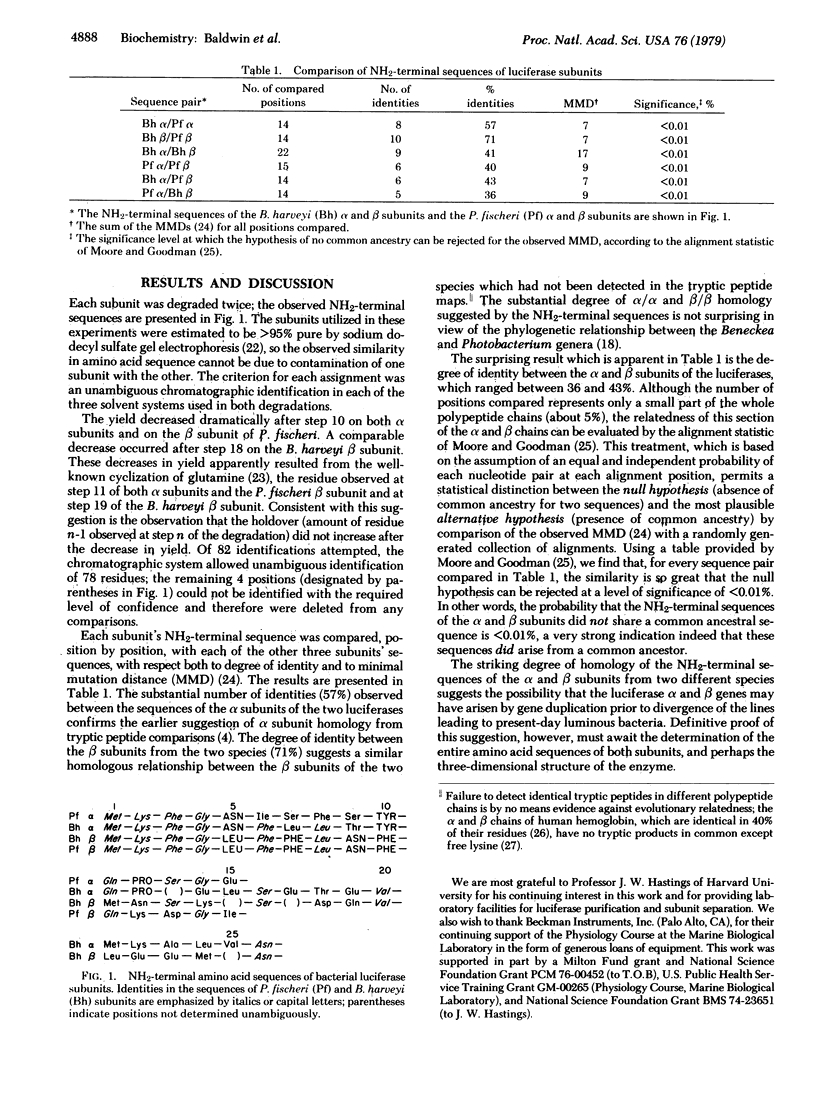
Abstract
The heterodimeric subunit structure of bacterial luciferase was demonstrated more than 10 years ago. The enzymes from both Beneckea harveyi and Photobacterium fischeri have since been studied in detail; they each consist of two nonidentical subunits, designated alpha and beta. Both are required for bioluminescence activity, with the active center apparently confined to the alpha subunit. Amino acid sequence analysis of the NH2 termini of the alpha and beta subunits of the B. harveyi and P. fischeri luciferases not only confirms the earlier observation that the alpha subunits are homologous but also demonstrates that the NH2-terminal sequences of the beta subunits of the luciferases from the two genera are homologous. Furthermore, within each luciferase, the NH2-terminal sequences of the alpha and beta subunits are similar, suggesting the possibility that the genes coding, for alpha and beta may have arisen by gene duplication, presumably prior to divergence of the lines leading to present-day luminous bacteria.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BRAUNITZER G., HILSE K., RUDLOFF V., HILSCHMANN N. THE HEMOGLOBINS. Adv Protein Chem. 1964;19:1–71. doi: 10.1016/s0065-3233(08)60188-6. [DOI] [PubMed] [Google Scholar]
- Baldwin T. O., Hastings J. W., Riley P. L. Proteolytic inactivation of the luciferase from the luminous marine bacterium Beneckea harveyi. J Biol Chem. 1978 Aug 25;253(16):5551–5554. [PubMed] [Google Scholar]
- Baldwin T. O., Nicoli M. Z., Becvar J. E., Hastings J. W. Bacterial luciferase. Binding of oxidized flavin mononucleotide. J Biol Chem. 1975 Apr 25;250(8):2763–2768. [PubMed] [Google Scholar]
- Baldwin T. O. The binding and spectral alterations of oxidized flavin mononucleotide by bacterial luciferase. Biochem Biophys Res Commun. 1974 Apr 23;57(4):1000–1005. doi: 10.1016/0006-291x(74)90795-5. [DOI] [PubMed] [Google Scholar]
- Becvar J. E., Hastings J. W. Bacterial luciferase requires one reduced flavin for light emission. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3374–3376. doi: 10.1073/pnas.72.9.3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline T. W., Hastings J. W. Mutationally altered bacterial luciferase. Implications for subunit functions. Biochemistry. 1972 Aug 29;11(18):3359–3370. doi: 10.1021/bi00768a008. [DOI] [PubMed] [Google Scholar]
- Cousineau J., Meighen E. Chemical modification of bacterial luciferase with ethoxyformic anhydride: evidence for an essential histidyl residue. Biochemistry. 1976 Nov 16;15(23):4992–5000. doi: 10.1021/bi00668a008. [DOI] [PubMed] [Google Scholar]
