Abstract
Objective
To determine whether expression of the stress response gene ATF3 and related members of activator protein complex-1, cJun and cFos, were altered in leiomyoma compared to myometrium, and whether this difference might correlate with leiomyoma size or race.
Design
Laboratory study.
Setting
University hospital.
Patients
Fifteen women undergoing hysterectomy for symptomatic leiomyoma.
Interventions
Tissue procurement, RNA isolation, reverse transcriptase PCR, real-time RT-PCR, immunohistochemistry, western blot.
Main Outcome Measures
Expression of mRNA and protein in leiomyoma and patient-matched myometrium.
Results
mRNA transcripts of ATF3 were decreased in leiomyoma compared to matched myometrium by both RT-PCR and real time RT-PCR. The decrease was greater than 5 fold in a majority of samples (P<0.05). The reduction seen in ATF3 mRNA expression did not show a correlation with race and leiomyoma size. Surprisingly, immunohistochemistry and western blot analysis demonstrated an elevation of ATF3 protein expression by a mean of 2.9 fold (P<0.05). Transcripts of related AP-1 genes, cJun and cFos, were significantly decreased by a mean of -29.57 (P<0.05) for cJun and -23.78 (P<0.05) for cFos, but there was no significant change in protein expression of the two transcription factors.
Conclusions
Alterations in ATF3 gene expression resemble the response to mechanical and ischemic stress reported in other tissues. Results suggested that ATF3 protein expression was increased in leiomyoma and may reflect increased tissue stress.
Keywords: fibroid, myometrium, jun, fos, ATF3, racial disparity, stress
Introduction
Leiomyoma are benign but potentially morbid uterine tumors that can be detected in the majority of women by the age of menopause (1). Leiomyoma cause a myriad of problems including pain, bleeding, incontinence, and infertility. Furthermore, leiomyoma are the most common indication for hysterectomy and represent a tremendous health care burden. Despite their prevalence, the etiology is poorly understood (2).
Several line of evidence suggest a genetic component to leiomyoma formation (3). Syndromes such as Alport syndrome (4), Bannayan-Zonana syndrome (due to tumor suppressor gene phosphatase and tensin homolog) (5), and hereditary leiomyomatosis and renal cell cancer (HLRCC) (6) feature leiomyomata as a facet of their syndromic phenotype. HLRCC is likely the same disease as Reed's syndrome (MCUL1) (3) with the etiology of both related to a loss of function in the fumarate hydratase (FH) gene. There is also strong epidemiological evidence that race in an important factor in the incidence of leiomyomata (1,7-9). Multiple studies have shown an increased prevalence of leiomyomata in African American women. Leiomyomata in African Americans occur at a younger age, grow faster, and cause more morbidity than in matched Caucasians (7). Distinct differences in expression of fumarate hydratase have been found between Caucasian and African-American women in relationship to leiomyoma disease (10). In order to search for specific genetic links with disease, microarray screens are a powerful tool. Previous microarray studies by our group (11-13), and by others (14-16), have demonstrated multiple differentially expressed genes in leiomyoma relative to surrounding normal myometrium.
Despite similar scientific strategy and methods, only eight differentially-expressed genes were shared across the majority of the studies (14, 15, 17). The gene Activating Transcription Factor-3 (ATF3) was one of the eight genes identified by multiple screening studies. In all 6 screening studies that evaluated ATF3, mRNA levels were reduced from 2.8 to 16.7 fold. ATF3 is particularly interesting because it is involved in the stress response of many tissues, including mechanical stress (18). Whereas stress is any adverse stimuli acting upon an organism or cell, mechanical stress is a specific type of stress, a mechanical stimuli that leads to a cellular response (19). Recent study has suggested that mechanical stress may play a role in the development of leiomyomata (20), and this may be related to the hypothesis that leiomyomata share features consistent with an altered response to wound healing (13, 21).
ATF3 belongs to a large family of over 20 proteins featuring a bZip DNA-binding domain, specifically the ATF/CREB, AP-1 (fos and jun proteins) and C/EBP families of related proteins. The consensus binding site for the ATF family (CREB/CREM, CRE-BP1 [ATF-2], ATF3, ATF4, ATF6 and ATF6 and B-ATF) is TGACGT(C/A)(G/A) and is identical to the CRE consensus TGACGTCA (22). Of note, ATF3 is a stress-inducible gene, induced by both oxidative stress and mechanical stress. Mechanotransduction is a means of solid-state signaling whereby mechanical stresses can activate cell surface proteins and induce signaling cascades. Although the signaling modules that regulate ATF3 expression are not fully elucidated, in many systems ATF3 induction occurs immediately and transiently after induced stress. Similarly, the AP-1 transcription factor complex regulates important processes, such as cell proliferation, differentiation, and cell death (23, 24). The exact mechanisms and pathways for the actions of AP-1 are tissue dependent (25). The function of ATF3 appears to be part of a cellular response that results in tissue destruction, in part through influence on apoptosis (26).
ATF3 mRNA has been shown to be decreased by microarray data, but these results must be considered preliminary requiring confirmatory testing (12). ATF3 protein has not been measured, nor have protein levels of related AP-1 family members been characterized in leiomoyoma and myometrium. In this study, we hypothesized that ATF3 expression would be altered in leiomyoma or might correlate with leiomyoma size and/or patient race. To test this hypothesis we collected leiomyomata from hysterectomy specimens and performed mRNA and protein analysis.
