Abstract
Recent data has identified STAG2, a core subunit of the multifunctional cohesin complex, as a highly recurrently mutated gene in several types of cancer. We sought to identify a therapeutic strategy to selectively target cancer cells harboring inactivating mutations of STAG2 using two independent pairs of isogenic glioblastoma cell lines containing either an endogenous mutant STAG2 allele or a wild-type STAG2 allele restored by homologous recombination. We find that STAG2 mutation is associated with significantly increased sensitivity to inhibitors of the DNA repair enzyme poly ADP-ribose polymerase (PARP). STAG2-mutated, PARP-inhibited cells accumulated in G2 phase and had a higher percentage of micronuclei, fragmented nuclei and chromatin bridges compared to wild-type STAG2 cells. We also observed more 53BP1 foci in STAG2-mutated glioblastoma cells suggesting that these cells have defects in DNA repair. Furthermore, cells with STAG2 mutations were more sensitive than cells with wild-type STAG2 when PARP inhibitors were used in combination with DNA damaging agents. These data suggest that PARP is a potential target for tumours harbouring inactivating mutations in STAG2, and strongly recommend that STAG2 status be determined and correlated with therapeutic response to PARP inhibitors, both prospectively and retrospectively, in clinical trials.
Keywords: poly (ADP-ribose) polymerase, STAG2, cohesin, olaparib, glioblastoma
INTRODUCTION
Inhibition of poly(ADP-ribose) polymerase (PARP) has emerged as a promising drug strategy for the treatment of cancers mutated for BRCA1/2 because of its ability to selectively kill cells through synthetic lethality (1, 2). More recently, PARP inhibitors have been shown to be effective in cells with defects in other genes involved in homologous recombination (HR) and the DNA damage response suggesting that PARP inhibitors may be effective in treating a wider range of tumours that do not have BRCA mutations (3–6). Identification of other tumour genotypes susceptible to PARP inhibition will expand the utility of these drugs.
The cohesin complex, named for its role in sister chromatid cohesion, is well-conserved across organisms (7). In humans, the core mitotic complex consists of four subunits: SMC1A, SMC3, RAD21 (also known as SCC1 or MCD1) and one of two possible stromal antigen proteins (STAG1 or STAG2). Together, these four subunits can encompass newly replicated sister chromatids and hold them in close proximity (8). Beyond its well-known function in chromosome segregation, cohesin has several additional roles in the cell. Similar to other genes sensitive to PARP inhibition, defects in cohesin components affect both replication fork integrity and HR repair (7, 9, 10). Cohesin is recruited to sites of replication fork pausing and double-strand breaks and has also been shown to promote replication fork restart and DNA repair through its interactions with other proteins (11–13). In addition, because of its ability to encircle sister chromatids, the cohesin complex is thought to promote error-free recombination repair with the neighbouring undamaged DNA strand in the S/G2 phases of the cell cycle (10). Supporting the idea that cells mutated for cohesin genes might be sensitive to PARP inhibition, we have shown that knockdown of three of the cohesin core components (SMC1, SMC3 and RAD21) can render cells sensitive to the PARP inhibitor olaparib (14).
Recently, the cohesin gene, STAG2, was discovered to be highly mutated in glioblastoma (GBM), Ewing’s sarcoma and melanoma cells (15). These mutations led to either truncation or functional inactivation of the STAG2 protein that is easily detected in cells or tissues by immunohistochemistry or Western blot using antibodies. Given the previous data that knockdown of cohesin components results in PARP inhibitor sensitivity (14), we wanted to determine if tumour cells with STAG2 mutations were susceptible to PARP inhibition. Here we show that GBM cell lines with mutations in STAG2 are significantly more sensitive to PARP inhibitors than matched, isogenic STAG2 wild-type lines. This proliferation defect results in an accumulation of cells in G2 and genome instability. Furthermore, STAG2-mutated cell lines demonstrate an increased sensitivity when combinations of DNA damaging chemotherapeutics and PARP inhibitors were used providing a therapeutic rationale for PARP inhibitors both as a single agent or in combination with other DNA damaging agents in STAG2-deficient tumours.
MATERIALS AND METHODS
Materials and cell culture
Olaparib (AZD2281), veliparib (ABT-888) and rucaparib (AG014699) were purchased from Selleck Chemicals; temozolomide and camptothecin were from Sigma-Aldrich. Antibodies used were anti-PAR (Trevigen), anti-STAG2 (Santa Cruz), and anti-SMC1, anti-SMC3, anti-pS10 Histone H3 (pH3), anti-53BP1anti-GAPD and anti-α-tubulin (all from Abcam). H4 and 42MGBA parental and STAG2 KI cell lines were described previously (15). H4 and 42MGBA cell lines obtained from Solomon et al. were from the American Type Culture Collection and DSMZ respectively and were cultured in DMEM + 10% FBS at 37°C and 5% CO2 for one to two months at a time before reinitiation from early passage, frozen stocks. Cell lines were checked regularly for the presence or absence of STAG2 by Western Blot (Supp. Fig. 1).