- Fitch W. M., Margoliash E. Construction of phylogenetic trees. Science. 1967 Jan 20;155(3760):279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Friedland J., Hastings J. W. Nonidentical subunits of bacterial luciferase: their isolation and recombination to form active enzyme. Proc Natl Acad Sci U S A. 1967 Dec;58(6):2336–2342. doi: 10.1073/pnas.58.6.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUIDOTTI G., HILL R. J., KONIGSBERG W. The structure of human hemoglobin. II. The separation and amino acid composition of the tryptic peptides from the alpha and beta chains. J Biol Chem. 1962 Jul;237:2184–2195. [PubMed] [Google Scholar]
- Gunsalus-Miguel A., Meighen E. A., Nicoli M. Z., Nealson K. H., Hastings J. W. Purification and properties of bacterial luciferases. J Biol Chem. 1972 Jan 25;247(2):398–404. [PubMed] [Google Scholar]
- Hastings J. W., Nealson K. H. Bacterial bioluminescence. Annu Rev Microbiol. 1977;31:549–595. doi: 10.1146/annurev.mi.31.100177.003001. [DOI] [PubMed] [Google Scholar]
- Hastings J. W., Weber K., Friedland J., Eberhard A., Mitchell G. W., Gunsalus A. Structurally distinct bacterial luciferases. Biochemistry. 1969 Dec;8(12):4681–4689. doi: 10.1021/bi00840a004. [DOI] [PubMed] [Google Scholar]
- Meighen E. A., Hastings J. W. Binding site determination from kinetic data. Reduced flavin mononucleotide binding to bacterial luciferase. J Biol Chem. 1971 Dec 25;246(24):7666–7674. [PubMed] [Google Scholar]
- Meighen E. A., Nicoli M. Z., Hastings J. W. Functional differences of the nonidentical subunits of bacterial luciferase. Properties of hybrids of native and chemically modified bacterial luciferase. Biochemistry. 1971 Oct 26;10(22):4069–4073. doi: 10.1021/bi00798a009. [DOI] [PubMed] [Google Scholar]
- Meighen E. A., Nicoli M. Z., Hastings J. W. Hybridization of bacterial luciferase with a variant produced by chemical modification. Biochemistry. 1971 Oct 26;10(22):4062–4068. doi: 10.1021/bi00798a008. [DOI] [PubMed] [Google Scholar]
- Meighen E. A., Smillie L. B., Hastings J. W. Subunit homologies in bacterial luciferases. Biochemistry. 1970 Dec 8;9(25):4949–4952. doi: 10.1021/bi00827a018. [DOI] [PubMed] [Google Scholar]
- Moore G. W., Goodman M. Alignment statistic for identifying related protein sequences. J Mol Evol. 1977 Apr 29;9(2):121–130. doi: 10.1007/BF01732744. [DOI] [PubMed] [Google Scholar]
- Nicoli M. Z., Meighen E. A., Hastings J. W. Bacterial luciferase. Chemistry of the reactive sulfhydryl. J Biol Chem. 1974 Apr 25;249(8):2385–2392. [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]