Material and Methods
Tissue Procurement
IRB approval was obtained at the National Naval Medical Center in Bethesda, Marlyand, where all surgeries were performed. Leiomyoma and myometrial tissue samples were then obtained from 15 patients undergoing hysterectomy for symptomatic leiomyomata. Tissue harvesting involved isolation of each leiomyoma as well as myometrium. Approximately 1 cm3 tissue pieces were taken when available. For some leiomyoma, this represented the entire specimen. For larger leiomyomata, tissue was taken from the periphery, beneath the capsule.
Tissue destined for RNA isolation was minced and immediately placed in RNAlater (Ambion, Inc., Austin, TX), tissue that was not to be used within 24 hours for RNA isolation was stored at -70C. The samples destined for IHC were cut into 5-mm3 section and stored in 4% formalin at the time of tissue harvesting. The tissue samples were then paraffin embedded and mounted on slides.
RNA Isolation
Leiomyoma and myometrium tissue samples were removed from the RNAlater (Ambion, Austin, TX) solution and re-minced. This tissue was then placed in 6ml TRIzol reagent (Invitrogen, Carlsbad, CA). Tissue homogenization was performed first with a scalpel and then with a Tissue Tearer, model 985-370 (Biospec Products, Inc.). The tissue homogenate was left at room temperature for 5 minutes, and then mixed with 1.2 ml of chloroform. This mixture was kept at room temperature for 15 minutes, and then centrifuged at 8500rpm for 20 minutes. The aqueous layer was transferred and mixed with 3ml isopropanol. This mixture was left at room temperature for 10 minutes, and then was centrifuged at 8500rpm for 20 minutes. The liquid was decanted, the pellet dried, and then the pellet was re-suspended in 10ml 75% ethanol and centrifuged at 8500rpm for 10 minutes. The liquid was decanted and the pellet re-suspended in DEPC-treated sterile water. This extract was then treated with DNA-free (Ambion) to ensure there was no DNA contamination. RNA integrity and concentration were confirmed by agarose electrophoresis and spectrophotometry readings measured at A260 and A280, the ratio of these readings was used to assess RNA purity.
Reverse Transcriptase PCR
Total RNA (leiomyoma or myometrium) was first diluted to obtain the following stock concentrations: 100ng/microliter, 10ng/microliter, and 1ng/microliter. Primers were designed using Beacon Designer (BioRad) so as to amplify a 90-150 bp section close to the 3′ end of the translated region (within 500bp). The sequences of the ATF3 primers were 5′-AAGTGAGTGCTTCTGCCATC-3′ (sense) and 5′-TTTCTTTCTCGTCGCCTCTTTT-3′ (anti-sense), for cFos 5′-GACCCTGAGCCCAAGCCC-3′ (sense) and 5′-GTAGGTGAAGACGAAGGAAGACG-3′ (anti-sense), and for cJun 5′-CCGTTTACACCAACCTCAGC-3′ (sense) and 5′-TCTGCGGTTCCTCCTTGAAG-3′ (anti-sense).
The primers were then generated on the 392 DNA/RNA Synthesizer (Applied Biosystems), and purified by HPLC. These were added to the reaction mix at 200nM concentration. Superscript One-Step RT-PCR with Platinum Taq (Invitrogen) was used, and the Gene Amp PCR System 9700 (Applied Biosystems) was used for thermal cycling. The reverse transcriptase was performed at 50 degrees Celsius for 30 minutes, followed by 94 degrees Celsius for 2 minutes. The samples then underwent 40 cycles at the following temperatures: 94 degrees Celsius for 15 seconds, 50-65 degrees Celsius (depending on optimal annealing temperatures for the designed primers) for 30 seconds, and 72 degrees Celsius for 3 minutes. Primers for 18s ribosomal RNA were included as an internal control. After completion of cycling, the samples were stored at 4 degrees Celsius. Leiomyoma and myometrial samples were then run side-by-side at the same starting concentration on a 2% agarose gel. They were run at 10ng concentrations both with and without 18s ribosomal RNA control gene primers to determine if there was competition between primer sets.
Real-Time RT-PCR
Total RNA (either from leiomyoma or myometrium) was diluted as described above. Primers were also designed and constructed as described above. Probes had been simultaneously designed with primers to ensure a good match. Initial experiments to determine the optimal concentration ratio between control and gene of interest primers, determined a concentration of 300nM for ATF3 primers and 12.5nM for 18s primers. The leiomyoma and myometrial samples were run simultaneously in four separate samples each. Thermal cycling occurred as described above on an iCycler IQ (BioRad).
The 18S RNA control was used to adjust for differences in starting RNA concentrations. Given the extremely low, and at times undetectable, levels of ATF3 gene product, it was necessary to run the samples with 10ng of RNA in order to reliably detect ATF3 signal. This is 100× higher concentration than normally required for real time analysis. Furthermore the ratio between ATF3 and 18s control primers had to be titrated to 24 in order to have the signal from the control not obscure the ATF3 signal. Data analysis was performed with iCycler (BioRad).