Cell counting experiments and Clonogenic assays
To assess cell number by nuclei counting, cells were plated in 96-well format with 6 technical replicates for each drug concentration. Twenty-four hours after plating, inhibitors or DMSO were diluted into DMEM and added to wells. Cells were fixed in 3.7% paraformaldehyde after four to five days and then stained with Hoechst 33342 before nuclei were counted on a Cellomics Arrayscan VTI.
For clonogenic assays, cells were plated at single cell density in 6-well dishes with three replicates per drug concentration. Drugs were added after twenty-four hours and cells allowed to grow for 10–14 days, changing drug media every 4–5 days. Colonies were then fixed and stained with 0.1% crystal violet in 95% ethanol for counting. Cell lines were all normalized to the DMSO control and compared using a two-tailed, unmatched Student’s t-test. Error bars represent standard error of the mean.
Immunoblotting and flow cytometry
Cells were grown with or without PARP inhibitor for three (H4) or four (42MGBA) days before all cells were collected by trypsinization and centrifugation. For immunoblotting, pellets were resuspended in 50mM Tris-HCl (pH 7.5), 150mM NaCl, 1% Triton X-100 and protease inhibitors (Roche). Cells were lysed by sonication and centrifuged to remove debris. Lysates were separated by SDS-PAGE, transferred to PVDF and blotted with the indicated antibodies.
For flow cytometry, cells were grown and harvested as above, before being fixed in cold 70% ethanol. Where indicated, cells were first stained with pH3 antibody followed by anti-rabbit conjugated to Alexa Fluor 488 (Jackson Laboratories), before being incubated with propidium iodide and RNase A. Cell cycle analysis was done using Flow Jo. Cell lines were compared using a one-tailed, matched Student’s t-test. Error bars represent standard error of the mean.
Immunofluorescence
Cells were grown on coverslips with and without PARP inhibitor for three (H4) or four (42MGBA) days before fixation in 1:1 methanol: acetone and permeabilization in 0.1% Triton X-100. Coverslips were incubated with anti-53BP1 and anti-rabbit conjugated to Cy3 (Jackson Laboratories) before being stained with DAPI and viewed on a Zeiss Axioplan 2 Fluorescence microscope. At least two hundred cells were counted for each experiment. For micronuclei, fragmented nuclei and chromatin bridges, cell lines were compared using a one-tailed, matched Student’s t-test. For 53BP1 foci, cell lines were compared using a Fisher’s exact test.
RESULTS
STAG2-mutated GBM cell lines are sensitive to PARP inhibition
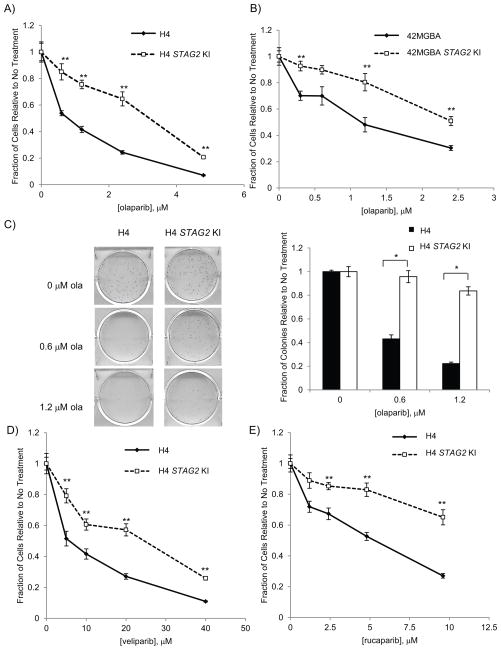
To determine whether STAG2 mutation causes PARP inhibitor sensitivity, we employed two paired sets of GBM cell lines described by Solomon and co-workers (15): H4 (which has a 25bp insertion in exon 12 of STAG2) and 42MGBA (which has a nonsense mutation in exon 20 of STAG2) which were each matched with STAG2 knock-in (KI) lines that have these mutations corrected via homologous recombination (H4 STAG2 KI and 42MGBA STAG2 KI, respectively). Using these two independent isogenic cell line pairs, we first looked at the proliferation of the H4 and 42MGBA cell lines in the presence of the PARP inhibitor, olaparib and found that over a range of concentrations both the H4 and 42MGBA STAG2-mutated cell lines showed significantly decreased cell number when compared with their STAG2 knock-in counterpart by nuclei-counting (Fig. 1A, B). STAG2-mutated cells treated with olaparib also resulted in fewer colonies compared to similarly treated STAG2 KI cells in clonogenic assays (Fig. 1C, Supp. Fig. 2A). Finally, when STAG2 was knocked down by shRNA in HCT116 cells, these cells decrease proliferation in the presence of olaparib similar to the GBM cell lines (Supp. Fig. 2B, C). These results are consistent with our previous findings for siRNA-mediated cohesin knockdown and PARP inhibition (14) and suggest that decreases in cohesin- both the tripartite ring components and the SCC3 ortholog STAG2- sensitize cells to olaparib.
Fig. 1.
