then cultured in vitro. Forty eight hours and seven days later, batches of schistosomules were harvested for analysis of (A) total RNA by RT-PCR and (B) aspartic protease activity. At 48 h following electroporation, knockdown of both SmCD transcripts and aspartic protease activity was apparent (not shown). By seven days, SmCD transcription was completely silenced (A) and protease activity profoundly suppressed (B). Panel A: RNA was 10-fold serially diluted for analysis by RT-PCR. The dilution series reflects input of 50.0, 5.0, 0.05 ng of total parasite RNAs. Two other genes, SmCB1 and GAPDH, were quantified by RT-PCR to monitor for possible off-target effects of dsRNA exposure. Panel B: SmCD protease activity was measured at pH 3.5 in soluble extracts of schistosomules using the cathepsin D-specific substrate MCA-Gly-Lys-Pro-Ile-Leu-Phe-Phe-Arg-Leu-Lys(DNP)-Arg. Extracts of SmCD-dsRNA treated schistosomules exhibited markedly less protease activity than the Luc-dsRNA treated controls. For all three age groups of the schistosomules, significant suppression of SmCD was evident (P ≤ 0.05 in each case), although there was a trend for diminished silencing with increasing age of the schistosomules for seven to nine to 11 days of age.
3.5. Infection of mice with RNAi-treated schistosomules suggest that SmCD may be essential for development and maturation
To investigate the longevity of the RNAi mediated gene silencing of SmCD, we infected mice with dsRNA-treated schistosomules. Within mice presumably nutrient levels are optimal and not limiting for the developing schistosomes in contrast to the culture conditions in vitro. For these mouse experiments, schistosomula were electroporated with SmCD 1.2 kb-dsRNA or Luc-dsRNA as an irrelevant control at 3 h post-transformation from cercariae, and injected in to the thigh muscles of female Balb/c mice. In addition, we targeted two other gut associated protease genes for inclusion as controls with which to compare the RNAi targeting SmCD. The two other genes were leucine aminopeptidases 1 and 2 (LAP1 and LAP2). These additional schistosomule groups were transformed with dsRNA-LAP1, dsRNA-LAP2 or both, dsRNA-LAP 1 + 2. The mice were euthanized 60 days after infection and schistosomes recovered thereafter by portal perfusion. In addition, the gross appearance of the liver was examined for schistosome egg induced granulomas, after which the livers were digested with collagenase to recover schistosome eggs. Adult worms were recovered from mice infected with schistosomules transformed with Luc-dsRNA, dsRNA-LAP1, dsRNA-LAP2 and dsRNA-LAP1+2. Livers from these mice exhibited hepatomegaly and the presence of granulomas. In addition, eggs were recovered from the digested livers. By contrast, no adult worms, schistosome egg granulomas or schistosome eggs were recovered from mice infected with the SmCD 1.2 kb-dsRNA transformed schistosomules (Table 1). These findings indicated that RNAi mediated suppression of SmCD activity lead to death of the parasites in vivo, and suggested that SmCD was an essential gene for the blood-stage schistosomes.
Table 1.
Fate of schistosomes and pathology associated with infection after infection of mice with three-hour-old schistosomula transformed with dsRNA. After electroporation with 10 μg dsRNA, schistosomules were injected into the thigh muscle of Balb/c mice (~500 parasites per mouse). The mice were euthanized and worms recovered by portal perfusion 60 days after infection. Livers were removed, the gross appearance examined, and the livers were digested with collagenase to recover schistosome eggs.