Immunohistochemistry
For immunohistochemistry, tissue samples stored in 4% formalin were placed in paraffin and sectioned onto slides. The tissue sections were deparaffinized using xylene and hydrated using graded ethanol solutions followed by distilled water. Slides were then treated with methanol/3% hydrogen peroxide solution for 15 minutes to block endogenous peroxidases, and treated with chondroitin ABC lyase at 0.15 U/ml for 15 minutes. Blocking was done with normal goat serum according to recommendations by Vectastain Elite ABC kit, PK-6101 (Vector Laboratories, Burlingame, CA). Slides were then treated with primary antibody [ATF-3] (C-19, Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at a dilution of 1:200 for 2 hours at room temperature. Biotinylated avidin-linked horseradish peroxidase secondary antibody was added according to guidelines of Vectastain Elite ABC kit, PK-6101 (Vector Laboratories). Tissue sections were then stained using 3, 3′-diaminobenzidine (DAB) substrate according to guidelines of DAB Substrate Kit for Peroxidase, PK-4100 (Vector Laboratories). Finally, slides were dehydrated and mounted using Hypermount (Shandon, Pittsburgh, PA). The negative controls used in this study were 3%BSA/0.1% Tween/TBS solution at the same dilution as the primary antibody as a replacement for the primary antibody on leiomyoma and myometrial tissues. This was also used as a replacement for the secondary antibody on leiomyoma and myometrial tissues. No staining was observed on the control slides of leiomyoma or myometrial tissues that were exposed to either no primary antibody, or no secondary antibody. Liver sections were used as positive controls; non-immune serum was used as a negative control.
Western Blot
Protein Isolation and Analysis
Protein extracts were prepared from the thawed myometrium and leiomyoma tissue samples. The finely minced tissue was extracted overnight at 4° Celsius in homogenizing buffer [250 mM Tris-HCl (pH 8.0), 200 μM sodium orthovanadate, 100 mM NaF, 1 mM EDTA, 120 mM NaCl, 0.5% Nonidet P-40 and one “complete” tablet (Roche Diagnostics, Indianapolis, IN)]. The homogenate was centrifuged at 14,000g for 30 minutes to pellet large cellular debris. The lysates were stored at -80°C. The standard bicinchoninic acid (BCA) assay (Pierce, Rockford, IL) was used to determine protein concentration. Based on a standard curve of absorbance (562nm) versus micrograms protein (0.2 to 50 μg protein standards), lysate concentrations were determined from the curve.
For Western blot analysis, 10 μg protein samples were denatured in 1× SDS buffer and heated at 100°C for 5 min. The samples were electrophoresed onto SDS-PAGE gels (Invitrogen) in Tris-glycine SDS running buffer. The separated proteins were electroblotted to Protran nitrocellulose (Schleicher and, Schuell, Keene, NH) and then probed with ATF-3 (C-19):sc-188 rabbit anti-human antibodies used at 1:200 from Santa Cruz Biotech. The primary antibodies were detected with the horseradish peroxidase (HRP)-conjugated anti-rabbit (Amersham Bioscience, UK) used at 1:500 in combination with the SuperSignal West Pico Chemiluminescent Substrate (Pierce). As an internal standard between the samples, HRP-conjugated anti-human beta-actin (sc1616, Santa Cruz) at a dilution of 1:50,000 was used. Western blots were quantitated with Quantity One (BioRad) software. A standard area on each gel was used for the blots and the intensity of signal was measured inside those areas. Each signal was normalized by dividing it by the matching beta-actin signal strength. The intensity of the protein band from leiomyoma was divided by the intensity of the patient matched band from the myometrial sample in order to achieve relative expression.
Statistical Analysis
Two tailed Student t-tests were used to test for significance where the distribution was found to be normal. In order to perform a t-test on data that was expressed as ratios, the ratios first were converted to logarithms, to correct for the fact that ratios are intrinsically an asymmetrical measure. Significance was defined as P<0.05.
The data for real time analysis of ATF3 was graphed and found to be nonparametric, with a bimodal distribution. For that dataset a Wilcoxon Signed Rank sum test was used for analysis of significance. Significance was defined as P<0.05.
Results
Of the fifteen patients included in the analysis, nine were African American, four Caucasian, one Hispanic, and one Asian (Table I). Patient ages ranged from 36 to 49 years. Leiomyoma size ranged from 0.5cm to 7.0cm in diameter. No patient was being treated with GnRH agonist (leuprolide acetate) or was post-menopausal at the time of hysterectomy.
Table 1. Patient Characteristics: Age, Race and Leiomyoma Diameter.
| Patient | Age | Race1 | Diameter (cm) |
|---|---|---|---|
| 1 | 43 | AA | 1 |
| 2 | 39 | AA | 1 |
| 3 | 37 | C | 2 |
| 4 | 36 | AA | 7.0 |
| 5 | 37 | AA | 1.6 |
| 6 | 46 | AA | 3.3 |
| 7a2 | 40 | AA | 3.0 |
| 7b | 0.5 | ||
| 8a | 44 | H | 5.7 |
| 8b | 0.5 | ||
| 9 | 40 | C | 3.0 |
| 10a | 36 | C | 4.8 |
| 10b | 0.5 | ||
| 11 | 49 | Asian | 7.0 |
| 12a | 46 | C | 4.2 |
| 12b | 2.3 | ||
| 12c | 0.5 | ||
| 12d | 0.5 | ||
| 12e | 0.5 | ||
| 13 | 46 | AA | 4.6 |
| 14 | 48 | AA | 3.3 |
| 15 | 44 | AA | 3.9 |
AA, African American; C, Caucasian; H, Hispanic.
a, b, c, d, and e represent different leiomyomata for the given patient.
ATF3 is critical for stress response regulation and was differentially expressed based on microarray studies, therefore we evaluated its expression in leiomyomata and patient matched myometrium. PCR amplification of reverse transcribed cDNA with ATF3 primers revealed only a faint band in leiomyoma tissue, in contrast to a strong band in myometrial tissue, suggesting a reduction of ATF3 message in leiomyoma relative to myometrium (Figure 1).
