Abstract
Mycobacterial diseases are a group of illnesses that cause a considerable number of deaths throughout the world, regardless of years of public health control efforts. Personalized medicine is a new but rapidly advancing field of healthcare. Personalized medicine in the field of mycobacteriology may be applied in the different levels of management such as prevention, diagnosis, treatment and prognosis. A genetic predisposition and a protein dysfunction study are recommended to tailor an individual approach in mycobacterial diseases.
Keywords: Mycobacterial, TB, NTM, Personalized medicine
1. Introduction
1.1. Mycobacterial disease
Mycobacterial diseases are a group of illnesses that are caused by the mycobacterium species. They include Mycobacterium tuberculosis (MTB) complex, nontuberculous mycobacterium (NTM), and Mycobacterium leprae.
MTB is a member of the MTB complex causes tuberculosis (TB); other members include Mycobacterium africanum and Mycobacterium bovis. In 2010, there were 8.8 million (range, 8.5–9.2 million) incident cases of TB worldwide; 1.1 million (range, 0.9–1.2 million) deaths from TB among human immunodeficiency virus (HIV)-negative people and an additional 0.35 million (range, 0.32–0.39 million) deaths from HIV-associated TB [1].
NTM are ubiquitous organisms that are prevalent in the environment. NTM has been isolated from domestic water supplies, workplaces, and hospitals [2]. Currently, more than 125 different species of NTM exist in the environment, many of which cause human illness [3]. The prevalence of NTM diseases was reported as 1.6–1.8 per 100,000 individuals in most industrialized countries. Recent studies proposed that NTM pulmonary disease is becoming increasingly prevalent in North America, with annual incidence rates of 13 cases per 100,000 in people ≥50 years of age [4]. It was speculated that the burden of NTM disease might soon exceed that of tuberculosis in industrialized nations [5].
M. leprae (the bacteria responsible for leprosy) grow slowly and mainly affect the skin, nerves, and mucous membranes. Leprosy is still a health problem in some parts of the world [6].
2. Personalized medicine
Personalized medicine is a new but rapidly advancing field of healthcare. Personalized medicine is about making the treatment as individualized as the disease. It involves identifying genetic, genomic, proteomic and clinical information in order to make accurate predictions about a person’s susceptibility to developing disease, the course of disease and its response to treatment.
In order for personalized medicine to be used effectively by healthcare providers and their patients, these findings should be transformed into precise diagnostic tests and targeted therapies. This has begun to happen in certain areas of infectious diseases, such as testing patients genetically to determine their likelihood of having a serious adverse reaction to HIV medications [7].
Personalized medicine should not be confused with “genetic medicine”. Genetics, a field more than 50 years old, is the study of inheritance of a gene or a group of genes. It investigates individual genes and their effects on health. Genetic diseases, seemingly “simple” hereditary disorders, can be influenced by other genes, as well as by environmental factors such as diet and exposure to toxins [8].
Genomic and personalized medicine aspire to target more complex diseases, such as cancer, heart disease, and now infectious diseases, which are primarily influenced by environmental factors and their interaction with the human genome. Because these diseases have strong multigene components—and in some cases the diseases might be caused by errors in the DNA between genes instead of within genes—they can be better evaluated using a whole-genome approach. Increasing concern about confidentiality of genetic data was addressed by the Genetic Information Non-Discrimination Act of 2008 (GINA) [9] that prohibits the use of genetic/genomic information by health insurance companies in order to determine a person’s eligibility for insurance, or to determine insurance premiums, as well as by employers in order to make decisions regarding hiring and firing, assigning jobs, and promoting and demoting. Because consumers should now be able to obtain their genome profiles and other genetic information without fear of retribution, GINA is reinvigorating the field of genomic testing.
Rapid and affordable single nucleotide polymorphism (SNP) genotyping over the whole genome is now possible, allowing for genome mapping uncovering novel associations not possible with a candidate gene approach.
The genome-wide association studies (GWAS) allocate the genotyping of the most frequent genetic polymorphism in the genome without hypothesizing about the genomic location of the causal variants. The completion of the human genome sequence, the deposition of SNPs into public databases, the rapid improvements in SNP genotyping methods and the International HapMap (haplotype map) project have allowed the genetic association field to progress to the stage that this approach is feasible. Previous investigations [10,11] and the International HapMap project have shown that most common variations in the genome can be represented by approximately 300,000 SNPs in White populations [12]. African and other populations with greater variation and less linkage disequilibrium need more SNPs to ensure coverage of the entire genome [13].