STAG2-mutated cell lines are more sensitive to PARP inhibitors. A) STAG2-mutated and STAG2 KI H4 glioblastoma cell lines were treated with increasing concentrations of olaparib in 96-well format and cell nuclei were counted after 4 days. B) 42MGBA glioblastoma cell lines were treated as in A) and cell nuclei were counted after 5 days. C) Clonogenic survival of H4 cells after olaparib treatment. The graph represents 3 technical replicates. D–E) STAG2-mutated and KI cell lines were treated with increasing concentrations of the PARP inhibitors D) veliparib and E) rucaparib as in A). ** p < 0.005, * p < 0.01
The cohesin complex contains one subunit each of SMC1, SMC3 and RAD21 as well as one of either STAG1 or STAG2. Excluding STAG2, we noted no difference in the levels of these proteins in the H4 parental and STAG2 KI cell lines (Supp. Fig. 1A). Upon immunoprecipitation of RAD21, there was evidence of an increase in STAG1-containing complex in the STAG2-mutated line suggesting compensation in the cells lacking STAG2 (Supp. Fig. 1A). Examination of poly ADP-ribose (PAR) levels, a measure of PARP activity, in cells by Western blot showed that PARylation in both the H4 and 42MGBA cell line pairs were high, and this was greatly decreased by treatment with the PARP inhibitor olaparib regardless of STAG2 status (Suppl Fig. 1 B, C). We also analyzed the protein levels of the core cohesin components SMC1, SMC3 and RAD21. As expected, the levels of SMC1, SMC3 and RAD21 were equivalent in untreated H4 STAG2-mutated and STAG2 KI cell lysates (Supp. Fig. 1D). Interestingly, treatment with olaparib caused a decrease in the protein levels of these core components, and this decrease was much more pronounced in the H4 STAG2-mutated cell line. The reason for these low levels is currently not known but suggests that the accessory protein STAG2 stabilizes the cohesin ring components when they are challenged with PARP inhibitor.
To ensure that the loss of survival we observed in STAG2-mutated cells was the result of PARP inhibition and not limited to the PARP inhibitor olaparib, we also treated H4 and 42MGBA paired cell lines with two other PARP inhibitors: veliparib, an oral inhibitor shown to cross the blood-brain barrier (16) and rucaparib, a potent inhibitor which targets a broad spectrum of PARP enzymes (17, 18). Both of these inhibitors were more effective on STAG2-mutated cells when compared with STAG2 KI cells (Fig. 1 D, E, Supp. Fig. 2D, E). From these data, we conclude that mutations in the cohesin component STAG2 render cells more sensitive to PARP inhibition.
PARP inhibition in STAG2-mutated cells is associated with an accumulation of cells in G2 phase and nuclear abnormalities
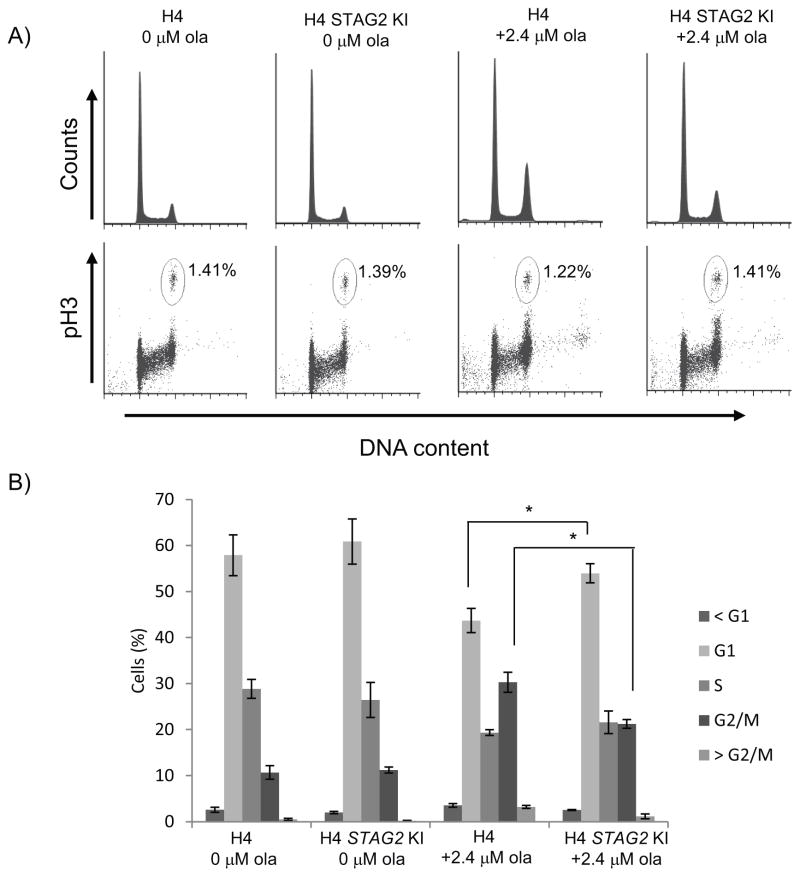
As a more robust proliferation defect was observed in STAG2-mutated cells after treatment with PARP inhibitor, we next sought to determine whether this could be attributed to a specific phase of the cell cycle. Analysis of olaparib-treated H4 cells showed an accumulation of STAG2-mutated cells in the G2/M phase (Fig. 2A,B). Since STAG2-cohesin is involved in cohesion of sister chromatids in both G2 and mitosis up until their segregation, we also used an antibody to histone H3 phosphorylated at S10 (pH3) as a mitotic marker to differentiate between the mitotic and G2 cells. Staining with this marker showed very little difference in the percentage of mitotic cells in treated and untreated cells (Fig. 2A), indicating that STAG2-mutated cells treated with olaparib accumulate in G2.