Figure 1.
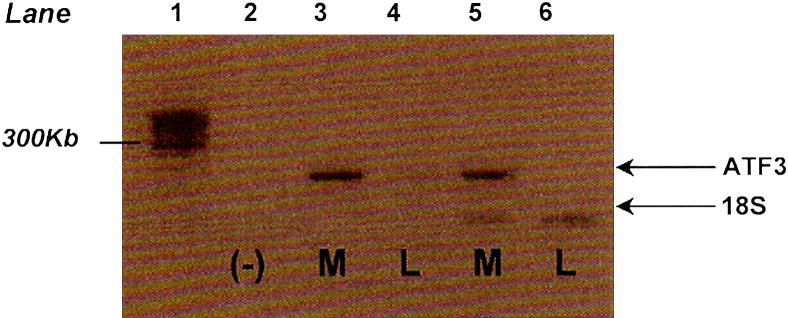
ATF3 mRNA expression in leiomyoma and myometrium
Representative reverse transcriptase PCR analyzing ATF3 levels in leiomyoma and myometrium samples (N=7). Lane 1: LM, lane marker. Lane 2: negative control, ATF3 and 18S primers (no mRNA). Lane 3: 10 μg myometrial mRNA, ATF3 primers. Lane 4: 10 μg leiomyoma mRNA, ATF3 primers. Lane 5: 10 μg myometrial mRNA, ATF3 and 18S primers. Lane 6: 10 μg leiomyoma mRNA, ATF3 and 18S primers.
In order to quantitate this difference, real time RT-PCR was then performed for ATF3 transcripts in samples from 13 patients and 19 different leiomyomata, comparing leiomyomata to matched myometrium (Table II). ATF3 mRNA was consistently under-expressed in all of the leiomyomata compared to their controls except in the one Hispanic patient whose leiomyoma and myometrial tissues showed no difference in ATF3 mRNA expression. The other twelve patients, however, showed variable under-expression of ATF3 mRNA which was noted to be in a bimodal distribution with one subgroup of patients demonstrating markedly lower expression than the other. Because of this distribution, and our interest in the possible genetic differences between races and leiomyoma size, we analyzed the real time data for ATF3 mRNA transcript based on leiomyoma size, fold under-expression, and race. There was no correlation between these variables with the number of patients studied.
Table 2. Realtime RT-PCR Analysis of ATF3 mRNA in Leiomyomata and Myometrium 1.
| Patient | ATF3 mRNA L:M |
|---|---|
| 1 | -19.3 ± .09 |
| 2 | -16.3 ± .33 |
| 3 | -18.6 ± .25 |
| 4 | -60.8 ± 13 |
| 5 | -1.8 ± .35 |
| 6 | -4.3 ± .23 |
| 7a2 | -3.0 ± 1.30 |
| 7b | -3.9 ± .22 |
| 8a | 1.4 ± .13 |
| 8b | 1.3 ± .18 |
| 9 | -3.5 ± .27 |
| 10a | -60.4 ± .13 |
| 10b | -71.0 ± .21 |
| 12a | -87.3 ± .27 |
| 12b | -91.0 ± .39 |
| 12c | -8.14 ± .24 |
| 12d | -7.13 ± .24 |
| 14 | -96 ± .42 |
| 15 | -62.5 ± .72 |
|
| |
| Mean3 | -32.3±8.2 |
Values are fold expression leiomyoma:myometrium determined by the Ct value plus or minus the standard error of the mean. Positive values indicate that mRNA expression was greater in leiomyoma than in myometrium. Fractional values were converted to their negative inverse to properly convey fold expression greater in myometrium than in leiomyoma.
a, b, c, and d indicate different leiomyomata from the same patient.
Fold mRNA expression of ATF3 (leiomyoma:myometrium) from real-time reverse transcriptase PCR. While the overall mean was -32.3±8.2, the data were found to be bimodal with means of -6.9 ± 2.1 and -75.5 ± 6.3. Differences in steady state mRNA between leiomyoma and myometrium were significant using a Wilcoxon Signed Rank Sum test (P<0.05).
To examine the protein expression, western blot of ATF3 was performed which revealed increased expression of ATF3 protein in the leiomyoma compared to myometrium (Figure 2A). Beta-actin was used as a control. The ATF3 protein product was over-expressed in all 8 patients with a mean of 2.9 (P<0.05) (Figure 2B). In order to localize the subcellular distribution of protein product, immunohistochemistry studies were performed which confirmed an intracellular location of ATF3 with concentration in the nucleus (Figure 2C).
Figure 2.