The development of the above techniques to accurately assess diagnostic and prognostic profiles and select patients with mycobacterial diseases who will respond to a given therapeutic agent is highly desirable and is the foundation for the future delivery of personalized medicine.
2.1. Personalized medicine in mycobacterial diseases
The idea that mycobacterial disease is not only influenced by bacterium but also by both genetic and environmental factors was formally stated 59 years ago [14]. At the time it was stated in reference to TB, but new studies have suggested that it applies to NTM as well.
While the exposure to NTM is universal and billions of people have latent tuberculosis infection (LTBI), only a few persons develop clinical disease [15]. It may be reasonable to state normal host defense mechanisms must be effective enough to prevent the infection. The interactions between mycobacterium and its environment have been suggested. In a minority of cases there is an obvious identifiable risk factor such as in the HIV population and immunocompromised patients, such as those undergoing transplants and chemotherapy [3,16]. A substantial proportion of these patients have no preexisting lung disease and no demonstrable immunodeficiency. A small number of NTM are predominantly nonsmoking elderly women [17]. In the remainder, a complex interaction of genetic and environmental factors causes the development of clinical mycobacterial disease. However, potential mycobacterium-host interaction at the gene level is still very limited according to recent studies.
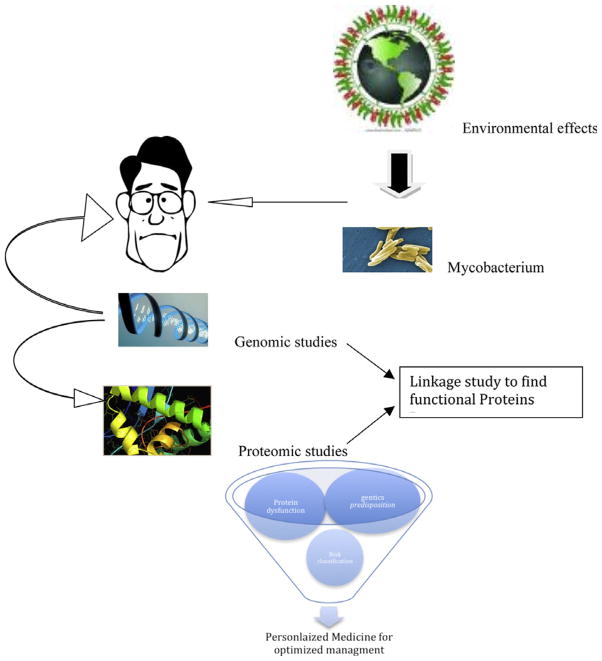
Personalized medicine in the field of mycobacteriology may be applied in the different levels of management, such as prevention, diagnosis, treatment and prognosis. The predisposition and protein dysfunction studies are recommended to tailor an individual approach to each patient (Fig. 1).
Fig. 1.
A strategy for personalized medicine in mycobacterial diseases.
Personalized medicine may answer some pertinent questions, such as:
Who is at risk of contracting LTBI after exposure to MTB?
Who will convert to an active case?
How can the diagnosis, treatment, and prognosis of mycobacterial infection be improved?
2.1.1. Who is at risk of contracting LTBI after exposure to MTB?
Indeed, the exposure to MTB happens in the environment, but the susceptibility to infection is a personal phenomenon.
Many investigations have confirmed that genetic factors are involved in whether or not one is susceptible to mycobacterial disease, and these investigations include results from adoption studies, twin studies, genome-wide linkage and population-based, case–control association studies [18].
Recently, an association was reported between NTM lung disease and the polymorphisms of the natural resistance-associated macrophage protein 1 gene [19].
This finding could raise the possibility of genetic susceptibility for patients with NTM lung disease. Afterward, toll-like receptors-2 (TLRs) [20], Interleukin-12 receptor beta-1 [21], interferon-gamma and interferon-gamma receptor-1 genes [22] were studied.
The identification of host genetic factors, such as human leukocyte antigens (HLA) of major histocompatibility complex and/or cytokines and related receptors associated with susceptibility or resistance to TB, may provide genetic markers to predict the development of mycobacterial disease after exposure to bacteria.
The immune mechanisms in TB have been investigated and it is known from the IL-12/IL-23/IFN-g pathway, together with other cytokines and factors, that they are vital for the immunologic response [23].