| dsRNA introduced | Number of schistosomula injected into each mouse | Number of mice infected | Total number of worms recovered | Survival percentage of schistosomes (%) | Gross liver pathology | Eggs recovered from liver |
|---|---|---|---|---|---|---|
| LAP1* | ~500 | 3 | ≥2 | 0.13 | hepatomegaly; granulomas | + |
| LAP2* | ~500 | 3 | ≥4 | 0.26 | hepatomegaly; granulomas | + |
| LAPs 1 and 2 | ~500 | 3 | ≥10 | 0.67 | hepatomegaly; granulomas | + |
| luciferase | ~500 | 3 | ≥7 | 0.46 | hepatomegaly; granulomas | + |
| cathepsin D | ~500 | 3 | 0 | 0 | normal | - |
LAP1 = leucine aminopeptidase 1; LAP2 = leucine aminopeptidase 2
4. Discussion
Increasing evidence indicates that loss-of-function genetic manipulation through RNA interference may be generally applicable for investigation of schistosome genes [15, 18], in contrast to the more problematical situation with parasitic nematodes where results have been much more inconsistent both between and within species [28]. Indeed, an orthologue of dicer – one of the central enzymes of the RNAi cascade - has been characterized in S. mansoni recently and, interestingly, in relation to the present study, its expression appears to be highest in the schistosomulum stage [13]. After processing of dsRNA by dicer, the resulting short interfering RNAs (siRNAs) join with the effector nuclease complex [29] which recognizes and attacks the homologous target mRNAs, leading to gene silencing. In schistosomes this can result in long-term knockdown and silencing of the target gene [18]. In terms of delivery of dsRNA, electroporation of schistosomula with dsRNA has proved to be an effective delivery method for gene silencing within this developmental stage of the parasite [15, 18]. Gene silencing by RNAi in schistosomes delivers a specific effect at the target gene and obvious or widespread non-specific effects on non-target genes have not been reported.
To investigate the involvement of the cathepsin D gene early stages of schistosome nutrition and growth, we performed a series of RNAi experiments, delivering two different length dsRNAs into schistosomules by square wave electroporation. Both long (full length, 1.2 kb) and a shorter (0.5 kb) form of dsRNA were effective in silencing the protease, without affecting the mRNA levels of two unrelated schistosome proteins, SmCB1 and GAPDH. The RNAi mediated reductions in transcripts were accompanied by suppression of SmCD enzyme activity. In like fashion to the findings of Delcroix et al. [6] who investigated SmCD gene suppression in adult schistosomes, we observed ~70% reduction in SmCD ascribable activity against a diagnostic peptide. In addition, visible RNAi phenotypes resulted. First, the growth of schistosomules in vitro was negatively affected; statistically significant differences in surface area were apparent at 14 days after transformation of three hour old somules with the SmCD 1.2 kb-dsRNA. Not all worms were affected to the same extent as evident from variation in the surface area occupied by parasites on two-dimensional photographic images. It is likely some schistosomules do not receive equivalent quantities of the dsRNA; such variation is consistent with results reported in model eukaryotes including Drosophila, C. elegans and Trypanosoma brucei [30-33] as well in cultured S. mansoni schistosomules [18]. Krautz-Peterson et al. [15] hypothesized that most dsRNA that is delivered to cultured seven day old or older schistosomules by electroporation enters the parasite through the mouth and gut. They considered that transit of dsRNA across the double lipid-bilayered tegument was unlikely. By contrast, since the mouth remains closed until around one week after transformation, entry of dsRNA through this orifice is not possible for younger parasites. Instead, the dsRNA likely enters young schistosomules through the acetabular glands which function in cercarial adhesion to and penetration of the mammalian hosts. Our experiment targeted dsRNA to three-hour old somules. If as hypothesized [15] the dsRNA enters through these glands of three-hour-old schistosomules, it may spread to the developing gut and gastrodermis where cathepsin D has been localized in older, blood stage schistosomes [6, 11].
Second, and more remarkable, there was a visible phenotype in which the guts of the treated worms appeared red in color in comparison to the (expected) black-colored guts of control schistosomules. Under normal conditions in vivo or in vitro, from about one week after cercarial transformation, the gut of schistosomules becomes patent and fills with an obvious black pigment, hemozoin (= hematin) which is the end product of hemoglobin proteolysis [18]. Aggregation of heme into an insoluble form – hemozoin - in the gut of S. mansoni, followed by its elimination through regurgitation, has evolved to defend against the toxicity of heme released from hemoglobin [34]. We hypothesize that the red-colored material was non-digested human hemoglobin. S. mansoni cathepsin D makes the initial cleavage of the hemoglobin monomer, after which other proteases including cathepsins B1, L1, L2 and C and leucine aminopeptidases digest the hemoglobin to short peptides or free amino acids [6, 11, 25]. In view of the markedly reduced protease activity in these schistosomules gene silenced with SmCD-dsRNA, the red-colored gut contents reflect disruption of hemoglobin proteolysis in cultured schistosomules. The requisite initial cleavages of the hemoglobin monomer that have been determined for this enzyme, e.g. α-chain Phe33-Phe34 and β-chain Phe41-Phe42 [6, 11], may not have occurred in the guts of SmCD dsRNA treated worms and that the hemoglobin protein remained intact or mostly intact, retaining its red hue. McKerrow and coworkers [6] reported that degradation cascades of blood protein substrates ingested by schistosomes are substrate-specific, wherein cathepsin D most efficiently produces the primary cleavage of hemoglobin although cysteine proteases provide some redundancy. By contrast, our current findings suggest that there is little redundancy at the apex of the hemoglobin degradation cascade given the observation of undigested hemoglobin in the worms and the clearance of SmCD dsRNA treated schistosomes from mice.