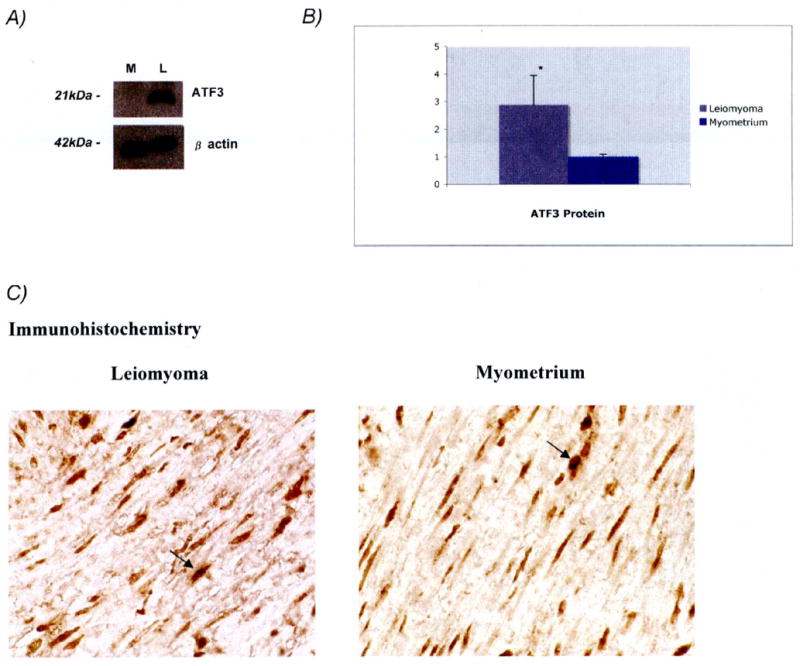
Assessment of ATF3 protein expression
A. Western Blot showing ATF3 protein expression (21kDa bands) using 10μg each of representative myometrium (M) and leiomyoma (L) lysates (upper panel). β actin (42kDa bands) was used as a control (lower panel). B. Graphical representation of mean fold protein expression (leiomyoma:myometrium) for ATF3 protein in leiomyoma (black bar) and myometrium (white bar) from 8 sample pairs determined by pixel intensity, P<0.05 (two-tailed Student's t-test). ATF3 was found to be more abundant in leiomyoma. C. Immunohistochemical images of ATF3 protein in leiomyoma and corresponding myometrium at 63× magnification. Intracellular staining of ATF3 is demonstrated by the arrows. Similar staining patterns were seen in 6 different sample pairs (not shown).
Because ATF-3 can dimerize with and be modulated by cJun and cFos, members of the AP-1 complex, levels of mRNA and protein of cFos and cJun were also assessed. Real time PCR for cFos and cJun showed a reduction in their mRNA transcripts in leiomyoma compared to myometrium. The mean fold expression in leiomyoma tissue relative to myometrial tissue for cFos was -23.78 (P<0.05) and for cJun was -29.57 (P<0.05) (Figure 3A). Western blot of cFos and cJun did not show significant differences in protein expression for these genes (Figure 3B).
Figure 3.
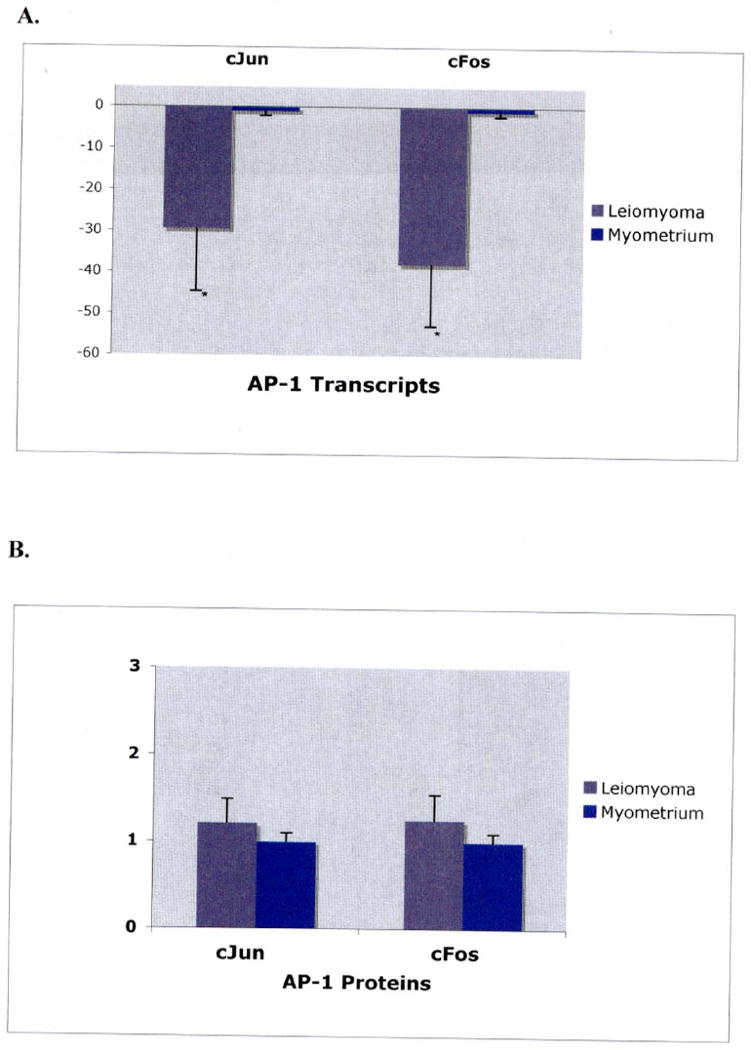
A. mRNA expression of AP-1 transcripts.
Graphical representation of mean fold mRNA expression (leiomyoma:myometrium) for cJun and cFos respectively, in leiomyoma (black bars) and myometrium (white bars) from 10 sample pairs. Expression values were determined by real-time reverse transcriptase PCR using 10ng of mRNA isolated from leiomyoma and myometrium samples, respectively. Significance was determined using a two-tailed t-test. The mean fold mRNA expression for Jun was found to be -29.56, SEM of 14.9 (p<0.05). Fos was found to be -37.97 with a SEM of 15.1 (p<0.05).