In previous studies, a candidate gene approach was carried out using SNPs associated with mycobacterial disease [15,24–26]. As a consequence, those studies had limited potential to uncover novel genomic regions that play a role in the etiology of mycobacterial disease.
Convincing evidence indicating the importance of IFN-g in particular in the control of mycobacterial infections has been found in both experimental and clinical studies [27,28]. IFN-g receptor genes may play a role here. However, the present study failed to show an association between IFN-gamma receptor 1 (IFNGR1) polymorphism and increasing susceptibility to TB [15], but the mutation in IFNGR2 increased the susceptibility in sporadic TB patients [29].
Polymorphism in IL-1 [30,31], IL-6 [32], IL-12 and its receptors [33,34], HLA [35,36], vitamin D receptor haplogroups [24,25], and mitochondrial DNA haplogroups [26] were extensively studied with some promising results.
2.1.2. Who will convert to an active case?
Susceptibility to developing TB disease after exposure is influenced by complex interactions between the host and the pathogen, and genetic and environmental factors [18].
Several genetic loci are involved in susceptibility to mycobacterial disease. Recently, it was shown that the mutation of a human gene encoding the chemokine (C-C motif) ligand 2 CCL2, which is essential for the recruitment of monocytes and T-cells, may increase the risk of developing active TB after exposure [37]. Recently a study showed new genetic targets which influence the risk of developing active TB among latently infected individuals by GWAS [38]. Finding the strong association between potential immune gene polymorphisms and LTBI will be progress with regard to patient selection for LTBI treatment in the future.
2.1.3. Personalized medicine may improve diagnosis of mycobacterial infection
Personalized medicine has improved the diagnostic approach to latent TB. Interferon-gamma release assays (IGRAs) can locate the patient’s immunologic response to mycobacterial tuberculosis-specific antigens. It is a starting point for a personalized diagnosis of mycobacterial diseases.
Finding the genetic susceptibility to facilitate the occurrence of drug-resistant mycobacterium might be another fascinating approach that has not yet been studied.
There is a significant demand for personalized biomarkers for the diagnosis of NTM infection in humans that needs to be addressed.
2.1.4 Personalized medicine may change treatment of mycobacterial infection
Finding personalized biomarkers to best define chemotherapy protocols, individualized treatment duration, and predict risk of adverse events with antibiotics would be other interesting markers for clinicians.
A study showed a polymorphism of the tumor necrosis factor gene (lymphotoxin A+252G/A genotype) in a pulmonary tuberculosis patient associated with impaired response to treatment [39].
Anti-TB drug-related hepatotoxicity is a serious worldwide medical problem among TB patients. It has been proposed that the production and elimination of the toxic metabolites depends on the activities of several enzymes, such as N-acetyl transferase 2, cytochrome P450 oxidase and glutathione S-transferase. It was shown that DNA sequence variations or polymorphisms at those enzymes loci could alter the activities of enzymes and increase the risk of hepatotoxicity. Since the prevalence of polymorphisms is different in worldwide populations, the risk of anti-TB hepatotoxicity varies in the populations. Thus, the knowledge of polymorphisms at those loci, prior to medication, may be a useful tool to evaluate the risk of anti-TB hepatotoxicity [40]. Another study showed that polymorphism of the N-acetyl transferase 2 gene is a risk factor for INH-induced hepatotoxicity [41].
2.1.5. Personalized medicine may improve prognosis of mycobacterial infection
The results of a recent study demonstrated that genetic polymorphisms of the IL-12+ IFN-g pathway may individually or jointly contribute to the prognosis of pulmonary TB [42].
Importantly, personalized medicine will help to bridge current treatment goals (bacteriologic cure) with patient cure. Currently, TB is treated and classified based on bacteriology response. However, the organ damage such as pulmonary fibrosis and its consequence effects are not explored enough. The current TB treatment must focus on a patient cure strategy other than cleaning mycobacterial agents from the body.
The biomarkers should be located to predict pulmonary fibrotic changes after TB treatment to reach this goal.
Conclusion
The future needs to include the stratification of mycobacterial-infected patients with personalized biomarkers in order to reach management goals. The inconsistent treatment results in different studies come from heterogeneity of patients with different genetic pools. Personalized medicine may organize the diagnosis and treatment strategies making it possible to deliver a better quality of healthcare to patients.
Footnotes
Conflict of Interest
There is no conflict of interest regarding this manuscript for the author.