In tandem with the impact of dsRNA on transcript levels, SmCD enzyme activity, growth and gut-localized hemoglobin proteolysis, the findings that SmCD 1.2 kb-dsRNA treated schistosomules did not survive in mice to maturity and reproduction together indicate that this schistosome protease plays a crucial function in the growth, development and maturation of these parasites. Moreover, they suggest that, at least in vivo, knockdown of the enzyme is lethal for the parasite.
Although there is redundancy in the protease enzyme cascade that performs hemoglobin proteolysis, for example in the substrates cleaved by cathepsin B1 and cathepsin L1, cathepsin D has the apical and perhaps unsupported role of initial cleavage of the hemoglobin monomer [6]. In this regard, cathepsin D represents a potential Achilles’ heel within this cascade. The aspartic protease of HIV-1 is the target of several therapeutic drugs including saquinavir, ritonavir and lopinavir and moreover these HIV protease inhibitors also show anti-malarial activity probably by targeting one of more of the plasmepsin aspartic proteases of Plasmodium falciparum (see [35]). In addition, the papain-like cysteine proteases of S. mansoni which like schistosome cathepsin D also participate in hemoglobin proteolysis are being developed for new chemotherapeutic interventions including the vinyl sulfone cysteine protease inhibitor K11777 [36]. Indeed, lopinavir blocks schistosome cathepsin D activity against hemoglobin within the schistosome gut [6], which represents a potentially valuable lead for novel anti-schistosomal drug design.
Acknowledgments
Schistosome infected snails and mice were supplied by Dr. Fred A. Lewis, Biomedical Research Institute, Rockville, Maryland through NIH contract NO155270. We thank Airat Agbetoba for expert technical assistance and Drs. Bernd Kalinna and Victoria Mann for critical review of the manuscript. The studies were supported in part by NIH-NIAID award number R01AI072773 (to PJB) and the Uruguayan Comision Sectorial de Investigacion Cientifica de la Universidad de la Republica and the Programa de Desarrollo Tecnologico (to GR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brindley PJ, Kalinna BH, Dalton JP, Day SR, Wong JY, Smythe ML, McManus DP. Proteolytic degradation of host hemoglobin by schistosomes. Mol Biochem Parasitol. 1997;1:1–9. doi: 10.1016/s0166-6851(97)00098-4. [DOI] [PubMed] [Google Scholar]
- 2.Tort J, Brindley PJ, Knox D, Wolfe KH, Dalton JP. Proteinases and associated genes of parasitic helminths. Adv Parasitol. 1999:161–266. doi: 10.1016/s0065-308x(08)60243-2. [DOI] [PubMed] [Google Scholar]
- 3.Wasilewski MM, Lim KC, Phillips J, McKerrow JH. Cysteine protease inhibitors block schistosome hemoglobin degradation in vitro and decrease worm burden and egg production in vivo. Mol Biochem Parasitol. 1996;2:179–89. doi: 10.1016/0166-6851(96)02703-x. [DOI] [PubMed] [Google Scholar]
- 4.Bogitsh BJ, Kirschner KF, Rotmans JP. Schistosoma japonicum: immunoinhibitory studies on hemoglobin digestion using heterologous antiserum to bovine cathepsin D. J Parasitol. 1992;3:454–9. [PubMed] [Google Scholar]
- 5.Williamson AL, Brindley PJ, Knox DP, Hotez PJ, Loukas A. Digestive proteases of blood-feeding nematodes. Trends Parasitol. 2003;9:417–23. doi: 10.1016/s1471-4922(03)00189-2. [DOI] [PubMed] [Google Scholar]
- 6.Delcroix M, Sajid M, Caffrey CR, Lim KC, Dvorak J, Hsieh I, Bahgat M, Dissous C, McKerrow JH. A multienzyme network functions in intestinal protein digestion by a platyhelminth parasite. J Biol Chem. 2006;51:39316–29. doi: 10.1074/jbc.M607128200. [DOI] [PubMed] [Google Scholar]