B. Protein expression cJun and cFos.
Graphical representation of mean fold protein expression (leiomyoma:myometrium) for cJun and cFos respectively, in leiomyoma (black bars) and myometrium (white bars) from 8 sample pairs. Values were determined by pixel intensity from western blot experiments. The mean fold protein expression for cJun and cFos were found to be 1.2 and 1.3, respectively. Protein levels between myometrium and leiomyoma did not differ significantly using a two-tailed t-test.
Discussion
ATF3 mRNA was reduced in leiomyoma compared to matched myometrial samples confirming our microarray results (11) and others (17). In contrast, protein expression for ATF3 was found to be increased in leiomyoma. This suggests that ATF3 has been activated in leiomyoma tissue, achieved relatively high and sustained levels of protein. Based on observations in other tissues (27), we suspect that the protein was repressing its own promoter, resulting in a low steady state level of mRNA. We were interested in whether ATF3 might vary with leiomyoma size and race, but our data did not show convincing evidence that ATF3 expression levels varied based on these factors. Related Activator Protein-1 Complex genes cFos and cJun had reduced mRNA transcript expression in leiomyoma, but no significant difference in protein expression.
ATF3 protein has been shown to be elevated after ischemia in cardiac muscle, mechanical or chemical injury in the liver, the post-seizure brain, and other tissues. In mice cardiomyocytes, ischemia and reperfusion injury have been shown to induce ATF3 protein (18). The increased expression of ATF3 in mice caused structural and functional disorders in the heart muscle such that the tissue became hypertrophied with disarrayed myofibrils, similar to that observed for leiomyoma structure particularly in collagen structures (28). Recent work (29) also suggested that mechanical stretch in murine cardiomyocytes may be a pathway through which ATF3 acts, in line with a recent finding of increased mechanical stiffness in human leiomyomata (20).
It has been hypothesized that failure of normal apoptosis plays a role in leiomyoma development (30). Activation of ATF3 has been shown to be involved in apoptotic responses to stress (26). However whether ATF3 activation ultimately leads to tissue destruction via apoptosis, or tissue growth via repression of apoptosis appears to be dependent on the environment and type of tissue. In most cells, ATF3 mRNA is almost undetectable (31). However, ATF3 mRNA levels increase rapidly in response to diverse stressors including ischemia, wounding, and various cytokines (22, 25), and fall soon after the stress event, likely because ATF3 can repress its own promoter (27).
ATF3 can function as a transcriptional repressor when it is a homodimer, but can be a transcriptional activator when it dimerizes with cJun (2) In fact, cJun seems to be a crucial component in the regulation of the AP-1 complex in cell proliferation and has been shown to have variable effects in different cell types (23). Our data suggests that cFos and cJun mRNA are downregulated in leiomyoma tissue, however, their protein concentration did not differ between myometrium and leiomyoma.
The mechanism for the development of leiomyomata is unknown. Epidemiologic data (32) suggest an increased risk of leiomyomata in women with hypertension and previous pelvic infections or infectious exposures. Both of these disease processes cause tissue stress with cell injury, inflammation, and cytokine release, which may also affect uterine smooth muscle and stimulate leiomyoma development. In fact, 8-hydroxy-2′-deoxyguanosine (8-OH-dG), a biomarker of oxidative stress, has previously been shown to be more abundant in leiomyoma than myometrium (33). Stress-responsive protein SRP27 has also been found in higher levels in leiomyoma than in myometrium in a pattern similar to that seen in breast cancer (34). In addition, the BCL-2 proto-oncogene, which produces a protein that can inhibit apoptosis, is overexpressed in leiomyoma compared to myometrium (35). It has further been shown that GnRHa inhibits leiomyoma cells in culture by inducing apoptosis (36). Together, these studies suggest that varying stresses to the uterus may lead to the development of leiomyoma, with inhibition of apoptosis as a biologically plausible mechanism for leiomyoma development.
Our results support the notion that leiomyoma share features of a disordered response to the stress of wound healing, or mechanical stress. ATF3 may be involved in leiomyoma development, providing a mechanistic link between the influence of irritants and stressors on leiomyoma development and the dysregulated apoptosis seen in these important tumors. Differing expression levels of ATF3 between leiomyoma and myometrium suggest a possible molecular mechanism whereby mechanical and ischemic stress, perhaps through the influence of apoptosis or disordered wound healing, could lead to the development of these common and troublesome uterine tumors.
Table 3. ATF3 mRNA fold expression by Race, Age, and Leiomyoma size1.
| Race | Least Difference (N=6) | Middle Difference (N=6) | Greatest Difference (N=7) |
|---|---|---|---|
| African American | 3 | 3 | 4 |
| Caucasian | 1 | 3 | 3 |
| Hispanic | 2 | 0 | 0 |
| Mean Age (yrs) | 40.8 | 41.9 | 42.7 |
| Mean Leiomyoma Diameter (cm) | 2.4 | 1.9 | 3.4 |
mRNA transcript expression were divided into 3 groups by fold expression leiomyoma to myometrium: least difference (1.4 to -4.3), middle difference (-7.1 to -19.3), greatest difference (-60.7 to -96.0). There was no apparent relationship between leiomyoma size or race and ATF3 expression.
Acknowledgments
The authors would like to thank the departments of Obstetrics and Gynecology of the Uniformed Services University of the Health Sciences and the National Naval Medical Center for their support of this project. Further they would like to thank the Reproductive Biology and Medicine Branch of the National Institutes of Health for ongoing support. Thanks to Claudia Massolo for laboratory assistance and Chantal Mayers who provided invaluable laboratory teaching and guidance throughout the project.