References
- 1.Global tuberculosis control: WHO report. 2011 Accessible in http://www.who.int/tb/publications/global_report/2011/gtbr11_full.pdf.
- 2.Cook JL. Nontuberculous mycobacteria: opportunistic environmental pathogens for predisposed hosts. British Medical Bulletin. 2010;96:45–59. doi: 10.1093/bmb/ldq035. [DOI] [PubMed] [Google Scholar]
- 3.Tabarsi P, Baghaei P, Farnia P, Mansouri N, Chitsaz E, Sheikholeslam F, et al. Nontuberculous mycobacteria among patients who are suspected for multidrug-resistant tuberculosis-need for earlier identification of nontuberculosis mycobacteria. The American Journal of the Medical Sciences. 2009;337(3):182–184. doi: 10.1097/maj.0b013e318185d32f. [DOI] [PubMed] [Google Scholar]
- 4.Winthrop KL, McNelley E, Kendall B, Marshall-Olson A, Morris C, Cassidy M, et al. Pulmonary nontuberculous mycobacterial disease prevalence and clinical features: an emerging public health disease. American Journal of Respiratory and Critical Care Medicine. 2010;182(7):977–982. doi: 10.1164/rccm.201003-0503OC. [DOI] [PubMed] [Google Scholar]
- 5.Adjemian J, Olivier K, Seitz AE, Holland SM, Prevots DR. Prevalence of nontuberculous mycobacterial lung disease in US medicare beneficiaries. American Journal of Respiratory and Critical Care Medicine. 2012 doi: 10.1164/rccm.201111-2016OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Beers SM, de Wit MY, Klatser PR. The epidemiology of Mycobacterium leprae: recent insight. FEMS Microbiology Letters. 1996;136(3):221–230. doi: 10.1111/j.1574-6968.1996.tb08053.x. [DOI] [PubMed] [Google Scholar]
- 7.Young B, Squires K, Patel P, Dejesus E, Bellos N, Berger D, et al. First large, multicenter, open-label study utilizing HLA-B*5701 screening for abacavir hypersensitivity in North America. AIDS. 2008;22(13):1673–1675. doi: 10.1097/QAD.0b013e32830719aa. [DOI] [PubMed] [Google Scholar]
- 8.Chang LJ, Chen SU, Tsai YY, Hung CC, Fang MY, Su YN, et al. An update of preimplantation genetic diagnosis in gene diseases, chromosomal translocation, and aneuploidy screening. Clinical and Experimental Reproductive Medicine. 2011;38(3):126–134. doi: 10.5653/cerm.2011.38.3.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laedtke AL, O’Neill SM, Rubinstein WS, Vogel KJ. Family physicians’ awareness and knowledge of the Genetic Information Non-Discrimination Act (GINA) Journal of Genetic Counseling. 2011 doi: 10.1007/s10897-011-9405-6. [DOI] [PubMed] [Google Scholar]
- 10.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nature Genetics. 2001;29(2):229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 11.Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, et al. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science. 2001;294(5547):1719–1723. doi: 10.1126/science.1065573. [DOI] [PubMed] [Google Scholar]
- 12.Balding DJ. A tutorial on statistical methods for population association studies. Nature Reviews Genetics. 2006;7(10):781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 13.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nature Reviews Genetics. 2005;6(2):95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 14.Dubos RJ, Dubos J. The White Plague: Tuberculosis, Man and Society Little. Brown & Co; Boston: 1952. pp. 188–189. [Google Scholar]
- 15.Mirsaeidi SM, Houshmand M, Tabarsi P, Banoei MM, Zargari L, Amiri M, et al. Lack of association between interferon-gamma receptor-1 polymorphism and pulmonary TB in Iranian population sample. The Journal of Infection. 2006;52(5):374–377. doi: 10.1016/j.jinf.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Mirsaeidi SM, Tabarsi P, Mardanloo A, Ebrahimi G, Amiri M, Farnia P, et al. Pulmonary mycobacterium Simiae infection and HTLV1 infection: an incidental co-infection or a predisposing factor? Monaldi archives for chest disease = Archivio Monaldi per le malattie del torace/Fondazione clinica del lavoro, IRCCS [and] Istituto di clinica tisiologica e malattie apparato respiratorio, Universita di Napoli. Secondo ateneo. 2006;65(2):106–109. doi: 10.4081/monaldi.2006.573. [DOI] [PubMed] [Google Scholar]