Fig. 2.
Treatment of STAG2-mutated H4 cells with olaparib results in an accumulation of G2 cells. A) Flow cytometry profiles of untreated and olaparib-treated H4 cell lines stained with propidium iodide for DNA content and pS10 Histone H3 antibody as a mitotic marker. B) Cell cycle distribution of H4 cell populations from three independent experiments. * p < 0.05
Similarly, 42MGBA STAG2-mutated cells treated with olaparib also show an accumulation of cells in G2/M (Supp. Fig. 3A, B). These results are consistent with H4 cells and suggest that glioblastoma cell lines treated with PARP inhibitor that are deficient for STAG2 show a more prolonged G2 delay when compared with those that express wild-type for STAG2. It should be noted that both H4 and 42MGBA STAG2 KI cell lines show sensitivity and G2/M accumulation at higher concentrations of olaparib. As glioblastoma cell lines in general can have multiple mutations and chromosomal abnormalities, it is possible these lines contain other defects in addition to STAG2 mutation. However, as growth differences are seen between mutated and KI cells across both H4 and 42MGBA lines, we believe STAG2 function to significantly contribute to PARP inhibitor sensitivity.
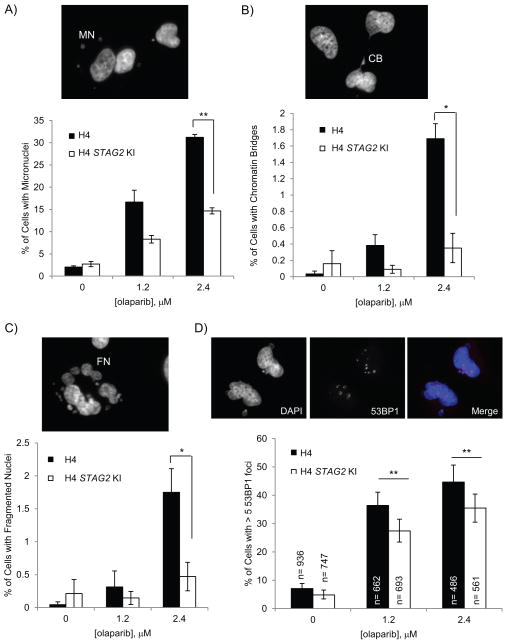
Given that we observed a consistent increase in both sub-G1 and >4N cells when STAG2-mutated lines were treated with olaparib (Fig. 2, Supp. Fig. 3), we next looked for differences in other genome instability and cell death phenotypes. Accordingly, we observed a higher percentage of cells with micronuclei (MN) in STAG2-mutated, olaparib-treated cells (Fig. 3A Suppl. Fig. 4, 5, 6A). We also observed a higher incidence in chromatin bridges (CB) in these cells when compared with STAG2 KI cells (Fig. 3B, Suppl. Fig. 6B). Both of these phenotypes are consistent with these cells having higher genome instability. In addition, both H4 and 42MGBA STAG2-mutated cell lines had a higher fraction of fragmented nuclei (FN) when treated with PARP inhibitor (Fig. 3C, Supp. Fig. 6C) which, along with higher percentages of sub-G1 cells in the flow cytometry profiles, suggest that these cells may be undergoing cell death. As we saw a large percentage of these fragmented nuclei (~12%) in 42MGBA cells treated with olaparib and acknowledge that fragmented nuclei can be a characteristic of apoptosis, we also looked for an increase in the levels of cleaved PARP, an indicator of apoptosis that is downstream of caspase-3/7 activation (19–21), in olaparib-treated cell lines. We did not, however, observe an increase in the levels of cleaved PARP (Supp. Fig. 1E). Therefore, we believe the olaparib-mediated cell death is unlikely to be apoptotic.
Fig. 3.