Supported in part by the intramural research program of the Reproductive Biology and Medicine Branch, NICHD, NIH.
Footnotes
National Naval Medical Center, National Institutes of Health, and the Uniformed Services University of the Health Sciences
Presented in part at the American Society for Reproductive Medicine, San Antonio, TX, 2003 and Montreal, CA, 2005.
Disclosure: The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the Department of Health and Human Services, the Department of the Army, or the Department of Defense.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baird DD, Dunson DB, Hill MC, Cousins D, Schectman JM. High cumulative incidence of uterine leiomyoma in black and white women: ultrasound evidence. Am J Obstet Gynecol. 2003;188:100–7. doi: 10.1067/mob.2003.99. [DOI] [PubMed] [Google Scholar]
- 2.Myers ER, Barber MD, Gustilo-Ashby T, Couchman G, Matcher DB, McCrory DC. Management of uterine leiomyomata: what do we really know? Obstet Gynecol. 2002;100:8–17. doi: 10.1016/s0029-7844(02)02019-7. [DOI] [PubMed] [Google Scholar]
- 3.Stewart EA, Morton CM. The Genetics of Uterine Leiomyoma. Obstet Gynecol. 2006;107:917–921. doi: 10.1097/01.AOG.0000206161.84965.0b. [DOI] [PubMed] [Google Scholar]
- 4.Zhou J, Mochizuki T, Smeets H, Antignac C, Laurila P, de Paepe A, et al. Deletion of the paired alpha 5(IV) and 6(IV) collagen genes in inherited smooth muscle tumors. Science. 1993;261:1167–9. doi: 10.1126/science.8356449. [DOI] [PubMed] [Google Scholar]
- 5.Marsh DJ, Dahia PL, Zheng Z. Germline Mutations in PTEN are present in BAnnayan-Zonana syndrome. Nature Genetics. 1997;16(4):333–334. doi: 10.1038/ng0897-333. [DOI] [PubMed] [Google Scholar]
- 6.Launonen V, Vierimaa O, Kiuru M, Isola J, Roth S, Pukkala E. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc Natl Acad Sci U S A. 2001;98:3387–92. doi: 10.1073/pnas.051633798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, Buck GM, Courey NG, Perez KM, Wactawski-Wende J. Risk Factors for Uterine Fibroids among Women Undergoing Tubal Sterilization. Am J Epidemiol. 2001;153:20–26. doi: 10.1093/aje/153.1.20. [DOI] [PubMed] [Google Scholar]
- 8.Wise L, Palmer J, Stewart EA, Rosenberg L. Age-Specific Incidence Rates for Self-Reported Uterine Leiomyomata in the Black Women's Health Study. Obstet Gynecol. 2005;105:563–568. doi: 10.1097/01.AOG.0000154161.03418.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faerstein E, Szklo M, Rosenshein N. Risk Factors for Uterine Leiomyoma: A Practice Based Case-Control Study. I. African-American Heritage, Reproductive History, Body Size, and Smoking 2001. Am J Epidemiol. 2001;153(1):1–10. doi: 10.1093/aje/153.1.1. [DOI] [PubMed] [Google Scholar]
- 10.Gross KL, Panhuysen CI, Kleinman MS, Goldhammer H, Jones ES, Nassery N, et al. Involvement of fumarate hydratase in nonsyndromic uterine leiomyomas: genetic linkage analysis and FISH studies. Genes Chrom Cancer. 2004;41:183–90. doi: 10.1002/gcc.20079. [DOI] [PubMed] [Google Scholar]
- 11.Catherino WH, Prupas C, Tsibris JC, Leppert PC, Payson M, Nieman LK, et al. Strategy for elucidating differentially expressed genes in leiomyomata identified by microarray technology. Fertil Steril. 2003;80:282–90. doi: 10.1016/s0015-0282(03)00953-1. [DOI] [PubMed] [Google Scholar]
- 12.Catherino WH, Segars JH. Microarray analysis in fibroids: which gene list is the correct list? Fertil Steril. 2003;80:293–4. doi: 10.1016/s0015-0282(03)00958-0. [DOI] [PubMed] [Google Scholar]
- 13.Catherino WH, Leppert PC, Stenmark MH, Payson M, Potlog-Nahari C, Nieman LK, et al. Decreased dermatopontin expression is a molecular link between uterine leiomyomata and keloids. Genes Chrom Cancer. 2004;40:204–217. doi: 10.1002/gcc.20035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsibris JCM, Segars JH, Coppola D, Mane S, Wilbanks GD, O'Brien WF, et al. Insights from gene arrays on the development and growth regulation of uterine leiomyomata. Fertil Steril. 2002;78:114–21. doi: 10.1016/s0015-0282(02)03191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skubitz KM, Skubitz AP. Differential gene expression in uterine leiomyoma. J Lab Clin Med. 2003;141:297–308. doi: 10.1016/S0022-2143(03)00007-6. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Mahadevappa M, Yamamoto K, Wen Y, Chen B, Warrington JA, et al. Distinctive proliferative phase differences in gene expression in human myometrium and leiomyomata. Fertil Steril. 2003;80:266–76. doi: 10.1016/s0015-0282(03)00730-1. [DOI] [PubMed] [Google Scholar]
- 17.Arslan AA, Gold LI, Mittal K, Suen T, Belitskaya-Levy I, Tang M, et al. Gene expression studies provide clues to the pathogenesis of uterine leiomyoma: new evidence and a systematic review. Hum Reprod. 2005;20:852–863. doi: 10.1093/humrep/deh698. [DOI] [PubMed] [Google Scholar]