- 17.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. American Journal of Respiratory and Critical Care Medicine. 2007;175(4):367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 18.Moller M, Hoal EG. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis. 2010;90(2):71–83. doi: 10.1016/j.tube.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Koh WJ, Kwon OJ, Kim EJ, Lee KS, Ki CS, Kim JW. NRAMP1 gene polymorphism and susceptibility to nontuberculous mycobacterial lung diseases. Chest. 2005;128(1):94–101. doi: 10.1378/chest.128.1.94. [DOI] [PubMed] [Google Scholar]
- 20.Ryu YJ, Kim EJ, Koh WJ, Kim H, Kwon OJ, Chang JH. Toll-like receptor 2 polymorphisms and nontuberculous mycobacterial lung diseases. Clinical and Vaccine Immunology: CVI. 2006;13(7):818–819. doi: 10.1128/CVI.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park HY, Kwon YS, Ki CS, Suh GY, Chung MP, Kim H, et al. Interleukin-12 receptor beta1 polymorphisms and nontuberculous mycobacterial lung diseases. Lung. 2008;186(4):241–245. doi: 10.1007/s00408-008-9096-4. [DOI] [PubMed] [Google Scholar]
- 22.Hwang JH, Kim EJ, Koh WJ, Kim SY, Lee SH, Suh GY, et al. Polymorphisms of interferon-gamma and interferon-gamma receptor 1 genes and non-tuberculous mycobacterial lung diseases. Tuberculosis. 2007;87(2):166–171. doi: 10.1016/j.tube.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Mansouri D, Adimi P, Mirsaeidi M, Mansouri N, Khalilzadeh S, Masjedi MR, et al. Inherited disorders of the IL-12-IFN-gamma axis in patients with disseminated BCG infection. European Journal of Pediatrics. 2005;164(12):753–757. doi: 10.1007/s00431-005-1689-9. [DOI] [PubMed] [Google Scholar]
- 24.Merza M, Farnia P, Anoosheh S, Varahram M, Kazampour M, Pajand O, et al. The NRAMPI, VDR and TNF-alpha gene polymorphisms in Iranian tuberculosis patients: the study on host susceptibility. The Brazilian Journal of Infectious Diseases: An Official Publication of the Brazilian Society of Infectious Diseases. 2009;13(4):252–256. doi: 10.1590/s1413-86702009000400002. [DOI] [PubMed] [Google Scholar]
- 25.Banoei MM, Mirsaeidi MS, Houshmand M, Tabarsi P, Ebrahimi G, Zargari L, et al. Vitamin D receptor homozygote mutant tt and bb are associated with susceptibility to pulmonary tuberculosis in the Iranian population. International Journal of Infectious Diseases: IJID: Official Publication of the International Society for Infectious Diseases. 2010;14(1):e84–5. doi: 10.1016/j.ijid.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Houshmand M, Banoei MM, Tabarsi P, Panahi MS, Kashani BH, Ebrahimi G, et al. Do mitochondrial DNA haplogroups play a role in susceptibility to tuberculosis? Respirology. 2007;12(6):823–827. doi: 10.1111/j.1440-1843.2007.01163.x. [DOI] [PubMed] [Google Scholar]
- 27.Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, Orme IM. Disseminated tuberculosis in interferon gamma gene-disrupted mice. The Journal of Experimental Medicine. 1993;178(6):2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirsch CS, Toossi Z, Othieno C, Johnson JL, Schwander SK, Robertson S, et al. Depressed T-cell interferon-gamma responses in pulmonary tuberculosis: analysis of underlying mechanisms and modulation with therapy. The Journal of Infectious Diseases. 1999;180(6):2069–2073. doi: 10.1086/315114. [DOI] [PubMed] [Google Scholar]
- 29.Vogt G, Chapgier A, Yang K, Chuzhanova N, Feinberg J, Fieschi C, et al. Gains of glycosylation comprise an unexpectedly large group of pathogenic mutations. Nature Genetics. 2005;37(7):692–700. doi: 10.1038/ng1581. [DOI] [PubMed] [Google Scholar]