- 7.Dalton JP, Brindley PJ. Schistosome asparaginyl endopeptidase SM32 in hemoglobin digestion. Parasitol Today. 1996;3:125. doi: 10.1016/0169-4758(96)80676-4. [DOI] [PubMed] [Google Scholar]
- 8.Brinkworth RI, Prociv P, Loukas A, Brindley PJ. Hemoglobin-degrading, aspartic proteases of blood-feeding parasites: substrate specificity revealed by homology models. J Biol Chem. 2001;42:38844–51. doi: 10.1074/jbc.M101934200. [DOI] [PubMed] [Google Scholar]
- 9.Koehler JW, Morales ME, Shelby BD, Brindley PJ. Aspartic protease activities of schistosomes cleave mammalian hemoglobins in a host-specific manner. Mem Inst Oswaldo Cruz. 2007;1:83–5. doi: 10.1590/s0074-02762007000100014. [DOI] [PubMed] [Google Scholar]
- 10.Wong JY, Harrop SA, Day SR, Brindley PJ. Schistosomes express two forms of cathepsin D. Biochim Biophys Acta. 1997;2:156–60. doi: 10.1016/s0167-4838(97)00019-8. [DOI] [PubMed] [Google Scholar]
- 11.Brindley PJ, Kalinna BH, Wong JY, Bogitsh BJ, King LT, Smyth DJ, Verity CK, Abbenante G, Brinkworth RI, Fairlie DP, Smythe ML, Milburn PJ, Bielefeldt-Ohmann H, Zheng Y, McManus DP. Proteolysis of human hemoglobin by schistosome cathepsin D. Mol Biochem Parasitol. 2001;1:103–12. doi: 10.1016/s0166-6851(00)00351-0. [DOI] [PubMed] [Google Scholar]
- 12.Morales ME, Kalinna BH, Heyers O, Mann VH, Schulmeister A, Copeland CS, Loukas A, Brindley PJ. Genomic organization of the Schistosoma mansoni aspartic protease gene, a platyhelminth orthologue of mammalian lysosomal cathepsin D. Gene. 2004;1:99–109. doi: 10.1016/j.gene.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Krautz-Peterson G, Skelly PJ. Schistosoma mansoni: The dicer gene and its expression. Exp Parasitol. 2007 doi: 10.1016/j.exppara.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skelly PJ, Da’dara A, Harn DA. Suppression of cathepsin B expression in Schistosoma mansoni by RNA interference. Int J Parasitol. 2003;4:363–9. doi: 10.1016/s0020-7519(03)00030-4. [DOI] [PubMed] [Google Scholar]
- 15.Krautz-Peterson G, Radwanska M, Ndegwa D, Shoemaker CB, Skelly PJ. Optimizing gene suppression in schistosomes using RNA interference. Mol Biochem Parasitol. 2007;2:194–202. doi: 10.1016/j.molbiopara.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Basch PF. Cultivation of Schistosoma mansoni in vitro. I. Establishment of cultures from cercariae and development until pairing. J Parasitol. 1981;2:179–85. [PubMed] [Google Scholar]
- 17.James ER, Taylor MG. Transformation of cercariae to schistosomula: a quantitative comparison of transformation techniques and of infectivity by different injection routes of the organisms produced. J Helminthol. 1976;4:223–33. doi: 10.1017/s0022149x0002664x. [DOI] [PubMed] [Google Scholar]
- 18.Correnti JM, Brindley PJ, Pearce EJ. Long-term suppression of cathepsin B levels by RNA interference retards schistosome growth. Mol Biochem Parasitol. 2005;2:209–15. doi: 10.1016/j.molbiopara.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Lazdins JK, Stein MJ, David JR, Sher A. Schistosoma mansoni: rapid isolation and purification of schistosomula of different developmental stages by centrifugation on discontinuous density gradients of Percoll. Exp Parasitol. 1982;1:39–44. doi: 10.1016/0014-4894(82)90090-x. [DOI] [PubMed] [Google Scholar]
- 20.Correnti JM, Jung E, Freitas TC, Pearce EJ. Transfection of Schistosoma mansoni by electroporation and the description of a new promoter sequence for transgene expression. Int J Parasitol. 2007 doi: 10.1016/j.ijpara.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 21.Dalton JP, Day SR, Drew AC, Brindley PJ. A method for the isolation of schistosome eggs and miracidia free of contaminating host tissues. Parasitology. 1997:29–32. doi: 10.1017/s0031182097001091. [DOI] [PubMed] [Google Scholar]