- 18.Okamoto Y, Chaves A, Chen J, Kelley R, Jones K, Weed HG, et al. Transgenic mice with cardiac specific expression of activating transcription factor 3, a stress inducible gene, have conduction abnormalities and contractile dysfunction. Am J Pathol. 2001;159:639–50. doi: 10.1016/S0002-9440(10)61735-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alenghat FJ, Ingber DE. Mechanotransduction: all signals point to cytoskeleton, matrix and integrins. Sci STKE k. 2002;119:Pe6. doi: 10.1126/stke.2002.119.pe6. [DOI] [PubMed] [Google Scholar]
- 20.Rogers R, Norian J, Malik M, Christman G, Mones A, Chen F, et al. Mechanical Homeostasis is Altered in Uterine Leiomyoma. Am J Obstet Gynecol. 2008;198:474.e1–11. doi: 10.1016/j.ajog.2007.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leppert PC, Catherino WH, Segars JH. A new hypothesis about the origin of uterine fibroids based on gene expression profiling with microarrays. Am J Obstet Gynecol. 2006;195:415–20. doi: 10.1016/j.ajog.2005.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hai T, Harmann M. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 23.Wagner E, Eferl R. AP-1: A double-edged sword in tumorigenesis. Nature Reviews Cancer. 2003 Nov;:859–68. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 24.Shaulian E, Karin M. AP-1: Linking hydrogen peroxide and oxidative stress to the control of cell proliferation and death. Life. 2001;52:17–24. doi: 10.1080/15216540252774711. [DOI] [PubMed] [Google Scholar]
- 25.Hess J, Angel P, Schorpp-Kistner M. AP-1 Subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–73. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 26.Hua B, Tamamori-Adachi M, Luo Y, Tamura K, Morioka M, Fukuda M, et al. A splice variant of stress response gene ATF3 counteracts NF-kB-dependent anti-apoptosis through inhibiting recruitment of CREB-binding protein/p300 coactivator. J Biol Chem. 2006;281:1620–9. doi: 10.1074/jbc.M508471200. [DOI] [PubMed] [Google Scholar]
- 27.Wolfgang C, Liang G, Yoshickika O, Allen A, Hai T. Transcriptional autorepression of the stress-inducible gene ATF3. J Biol Chem. 2000;275:16865–70. doi: 10.1074/jbc.M909637199. [DOI] [PubMed] [Google Scholar]
- 28.Leppert PC, Baginski T, Prupas C, Catherino WH, Pletch S, Segars JH. Comparative ultrastructure of collagen fibrils in uterine leiomyomas and normal myometrium. Fertil Steril. 2004;82(Suppl 3):1182–7. doi: 10.1016/j.fertnstert.2004.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kehat I, Hasin T, Aronheim A. The Role of Basic Leucine Zipper Protein-Mediated Transcription in Physiologic and Pathologic Myocardial Hypertrophy. Ann N Y Acad Sci. 2006;1080:97–109. doi: 10.1196/annals.1380.009. [DOI] [PubMed] [Google Scholar]
- 30.Martel K, Ko A, Christman G, Stribley J. Apoptosis in human uterine leiomyomas. Semin Repro Med. 2004;22:91–103. doi: 10.1055/s-2004-828615. [DOI] [PubMed] [Google Scholar]
- 31.Chen B, Wolfgang C, Hai T. Analysis of ATF3, a transcription factor induced by physiologic stresses and modulated by gad 153/chop10. Mol Cell Biol. 1996;16:1157–68. doi: 10.1128/mcb.16.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Payson M, Leppert P, Segars J. Epidemiology of myomas. Obstet Gynecol Clin North Am. 2006;33:1–11. doi: 10.1016/j.ogc.2005.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foksinski M, Kotzbach R, Szymanski W, Olinski R. The level of typical biomarker of oxidative stress 8-hydroxyl-2′-deoxyguanosine is higher in uterine myomas than in control tissues and correlates with the size of the tumor. Free Radic Biol Med. 2000;29:597–601. doi: 10.1016/s0891-5849(00)00358-0. [DOI] [PubMed] [Google Scholar]
- 34.Navarro D, Cabrero JJ, Falcon O, Jimenez P, Ruiz A, Chrino R, et al. Monoclonal antibody characterization of progesterone receptors, estrogen receptors, and the stress responsive protein of 27 kDA (SRP27) in human uterine leiomyoma. J Steroid Biochem Mol Biol. 1989;34:491–8. doi: 10.1016/0022-4731(89)90133-7. [DOI] [PubMed] [Google Scholar]
- 35.Maruo T, Matsuo H, Samoto T, Shimomua Y, Kurachi O, Gao Z, et al. Effects of progesterone on uterine leiomyoma growth and apoptosis. Steroids. 2000;65:585–92. doi: 10.1016/s0039-128x(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Matsuo H, Kurachi O, Maruo T. Down-regulation of proliferation and up-regulation of apoptosis by gonadotropin-releasing hormone agonist in cultured uterine leiomyoma cells. Eur J Endocrinol. 2002;146:447–56. doi: 10.1530/eje.0.1460447. [DOI] [PubMed] [Google Scholar]