Cells treated with PARP inhibitor show more genome instability and DNA repair defects. A–C) H4 cells grown with or without olaparib for three days were scored for the presence of A) micronuclei (MN), B) chromatin bridges (CB) and C) fragmented nuclei (FN). Graphs represent cell counts from three independent experiments. D) Presence of 53BP1 foci in paired H4 cell lines untreated and treated with olaparib for three days. Error bars in D) are 95% confidence intervals. ** p < 0.005, * p < 0.05
PARP inhibitor sensitization is characterized by increased levels of DNA damage
As PARP inhibitor-treated cells showed a delay in G2 phase and eventual genome instability, we hypothesized that these cells may be responding to increased DNA damage. To further examine this, we stained cells for the DNA damage response protein 53BP1, which rapidly forms foci upon DNA damage, and found that more STAG2-mutated cells have > 5 53BP1 foci after olaparib treatment than similarly treated STAG2 KI cells (Fig. 3D, Supp. Fig. 6D). In fact, at 2.4 μM olaparib, the STAG2-mutated H4 line had an increase of ~10% of cells with > 5 53BP1 foci over its STAG2 KI counterpart (Fig. 3D). This is remarkably similar to the ~10% increase in G2 cells seen at the same concentration over the same time period (Fig. 2B). Together, these results suggest that the lower survival rate of STAG2-mutated cells after treatment with PARP inhibitor may be due to increased levels of DNA damage which leads to accumulation of cells in G2, genome instability and cell death.
Combining PARP inhibition with camptothecin or temozolomide is more synergistic in STAG2-mutated GBM cells
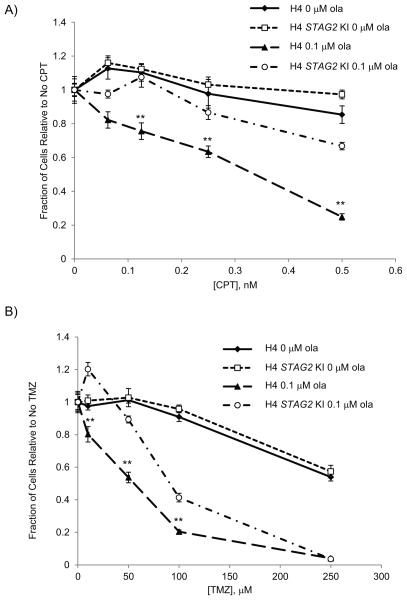
PARP inhibitors have been used extensively to potentiate the toxicity of several chemotherapeutic agents by increasing the DNA damage of these agents (6). Since our paired cell lines showed an increase of 53BP1 foci, a marker for DNA damage, in STAG2-mutated cells, we wanted to determine the synergistic effect of PARP inhibition with DNA damaging agents in STAG2-mutated and KI cells. To this end, we tested the effect of the topoisomerase I poison camptothecin (CPT), which causes DNA lesions in replicating cells, alone and in combination with olaparib. We found that STAG2-mutated H4 cells were more sensitive than H4 STAG2 KI cells to CPT even in the absence of olaparib (Supp. Fig. 7A). In combination with low doses of olaparib, however, cells were sensitized to a much lower dose of CPT than when CPT was used alone with a large differential seen between STAG2-mutated and STAG2 KI cells (Fig. 4A). Furthermore, 42MGBA cells were significantly more sensitive than 42MGBA STAG2 KI cells when treated with both CPT and olaparib (Supp. Fig. 7B).
Fig. 4.
Combinations of PARP inhibitor and known chemotherapeutics are more effective in STAG2-mutated H4 cells. Paired H4 cell lines were treated with either A) camptothecin (CPT) and olaparib or B) temozolomide (TMZ) and olaparib and cell number was determined after 4 days using nuclei-counting. ** p < 0.005
We also used a combination of olaparib and temozolomide (TMZ), an alkylating agent that has previously shown robust synergy with PARP inhibitors and is currently used to treat GBMs. These two drugs together had similar results to those with olaparib and CPT, showing increased sensitivity in STAG2-mutated compared to STAG2 KI lines (Fig. 4B). As both CPT and TMZ are known to be involved in generating lesions that affect DNA replication and repair, our results suggest that the response to these lesions involves not only PARP activity, but also STAG2.
DISCUSSION
The concept of synthetic lethality holds the promise of chemotherapeutics that specifically target tumour cells for killing. Key to the development of synthetic lethal therapeutics is the identification of synthetic lethal interactions between mutations frequently observed in tumours and small molecule inhibitors. PARP inhibitors are a promising class of small molecules that are currently in multiple clinical trials for cancer. The goal of this study was to determine if mutations in the cohesin complex gene STAG2, which is frequently mutated in several tumour types and is easily assayed using immunohistochemistry, resulted in sensitivity to PARP inhibitors. In this study, we demonstrated that STAG2-mutated glioblastoma cell lines were more sensitive to PARP inhibition than paired cell lines that contained wild-type STAG2. This increase in sensitivity in STAG2-mutated cells was characterized by increased DNA damage, an accumulation of cells in G2 phase, and nuclear abnormalities such as chromatin bridges, micronuclei, and fragmented nuclei. PARP inhibition also increased the sensitivity of STAG2-mutated cells to the topoisomerase poison camptothecin and the DNA alkylating agent temozolomide, suggesting that PARP inhibitors could be used in combination with DNA damaging agents to affect STAG2-mutated tumour cell killing.
Cohesin and cohesin-associated genes are frequently mutated in a number of solid tumour types and leukemias (15, 22–25). More specifically, loss of STAG2 expression has been shown by immunohistochemistry to be common in a significant number of solid tumour types including GBM (19%), Ewing’s sarcoma (21%), and melanoma (19%) (15). STAG2 truncating mutations have also recently been found in bladder cancer (26–28). A large majority of the glioblastoma, melanoma and Ewing’s sarcoma tumours had little intratumoural heterogeneity (15), suggesting that sensitivity to PARP inhibitors may be especially relevant in these tumour types.