- 22.Yasuda Y, Kageyama T, Akamine A, Shibata M, Kominami E, Uchiyama Y, Yamamoto K. Characterization of new fluorogenic substrates for the rapid and sensitive assay of cathepsin E and cathepsin D. J Biochem (Tokyo) 1999;6:1137–43. doi: 10.1093/oxfordjournals.jbchem.a022396. [DOI] [PubMed] [Google Scholar]
- 23.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;7:36–42. [Google Scholar]
- 24.Tzanetakis IE, Keller KE, Martin RR. The use of reverse transcriptase for efficient first- and second-strand cDNA synthesis from single- and double-stranded RNA templates. J Virol Methods. 2005;1-2:73–7. doi: 10.1016/j.jviromet.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 25.McCarthy E, Stack C, Donnelly SM, Doyle S, Mann VH, Brindley PJ, Stewart M, Day TA, Maule AG, Dalton JP. Leucine aminopeptidase of the human blood flukes, Schistosoma mansoni and Schistosoma japonicum. Int J Parasitol. 2004;6:703–14. doi: 10.1016/j.ijpara.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Kimber MJ, McKinney S, McMaster S, Day TA. CC Fleming, and AG Maule, flp gene disruption in a parasitic nematode reveals motor dysfunction and unusual neuronal sensitivity to RNA interference. Faseb J. 2007;4:1233–43. doi: 10.1096/fj.06-7343com. [DOI] [PubMed] [Google Scholar]
- 27.Boyle JP, Wu XJ, Shoemaker CB, Yoshino TP. Using RNA interference to manipulate endogenous gene expression in Schistosoma mansoni sporocysts. Mol Biochem Parasitol. 2003;2:205–15. doi: 10.1016/s0166-6851(03)00078-1. [DOI] [PubMed] [Google Scholar]
- 28.Knox DP, Geldhof P, Visser A, Britton C. RNA interference in parasitic nematodes of animals: a reality check? Trends Parasitol. 2007;3:105–7. doi: 10.1016/j.pt.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Tavernarakis N, Wang SL, Dorovkov M, Ryazanov A, Driscoll M. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat Genet. 2000;2:180–3. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- 30.Grant B, Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell. 1999;12:4311–26. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashmi S, Zhang J, Oksov Y, Lustigman S. The Caenorhabditis elegans cathepsin Z-like cysteine protease, Ce-CPZ-1, has a multifunctional role during the worms’ development. J Biol Chem. 2004;7:6035–45. doi: 10.1074/jbc.M312346200. [DOI] [PubMed] [Google Scholar]
- 33.Ullu E, Tschudi C, Chakraborty T. RNA interference in protozoan parasites. Cell Microbiol. 2004;6:509–19. doi: 10.1111/j.1462-5822.2004.00399.x. [DOI] [PubMed] [Google Scholar]
- 34.Oliveira MF, d’Avila JC, Tempone AJ, Soares JB, Rumjanek FD. A Ferreira-Pereira, ST Ferreira, and PL Oliveira, Inhibition of heme aggregation by chloroquine reduces Schistosoma mansoni infection. J Infect Dis. 2004;4:843–52. doi: 10.1086/422759. [DOI] [PubMed] [Google Scholar]
- 35.Andrews KT, Fairlie DP, Madala PK, Ray J, Wyatt DM, Hilton PM, Melville LA, Beattie L, Gardiner DL, Reid RC, Stoermer MJ, Skinner-Adams T, Berry C, McCarthy JS. Potencies of human immunodeficiency virus protease inhibitors in vitro against Plasmodium falciparum and in vivo against murine malaria. Antimicrob Agents Chemother. 2006;2:639–48. doi: 10.1128/AAC.50.2.639-648.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdulla MH, Lim KC, Sajid M, McKerrow JH, Caffrey CR. Schistosomiasis mansoni: novel chemotherapy using a cysteine protease inhibitor. PLoS Med. 2007;1:e14. doi: 10.1371/journal.pmed.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]