- 30.Awomoyi AA, Charurat M, Marchant A, Miller EN, Blackwell JM, McAdam KP, et al. Polymorphism in IL1B: IL1B-511 association with tuberculosis and decreased lipopolysaccharide-induced IL-1beta in IFN-gamma primed ex-vivo whole blood assay. Journal of Endotoxin Research. 2005;11(5):281–286. doi: 10.1179/096805105X58706. [DOI] [PubMed] [Google Scholar]
- 31.Gomez LM, Camargo JF, Castiblanco J, Ruiz-Narvaez EA, Cadena J, Anaya JM. Analysis of IL1B, TAP1, TAP2 and IKBL polymorphisms on susceptibility to tuberculosis. Tissue Antigens. 2006;67(4):290–296. doi: 10.1111/j.1399-0039.2006.00566.x. [DOI] [PubMed] [Google Scholar]
- 32.Amirzargar AA, Rezaei N, Jabbari H, Danesh AA, Khosravi F, Hajabdolbaghi M, et al. Cytokine single nucleotide polymorphisms in Iranian patients with pulmonary tuberculosis. European Cytokine Network. 2006;17(2):84–89. [PubMed] [Google Scholar]
- 33.Remus N, El Baghdadi J, Fieschi C, Feinberg J, Quintin T, Chentoufi M, et al. Association of IL12RB1 polymorphisms with pulmonary tuberculosis in adults in Morocco. The Journal of Infectious Diseases. 2004;190(3):580–587. doi: 10.1086/422534. [DOI] [PubMed] [Google Scholar]
- 34.Akahoshi M, Nakashima H, Miyake K, Inoue Y, Shimizu S, Tanaka Y, et al. Influence of interleukin-12 receptor beta1 polymorphisms on tuberculosis. Human Genetics. 2003;112(3):237–243. doi: 10.1007/s00439-002-0873-5. [DOI] [PubMed] [Google Scholar]
- 35.Singh SP, Mehra NK, Dingley HB, Pande JN, Vaidya MC. Human leukocyte antigen (HLA)-linked control of susceptibility to pulmonary tuberculosis and association with HLA-DR types. The Journal of Infectious Diseases. 1983;148(4):676–681. doi: 10.1093/infdis/148.4.676. [DOI] [PubMed] [Google Scholar]
- 36.Tajik N, Shah-hosseini A, Mohammadi A, Jafari M, Nasiri M, Radjabzadeh MF, et al. Susceptibility to pulmonary tuberculosis in Iranian individuals is not affected by compound KIR/HLA genotype. Tissue Antigens. 2012;79(2):90–96. doi: 10.1111/j.1399-0039.2011.01812.x. [DOI] [PubMed] [Google Scholar]
- 37.Feng WX, Flores-Villanueva PO, Mokrousov I, Wu XR, Xiao J, Jiao WW, et al. CCL2-2518 (A/G) polymorphisms and tuberculosis susceptibility: a meta-analysis. International Journal of Tuberculosis and Lung Disease. 2012;16(2):150–156. doi: 10.5588/ijtld.11.0205. [DOI] [PubMed] [Google Scholar]
- 38.Png E, Alisjahbana B, Sahiratmadja E, Marzuki S, Nelwan R, Balabanova Y, et al. A genome wide association study of pulmonary tuberculosis susceptibility in Indonesians. BMC Medical Genetics. 2012;13(1):5. doi: 10.1186/1471-2350-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Elorriaga G, Carrillo-Montes G, Mendoza-Aguilar M, Gonzalez-Bonilla C. Polymorphisms in tumor necrosis factor and lymphotoxin A in tuberculosis without and with response to treatment. Inflammation. 2010;33(4):267–275. doi: 10.1007/s10753-010-9181-8. [DOI] [PubMed] [Google Scholar]
- 40.Roy PD, Majumder M, Roy B. Pharmacogenomics of anti-TB drugs-related hepatotoxicity. Pharmacogenomics. 2008;9(3):311–321. doi: 10.2217/14622416.9.3.311. [DOI] [PubMed] [Google Scholar]
- 41.Ben Mahmoud L, Ghozzi H, Kamoun A, Hakim A, Hachicha H, Hammami S, et al. Polymorphism of the N-acetyltransferase 2 gene as a susceptibility risk factor for antituberculosis drug-induced hepatotoxicity in Tunisian patients with tuberculosis. Pathologie-Biologie. 2011 doi: 10.1016/j.patbio.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Tang S, Shen H. Association of genetic polymorphisms in the IL12-IFNG pathway with susceptibility to and prognosis of pulmonary tuberculosis in a Chinese population. European Journal of Clinical Microbiology & Infectious Diseases: Official Publication of the European Society of Clinical Microbiology. 2010;29(10):1291–1295. doi: 10.1007/s10096-010-0985-0. [DOI] [PubMed] [Google Scholar]