One explanation for the high frequency loss of STAG2 expression is the location of STAG2 on the X chromosome, meaning that only a single mutation is needed to inactivate it. Furthermore, unlike the core cohesin components SMC1A, SMC3 and RAD21, which are essential for cell survival, somatic cells have a mitotic STAG2 paralog, STAG1, which may share a level of functional redundancy with STAG2. STAG1 can also form a functional cohesin complex, but unlike STAG2, it has not been found to be lost or mutated in glioblastoma lines (15). The presence of STAG1 may explain why truncating mutations and loss of expression of STAG2 are well-tolerated in cells. In support of this, we have found that more STAG1 associates with the cohesin complex in STAG2-mutated cells compared to STAG2 KI cells in immunoprecipitation experiments (Supp. Fig. 1A). We show here, however, that STAG1 is not sufficient for survival in STAG2-mutated cells upon exposure to PARP inhibitors (Fig. 1) and suggest that STAG2 status in tumours may be a marker for PARP inhibitor sensitivity.
The cohesin complex is multifunctional and has a known role in G2 DNA repair that has mainly been attributed to its physical ability to hold sister chromatids in close proximity after replication for efficient error-free homology searching before recombination or template switching (10, 29). Consistent with a DNA repair function, cohesin components have been found to localize to DNA double-strand breaks (DSBs) in human cells (11, 30). Furthermore, Bauerschmidt and co-workers have demonstrated that depletion of SMC1 impairs the repair of radiation-induced DSBs as measured by the increase in γH2AX and 53BP1 foci in G2 cells (30). Our results show that in the absence of PARP inhibition, STAG2-mutated cells have slightly increased 53BP1 foci compared with STAG2 KI (Fig. 3D, Supp. Fig. 6D), suggesting that loss of STAG2 results in increased DNA damage. Inhibition of PARP activity increased the 53BP1 foci differential between the STAG2- mutated and KI cells further and also led to the formation of micronuclei and fragmented nuclei (Fig. 3, Supp. Fig. 6). This suggests that prolonged or less accurate DNA repair in STAG2-mutated cells after PARP inhibitor treatment can result in accumulation of cells in G2 phase, and genome instability or cell death.
Regulation of cohesin dynamics on DNA is controlled by different post-translational modifications to the complex (31). For instance, both SMC1 and SMC3 are phosphorylated in an ATM-dependent manner and this phosphorylation may be required for efficient mobilization of the complex upon DNA damage (32–34). Similarly, STAG2 is modified by phosphorylation to promote separase-independent dissociation of cohesin from chromosome arms during early mitosis (35) thereby demonstrating that cohesin localization and function can be influenced by STAG2 post-translational modification. The contribution of STAG2 phosphorylation to the mobilization of cohesin in response to DNA damage, if any, has yet to be determined. In addition to phosphorylation, PARylation by PARP is a post-translational modification that occurs at sites of DNA damage and is known to affect both chromatin architecture and the recruitment of DNA repair factors (36). It is not known if cohesin is directly PARyled in response to replication stress or DNA damage, but cohesin components have recently been shown to immunoprecipitate with a PAR antibody after treatment with the DNA alkylating agent, methylnitronitrosoguanidine (MNNG), suggesting an interaction of PAR and cohesin under certain conditions (37). Our results and those of others, that have been obtained using PARP inhibitors on cells depleted of cohesin components (14, 38, 39), provide additional evidence of a link between cohesin and PARP activity.
Several chemotherapeutic agents including TMZ and CPT are currently in clinical trials with PARP inhibitors as it has been proposed that combining PARP inhibition with DNA damaging agents will exacerbate their effects (40). Both TMZ and topoisomerase poisons like CPT show increased toxicity in tumours when combined with PARP inhibitors (41–44) and we confirm this synergy in our GBM cell lines (Fig. 4, Supp. Fig. 7). Several reports have suggested that the potentiation of DNA damaging agents by PARP inhibition results in an increased need for double-strand break repair and HR. For example, combining either TMZ or CPT with PARP inhibitors leads to an increase in double-strand breaks (45, 46). Furthermore, resistance to TMZ and veliparib in HCT116 cells has been attributed, at least in part, to an increase in Rad51-dependent HR (47). Other reports have shown that sensitization to alkylating agents by PARP inhibitors is enhanced in cells downregulated for HR and DSB repair pathway components (48, 49). Our results show that STAG2 deficiency is another condition that can further sensitize cells to combinations of PARP inhibitors and DNA damaging agents. Given its mutation or loss of expression in ~20% of GBMs as well as several other tumour types (15), STAG2 shows potential as a marker of sensitivity not just to PARP inhibitor monotherapy, but also to combination therapy with CPT or TMZ. Consequently, the status of STAG2 in tumours should be considered as PARP inhibitors move forward in clinical trials.
Supplementary Material
Acknowledgments
Financial Support: P. Hieter acknowledges support from the National Institutes of Health (R01CA158162) and Canadian Institutes of Health Research (MOP-38096). M.L. Bailey is funded by a fellowship from the Michael Smith Foundation for Health Research.
Abbreviations Used
- GBM
Glioblastoma
- PARP
poly (ADP-ribose) polymerase
- CPT
camptothecin
- TMZ
temozolomide
Footnotes
The authors disclose no potential conflicts of interest
References
- 1.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 2.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 3.Gottipati P, Vischioni B, Schultz N, Solomons J, Bryant HE, Djureinovic T, et al. Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res. 2010;70:5389–98. doi: 10.1158/0008-5472.CAN-09-4716. [DOI] [PubMed] [Google Scholar]
- 4.McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–15. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 5.Aguilar-Quesada R, Munoz-Gamez JA, Martin-Oliva D, Peralta A, Valenzuela MT, Matinez-Romero R, et al. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol Biol. 2007;8:29. doi: 10.1186/1471-2199-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Weaver DT. The ups and downs of DNA repair biomarkers for PARP inhibitor therapies. Am J Cancer Res. 2011;1:301–27. [PMC free article] [PubMed] [Google Scholar]
- 7.Dorsett D, Strom L. The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr Biol. 2012;22:R240–50. doi: 10.1016/j.cub.2012.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nasmyth K, Haering CH. Cohesin: Its roles and mechanisms. Annu Rev Genet. 2009;43:525–58. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 9.O’Neil NJ, van Pel DM, Hieter P. Synthetic lethality and cancer: Cohesin and PARP at the replication fork. Trends Genet. 2013;29:290–7. doi: 10.1016/j.tig.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu N, Yu H. The smc complexes in DNA damage response. Cell Biosci. 2012;2(5):3701-2-5. doi: 10.1186/2045-3701-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caron P, Aymard F, Iacovoni JS, Briois S, Canitrot Y, Bugler B, et al. Cohesin protects genes against gammaH2AX induced by DNA double-strand breaks. PLoS Genet. 2012;8:e1002460. doi: 10.1371/journal.pgen.1002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strom L, Lindroos HB, Shirahige K, Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16:1003–15. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 13.Tittel-Elmer M, Lengronne A, Davidson MB, Bacal J, Francois P, Hohl M, et al. Cohesin association to replication sites depends on rad50 and promotes fork restart. Mol Cell. 2012;48:98–108. doi: 10.1016/j.molcel.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLellan JL, O’Neil NJ, Barrett I, Ferree E, van Pel DM, Ushey K, et al. Synthetic lethality of cohesins with PARPs and replication fork mediators. PLoS Genet. 2012;8:e1002574. doi: 10.1371/journal.pgen.1002574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–43. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donawho CK, Luo Y, Luo Y, Penning TD, Bauch JL, Bouska JJ, et al. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res. 2007;13:2728–37. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 17.Thomas HD, Calabrese CR, Batey MA, Canan S, Hostomsky Z, Kyle S, et al. Preclinical selection of a novel poly(ADP-ribose) polymerase inhibitor for clinical trial. Mol Cancer Ther. 2007;6:945–56. doi: 10.1158/1535-7163.MCT-06-0552. [DOI] [PubMed] [Google Scholar]
- 18.Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol. 2012;30:283–8. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- 19.Duriez PJ, Shah GM. Cleavage of poly(ADP-ribose) polymerase: A sensitive parameter to study cell death. Biochem Cell Biol. 1997;75:337–49. [PubMed] [Google Scholar]
- 20.Kaufmann SH, Desnoyers S, Ottaviano Y, Davidson NE, Poirier GG. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: An early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–85. [PubMed] [Google Scholar]
- 21.Tewari M, Quan LT, O’Rourke K, Desnoyers S, Zeng Z, Beidler DR, et al. Yama/CPP32 beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 1995;81:801–9. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- 22.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–10. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–78. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y, et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet. 2013;45:1232–7. doi: 10.1038/ng.2731. [DOI] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solomon DA, Kim JS, Bondaruk J, Shariat SF, Wang ZF, Elkahloun AG, et al. Frequent truncating mutations of STAG2 in bladder cancer. Nat Genet. 2013;45:1428–30. doi: 10.1038/ng.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balbas-Martinez C, Sagrera A, Carrillo-de-Santa-Pau E, Earl J, Marquez M, Vazquez M, et al. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat Genet. 2013;45:1464–9. doi: 10.1038/ng.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo G, Sun X, Chen C, Wu S, Huang P, Li Z, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45:1459–63. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sjogren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in saccharomyces cerevisiae. Curr Biol. 2001;11:991–5. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 30.Bauerschmidt C, Arrichiello C, Burdak-Rothkamm S, Woodcock M, Hill MA, Stevens DL, et al. Cohesin promotes the repair of ionizing radiation-induced DNA double-strand breaks in replicated chromatin. Nucleic Acids Res. 2010;38:477–87. doi: 10.1093/nar/gkp976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudra S, Skibbens RV. Cohesin codes - interpreting chromatin architecture and the many facets of cohesin function. J Cell Sci. 2013;126:31–41. doi: 10.1242/jcs.116566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo H, Li Y, Mu JJ, Zhang J, Tonaka T, Hamamori Y, et al. Regulation of intra-S phase checkpoint by ionizing radiation (IR)-dependent and IR-independent phosphorylation of SMC3. J Biol Chem. 2008;283:19176–83. doi: 10.1074/jbc.M802299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 2004;18:1423–38. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bauerschmidt C, Woodcock M, Stevens DL, Hill MA, Rothkamm K, Helleday T. Cohesin phosphorylation and mobility of SMC1 at ionizing radiation-induced DNA double-strand breaks in human cells. Exp Cell Res. 2011;317:330–7. doi: 10.1016/j.yexcr.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107:18475–80. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gagne JP, Pic E, Isabelle M, Krietsch J, Ethier C, Paquet E, et al. Quantitative proteomics profiling of the poly(ADP-ribose)-related response to genotoxic stress. Nucleic Acids Res. 2012;40:7788–805. doi: 10.1093/nar/gks486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brough R, Bajrami I, Vatcheva R, Natrajan R, Reis-Filho JS, Lord CJ, et al. APRIN is a cell cycle specific BRCA2-interacting protein required for genome integrity and a predictor of outcome after chemotherapy in breast cancer. EMBO J. 2012;31:1160–76. doi: 10.1038/emboj.2011.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yadav S, Sehrawat A, Eroglu Z, Somlo G, Hickey R, Yadav S, et al. Role of SMC1 in overcoming drug resistance in triple negative breast cancer. PLoS One. 2013;8:e64338. doi: 10.1371/journal.pone.0064338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basu B, Sandhu SK, de Bono JS. PARP inhibitors: Mechanism of action and their potential role in the prevention and treatment of cancer. Drugs. 2012;72:1579–90. doi: 10.2165/11635510-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 41.Barazzuol L, Jena R, Burnet NG, Meira LB, Jeynes JC, Kirkby KJ, et al. Evaluation of poly (ADP-ribose) polymerase inhibitor ABT-888 combined with radiotherapy and temozolomide in glioblastoma. Radiat Oncol. 2013;8:65, 717X–65. doi: 10.1186/1748-717X-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plummer R, Lorigan P, Steven N, Scott L, Middleton MR, Wilson RH, et al. A phase II study of the potent PARP inhibitor, rucaparib (PF-01367338, AG014699), with temozolomide in patients with metastatic melanoma demonstrating evidence of chemopotentiation. Cancer Chemother Pharmacol. 2013;71:1191–9. doi: 10.1007/s00280-013-2113-1. [DOI] [PubMed] [Google Scholar]
- 43.Kummar S, Chen A, Ji J, Zhang Y, Reid JM, Ames M, et al. Phase I study of PARP inhibitor ABT-888 in combination with topotecan in adults with refractory solid tumors and lymphomas. Cancer Res. 2011;71:5626–34. doi: 10.1158/0008-5472.CAN-11-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palma JP, Wang YC, Rodriguez LE, Montgomery D, Ellis PA, Bukofzer G, et al. ABT-888 confers broad in vivo activity in combination with temozolomide in diverse tumors. Clin Cancer Res. 2009;15:7277–90. doi: 10.1158/1078-0432.CCR-09-1245. [DOI] [PubMed] [Google Scholar]
- 45.Liu X, Shi Y, Guan R, Donawho C, Luo Y, Palma J, et al. Potentiation of temozolomide cytotoxicity by poly(ADP)ribose polymerase inhibitor ABT-888 requires a conversion of single-stranded DNA damages to double-stranded DNA breaks. Mol Cancer Res. 2008;6:1621–9. doi: 10.1158/1541-7786.MCR-08-0240. [DOI] [PubMed] [Google Scholar]
- 46.Zhang YW, Regairaz M, Seiler JA, Agama KK, Doroshow JH, Pommier Y. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res. 2011;39:3607–20. doi: 10.1093/nar/gkq1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, Han EK, Anderson M, Shi Y, Semizarov D, Wang G, et al. Acquired resistance to combination treatment with temozolomide and ABT-888 is mediated by both base excision repair and homologous recombination DNA repair pathways. Mol Cancer Res. 2009;7:1686–92. doi: 10.1158/1541-7786.MCR-09-0299. [DOI] [PubMed] [Google Scholar]
- 48.Loser DA, Shibata A, Shibata AK, Woodbine LJ, Jeggo PA, Chalmers AJ. Sensitization to radiation and alkylating agents by inhibitors of poly(ADP-ribose) polymerase is enhanced in cells deficient in DNA double-strand break repair. Mol Cancer Ther. 2010;9:1775–87. doi: 10.1158/1535-7163.MCT-09-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quiros S, Roos WP, Kaina B. Rad51 and BRCA2--new molecular targets for sensitizing glioma cells to alkylating anticancer drugs. PLoS One. 2011;6:e27183. doi: 10.1371/journal.pone.0027183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.